Abstract
Background
The genus Nebo has been identified as a medically important scorpion species distributed across Arabia and the Middle East. However, its taxonomic status remains unclear.
Aim
The molecular phylogeny of two Nebo species from Saudi Arabia and comparative sequences from Palestine is presented based on the mitochondrial cytochrome oxidase subunit I (COI) gene.
Methodology
Scorpion specimens were collected from two different localities, mainly the Southern part of Saudi Arabia. Then, DNA was extracted, amplified using invertebrate universal primers, and sequenced to identify the COI gene. The obtained sequences were analyzed, and phylogenetic trees based on maximum parsimony, neighbor-joining, and Bayesian inference were constructed.
Results
The inferred phylogeny indicates the monophyletic status of the family Diplocentridae and its subfamily Nebinae and Diplocentrinae. Also, the phylogenetic analyses support the existence of interspecific and intraspecific variations among/ within Nebo hierichonticus and Nebo yemenensis which may indicate distinct species.
Conclusion
Further morphological studies with additional specimens from the Arabian Peninsula may reveal possible undiscovered and cryptic species.
Similar content being viewed by others
Introduction
Arthropod is one of the diverse group on earth [1, 2] and Arachnida is one of the important class belonging to it. With more than 2,700 species, scorpions are a category of arachnids that have significant evolutionary success within invertebrates [3]. With about 138 species in 10 genera, Diplocentridae Karsch, 1880 is considered native to the New World and Middle East [4]. After revising higher scorpion systematics, Soleglad & Fet [5] abolished the family Diplocentridae and merged all genera into the Scorpionidae family. Despite being closely related to Scorpionidae, Diplocentridae differ from them because they have subaculear tubercles on the telsons [6]. Many researchers are considering Diplocentridaee as a valid family [7, 8].
There are several species belonging family Diplocentridae that have been identified and reported as medically significant scorpion species [9]. The genus Nebo contains nine species distributed across Arabia and the Middle East [10,11,12,13,14]. This genus has been difficult to identify taxonomically because of the limited number of specimens available, morphometric ratios used and other morphological characters such as carination, trichobothrial patterns, pectinal tooth counts, and tarsomere II spine formulas [15,16,17]. Kinzelbach [18] and Vachon & Kinzelbach [19] proposed to treat all taxa belong genus Nebo from full species status to subspecies of N. hierichonticus (Simon, 1872). However, Francke [15] and Sissom [16] considered allopatric populations to have consistent morphological characters, morphometric ratios and thus considered them to be valid species. In Saudi Arabia, two species of Nebo have been reported; N. hierichonticus and N. yemenensis [15, 20, 21]. Based on the specimens with very limited sample size studied by [17], some of their diagnostic characteristics were closer to N. hierichonticus while others closer to N. yemenensis.
In addition, Arachnid orders like scorpions are understudied in comparison to other arachnid families, especially when it comes to their systematics and taxonomy [22]. Many scorpion taxa are characterized by high levels of morphological uniformity and conservatism, which hampers straightforward species delimitation [23]. More insights were gained later from studies of mitochondrial and nuclear gene variation [24]. Recent research has employed the molecular phylogeny to interpret the evolutionary relationships of numerous populations of scorpions [25,26,27,28];. Based on morphological studies Nebo yemenensis reported for first time from the Southwestern highlands of Asir and Jizan Provinces, Saudi Arabia by [21]. To evaluate the taxonomical status and the occurrence of N. yemenensis, with additional comparative sequence data of N. hierichonticus from Palestine retrieved from the Genbank.
Material and methods
Biological material
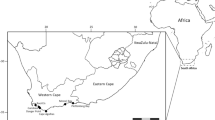
Four adult specimens belonging to N. yemenenesis scorpion species were collected from two different regions in Saudi Arabia (Fig. 1; Table 1). The scorpions were collected mainly at night using ultraviolet from September 2021 to July 2022 and preserved according to the methods described [14]. The climate and habitat of this species were summarized and described by [21].
Collection localities of Nebo yemenensis samples collected from Saudi Arabia that are given in Table 1
DNA extraction, amplification, and sequencing
The whole genomic DNA was extracted from freshly preserved (96% ethanol) scorpion specimens using Qiagen extraction kit (Qiagen) according to its manufacturer’s instructions. A fragment of COI gene was amplified via standard polymerase chain reaction (PCR) using invertebrate universal primers (LCO1490 and HCO2198) as determined by [29]. The amplified products of COI gene were checked, purified, and sequenced on an ABI 3500 automated sequencer (Applied Biosystems Inc., USA). The obtained sequences were deposited in GenBank (Table 1) at https://www.ncbi.nlm.nih.gov/genbank/ with accession numbers (OP970165, OP970166, OP970167, OP970168).
Phylogenetic analysis
The obtained Sequences were screened and analyzed by eye using Finch TV 1. 4. 0 (Geospiza, Inc., USA; http://www.geospiza.com). Also, additional comparative sequences for the subfamily Nebianae represented by Nebo hierichonticus from Palestine (MT418015.1) and Diplocentridae were retrieved from GenBank as an ingroup. Also, Scorpio palmatus was downloaded as outgroup (KT188367.1). The sequence data was aligned using the default settings of ClustalW [30]. The nucleotide composition was calculated based on only the sequences within each ingroup. The pairwise genetic distances (p-distances) were calculated for the whole data set using Mega 6 [30]. All phylogenetic analyses were performed using three different methods based on the COI data set (n = 19), including maximum-parsimony, neighbor-joining, and Bayesian inference as described by [26].
Results
Genetic data
A total of 590 aligned nucleotides from the COI data set were analyzed. There were 379 (64.23%) constant bases, 211 (35.67%) variable bases, and 164 (27.79%) parsimonious bases. The composition of nucleotides was highly biased towards A–T bases. Among the sequence data, T, C, A, and G had mean values of 44.3, 13.3, 21.4, and 21.0%, respectively. It was found that 47 polymorphic segregating sites were detected in the 590 bp region. The genetic distance between N. yemenensis and the others of genus Nebo ranged from 4.0 to 6.0%. while the divergences among other diplocentrid taxa ranged from 0.12 to 0.17 (Table 2).
Phylogenetic analyses
We performed the maximum parsimony analysis within 590 bp in the length of the sequences, the gaps treated as missing which produced two most-parsimonious trees with a length of 625 steps (homoplasy index = 0.5152; consistency index = 0.4848; retention index = 0.5589). The resulting tree showed two major supported clades which are represented by Diplocentridae related taxa (Fig. 2). A first clade includes all taxa belonging to the subfamily Nobinae. This clade is split into two subclades, including N. hierichonticus from Palestine (MT418015.1) as a basal clade to N. yemenensis from Saudi Arabia. The later also divided into two subclades, including N. yemenensis sample from Thaqeef, Mecca (OP970165), as a basal clade the samples from Southwestern Saudi Arabia (OP970166- OP970168) which grouped as a sister group. While the second clade encompassed all taxa belonging to the family Diplocentrinae. Similarly, the neighbor-joining analysis generated a tree which showed the results of the neighbor-joining analysis (Fig. 3). The resulting tree has a general topology nearly identical to the maximum parsimony tree as shown in Fig. 2. The general topology of the Bayesian inference tree shown in Fig. 4 is very similar to both the maximum parsimony (Fig. 2) and neighbor-joining trees (Fig. 3). This analysis clearly showed that the clustering of subfamily Nebinae which represented samples belong genus Nebo from Palestine and Saudi Arabia, and the diplocentrids related taxa. The first clade is supported by a posterior probability value 1. While the second clade is weakly supported by posterior probability value 0.73.
Discussion
The phylogenetic analyses based on maximum-parsimony, neighbor-joining, and Bayesian inference strongly to support the monophyly of family Diplocentridae based on COI gene as represented by different species of Diplocentridae taxa (Figs. 2, 3 and 4). Prendini [7] validated the monophyly of Diplocentridae due to three features: “a fused lamellar hook and a median lobe of the hemispermatophore, subaculear tubercle, and the red venom coloration”. Also, our analyses support the monophyly of the subfamily Nebinae and subfamily Diplocentrinae. It was also revealed that Nebinae is monophyletic since just one character (trichobotrium position it distal to ib) supports this hypothesis. However, internal relationships have not been resolved completely, revealing that some genera are paraphyletic. As a result of various diagnostic characteristics that split Nebinae and Diplocentrinae, Santibáñez-López et al. [8] concluded that the Nebinae subfamily is a synonym for Diplocentrinae, based on multilocus morphological and molecular phylogenetic analysis.
Within the Nebinae clade, N. hierichonticus was basal to the N. yemenensis from Saudi Arabia. N. hierichonticus, differs from the above in that it has diagnostically relevant morphometric ratios indicated by its holotype and paratype, except the length/width of the pedipalp chela is slightly higher than that determined by [15]. N. hierichonticus, a fossorial scorpion species found under self-dug deep caves in the deserts and arid to semiarid mountainous regions across Egypt (Sinai), Palestine, Jordan, Syria and Lebanon [31]. The genetic distance between N. yemenensis and the others of the genus Nebo ranged from 4.0 to 6.0% (Table 2). It has been reported previously that several studies on allied genera have yielded similar results (e.g. Androctonus, Buthus, Buthacus Hottentotta, Leiurus, and Scorpio), [26,27,28, 32,33,34,35,36,37,38,39].
Within the Saudi Arabian N. yemenensis, the sister group relationship is strongly supported between the samples collected from Mecca (OP970165) and Jizan (OP970166- OP970168). However, the divergence between N. yemenensis obtained from Mecca and those obtained Jizan, in Southwestern Saudi Arabia, ranged from 3 to 5%. The existence of cryptic species between these populations could explain this. Kinzelbach [18] have regarded several species in the genus and [19] as subspecies of N. hierichonticus (Simon, 1872). However, Francke [15] and Sissom [16] argued that these morphometric ratios were reliable and nonoverlapping among populations and therefore referred to them as valid species. The male hemispermatophore was also found to possess other useful characteristics consistent with Francke’s “morphometric” species. Francke [15] was unable to assign Nebo from Saudi Arabia to any taxon due to the very limited sample size (some characters resembled N. hierichonticus, while others resembled N. yemenensis). However, it was suggested that their eventual location and determination were significant because it reduced the large discontinuity between N. hierichonticus in the north and the other species in the south (Saudi Arabia, Yemen, and Oman). [39,40,41] referred to the existence of three distinct clusters of Androctonus crassicauda populations collected from different ecogeographical regions in Saudi Arabia based on molecular and morphological investigations. In other words, the variation between species in the genus Nebo may reflect vicariances and dispersals caused by climatic and geologic changes that have shaped the Arabian landscape during the past few million years.
Conclusion
In conclusion, this study demonstrated a clear intraspecific and interspecific variation among two species of genus Nebo from Saudi Arabia and Palestine, with strong support for a monophyletic relationship of the subfamily Nebinae and Diplocentrinae pending additional analyses with more representative species. The relationships between Saudi N. yemenensis and other species of this genus need to be clarified, which will require additional in-depth morphological and molecular revisions.
Data availability
The dataset generated and analyzed during the current study in the https://www.ncbi.nlm.nih.gov/genbank/ with accession numbers (OP970165, OP970166, OP970167, OP970168).
References
Rana N, Saleem M, Majeed W, Jalal F, Ehsan N, Nargis S. Diversity of arthropods regarding habitat specialty in agro-ecosystem of Faisalabad, Pakistan. GSC Biol Pharm Sci. 2019;6:01–8.
Majeed W, Rana N, de Azevedo Koch EB, Nargis S. Seasonality and climatic factors affect diversity and distribution of Arthropods around Wetlands. Pak J Zool. 2020;52:2135–44.
Moreno-Carmona M, Montaña-Lozano P, Prada Quiroga CF, Baeza JA. Comparative analysis of mitochondrial genomes reveals family-specific architectures and molecular features in scorpions (Arthropoda: Arachnida: Scorpiones). Gene. 2023;859:147189.
Rein JO. Scorpion files. Family Diplocentridae.https://www.ntnu.no/ub/scorpion-files/diplocentridae.phpAccessed on 15.06.2022. 2022.
Soleglad ME, Fet V. High-level systematics and phylogeny of the extant scorpions (Scorpiones: Orthosterni). Euscorpius. 2003;2003:1–56.
Francke OF. Scorpions of the genus Diplocentrus Peters from Oaxaca, Mexico. J Arachnol. 1977;4:145–200.
Prendini L. Phylogeny and classification of the superfamily Scorpionoidea Latreille 1802 (Chelicerata, Scorpiones): an exemplar approach. Cladistics. 2000;16:1–78.
Santibáñez-López CE, Kriebel R, Sharma PP. Eadem figura manet: measuring morphological convergence in diplocentrid scorpions (Arachnida:Scorpiones:Diplocentridae) under a multilocus phylogenetic framework. Invertebr Syst. 2017;31:233–48.
Ward MJ, Ellsworth SA, Rokyta DR. Venom-gland transcriptomics and venom proteomics of the Hentz striped scorpion (Centruroides hentzi; Buthidae) reveal high toxin diversity in a harmless member of a lethal family. Toxicon. 2018;142:14–29.
Birula AA. Ueber Scorpio maurus Linné und seine unterarten. Horae Soc Entomol Ross. 1910;35:115–92.
Pallary P. Les scorpions du Sahara central. Bull la Société d’Histoire Nat l’Afrique du Nord. 1929;20:133–41.
Werner F. Neu-Eingänge von Skorpionen im Zoologischen Museum in Hamburg. Festschrift zum 60. Geburtstage von Profr Dr Embrik strand. Riga. 1936;2:171–93.
Fet V. Family Scorpionidae Latreille, 1802. In: F V, S WD, L G, M.E. B, editors. Catalog of the Scorpions of the World (1758–1998). New York; New York Entomological Society.; 2000. pp. 427–86.
Prendini L, Crowe TM, Wheeler WC. Systematics and biogeography of the family Scorpionidae (Chelicerata: Scorpiones), with a discussion on phylogenetic methods. Invertebr Syst. 2003;17:185–259.
Francke OF. Revision of the Genus Nebo Simon (Scorpiones: Diplocentridae). J Arachnol. 1980;8:35–52.
Sissom WD. Descriptions of New and poorly known Scorpions of Yemen (Scorpiones: Buthidae, Diplocentridae, Scorpionidae). Fauna Saudi Arab. 1994;14:3–39.
Hendrixson BE. Buthid scorpions of Saudi Arabia, with notes on other families (Scorpiones: Buthidae, Liochelidae, Scorpionidae). Fauna Arab. 2006;21:33–120.
Kinzelbach R. Vorderer Orient. Skorpione. (Arachnida: Scorpiones). Tübinger Atlas der Vor Orients (TAV),Karte Nr A. 1985;VI 14.2.
Vachon M, Kinzelbach R. On the taxonomy and distribution of the scorpions of the Middle East. pp. 91–103. Proceeding Symp Fauna Zoogeography Middle East, Mainz, 1985 (Beihefte zum Tübingen Atls des Vor Orients). 1987;28 (Beihef:338.
Alqahtani AR, Badry A. A contribution to the scorpion fauna of Saudi Arabia, with an identification key (Arachnida: Scorpiones). J King Saud Univ - Sci. 2021;33:101396.
Alqahtani AR. The first record of scorpion Nebo yemenensis Francke, 1980 in Saudi Arabia. Arthropoda Sel. 2022;31:299–304.
Beron P. Review of the Classification of the Orders of Arachnida. 2018. p. 135–317.
Štundlová J, Šmíd J, Nguyen P, Šťáhlavský F. Cryptic diversity and dynamic chromosome evolution in Alpine scorpions (Euscorpiidae: Euscorpius). Mol Phylogenet Evol. 2019;134:152–63.
Gantenbein B, Fet V, Largiader CR, Scholl A. First DNA pylogeny of Euscorpius Thorell, 1876 (Scorpiones, Euscorpiidae) and its bearing on taxonomy and biogeography of the genus. Biogeographica. 1999;75:49–65.
Ayhan H, Atay E, Çarhan A, Yağmur EA, Özkan Ö, Çelebi B. Phylogenetic relationships among the Leiurus abdullahbayrami (Scorpiones: Buthidae) populations in Turkey inferred from mitochondrial DNA sequence data. Biharean Biol. 2018;12:65–9.
Alqahtani AR, Badry A. Interspecific phylogenetic relationship among different species of the genus Buthacus(Scorpiones: Buthidae) inferred from 16S rRNA in Egypt and Saudi Arabia. Zool Middle East. 2020;66:178–85.
Alqahtani AR, Badry A. Genetic diversity among different species of the genus Leiurus (Scorpiones: Buthidae) in Saudi Arabia and the Middle East. Saudi J Biol Sci. 2020;27:3348–53.
Sarhan M, Badry A, Younes M, Saleh M. Genetic diversity within Leiurus quinquestriatus (Scorpiones: Buthidae) populations in Egypt as inferred from 16S mDNA sequence analysis.Zool Middle East. 2020;:269–76.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–9.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9.
Levy G, Amitai P. Fauna Palaestina. Arachnida I: Scorpiones. 1980.
Gantenbein B, Kropf C, Largiadèr CR, Scholl A. Molecular and morphological evidence for the presence of a new Buthid taxon (Scorpiones: Buthidae) on the island of Cyprus. Rev Suisse Zool. 2000;107:213–32.
Froufe E, Alekseyev S, Alexandrino P, Weiss S. The evolutionary history of sharp- and blunt-snouted lenok (Brachymystax lenok (Pallas, 1773)) and its implications for the paleo-hydrological history of Siberia.BMC Evol Biol. 2008;8.
Ben Othmen A, Said K, Mahamdallie SS, Testa JM, Haouas Z, Chatti N, et al. Phylogeography of Androctonus species (Scorpiones: Buthidae) in Tunisia: diagnostic characters for linking species to scorpionism. Acta Trop. 2009;112:77–85.
Ozkan O, Ahmet C, Zafer K. A study on the genetic diversity of Androctonus crassicauda (olivier, 1807; scorpiones: Buthidae) from Turkey. J Venom Anim Toxins Incl Trop Dis. 2010;16:599–606.
Sousa P, Froufe E, James Harris D, Alves PC, van der Meijden A. Genetic diversity of maghrebian Hottentotta (Scorpiones: Buthidae) scorpions based on CO1: new insights on the genus phylogeny and distribution. Afr Invertebr. 2011;52:135–43.
Coelho P, Sousa P, Harris DJ, Van der Meijden A. Deep intraspecific divergences in the medically relevant fat-tailed scorpions (Androctonus, Scorpiones). Acta Trop. 2014;134:43–51.
Toprak S, Parmaksiz A, Aslan N. Mitochondrial cytochrome oxidase subunit I-sequence variation in three scorpion species from southeast Turkey. Indian J Anim Res. 2019;53:187–91.
Alqahtani AR, Badry A, Amer SAM, Al Galil FMA, Ahmed MA, Amr ZS. Intraspecific molecular variation among Androctonus crassicauda (Olivier, 1807) populations collected from different regions in Saudi Arabia. J King Saud Univ - Sci. 2022;34:101998.
Alqahtani AR, Badry A, Aly H, Amer SAM, Al Galil FMA, Ahmed MA, et al. Genetic diversity and population structure of Androctonus crassicauda (Scorpiones: Buthidae) in different ecogeographical regions of Saudi Arabia and Iran. Zool Middle East. 2022;68:171–9.
Alqahtani AR, Badry A, Al Galil FMA, Amr ZS. Morphometric and meristic diversity of the species Androctonus crassicauda (Olivier, 1807) (Scorpiones: Buthidae) in Saudi Arabia. PeerJ. 2022;10.
Acknowledgements
The authors are thankful to the Deanship of Scientific Research at University of Bisha for supporting this work through the Fast-Track Research Support Program.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
ARA designed the Study. NJA, HA, AB and ARA wrote the manuscript. NJA, HA and AB analyzed the data statistically. AB, ARA, HA review the article and AB, NJA, HA edited final manuscript. All authors approved final version of the manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
The Faculty of Science, Department of Biology certifies that, this work was carried out according to the ethical framework of laws of wildlife protection in the Kingdom of Saudi Arabia.
Consent to publish
Not applicable.
Conflict of interest
All authors declare that they have no conflict of interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Alqahtani, A.R., Alotaibi, N.J., Aly, H. et al. The phylogenetic relationship among two species of genus Nebo (Scorpiones: Diplocentridae) from Saudi Arabia and Middle East. BMC Zool 8, 4 (2023). https://doi.org/10.1186/s40850-023-00166-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40850-023-00166-9