Abstract
Background
Neonatal diet impacts many physiological systems and can modify risk for developing metabolic disease and obesity later in life. Less well studied is the effect of postnatal diet (e.g., comparing human milk (HM) or milk formula (MF) feeding) on mitochondrial bioenergetics. Such effects may be most profound in splanchnic tissues that would have early exposure to diet-associated or gut microbe-derived factors.
Methods
To address this question, we measured ileal and liver mitochondrial bioenergetics phenotypes in male piglets fed with HM or MF from day 2 to day 21 age. Ileal and liver tissue were processed for mitochondrial respiration (substrate only [pyruvate, malate, glutamate], substrate + ADP, and proton “leak” post-oligomycin; measured by Oroboros methods), mitochondrial DNA (mtDNA) and metabolically-relevant gene expression analyses.
Results
No differences between the diet groups were observed in mitochondrial bioenergetics indices in ileal tissue. In contrast, ADP-dependent liver Complex I-linked OXPHOS capacity and Complex I + II-linked OXPHOS capacity were significantly higher in MF animals relative to HM fed piglets. Interestingly, p53, Trap1, and Pparβ transcript abundances were higher in MF-fed relative to HM-fed piglets in the liver. Mitochondrial DNA copy numbers (normalized to nuclear DNA) were similar within-tissue regardless of postnatal diet, and were ~ 2–3 times higher in liver vs. ileal tissue.
Conclusion
While mechanisms remain to be identified, the data indicate that neonatal diet can significantly impact liver mitochondrial bioenergetics phenotypes, even in the absence of a change in mtDNA abundance. Since permeabilized liver mitochondrial respiration was increased in MF piglets only in the presence of ADP, it suggests that formula feeding led to a higher ATP turnover. Specific mechanisms and signals involved with neonatal diet-associated differences in liver bioenergetics remain to be elucidated.
Similar content being viewed by others
Background
Breastfeeding has been shown to have positive impact on the body’s physiological systems, including the immune system and metabolically-important tissues such as liver, adipose, and cognitive centers in the brain [1, 2]. Recent studies have reported that milk formula-fed (MF) infants present more rapid weight gain during the first weeks of life compared to breastfed infants, and this appears to be associated with weight gain later in life [3,4,5]. The nutrient composition of human milk (HM) in comparison to milk formulas may play a significant role in the observed metabolic outcomes and the reported health differences when comparing these two neonatal diets [6,7,8,9,10,11]. Mitochondrial function and energy homeostasis impinge on all of these systems, but the role of infant diet and programming of cellular bioenergetics remains largely unexplored.
A previous study in 8 week old rats fed with HM or donkey milk (DM) in comparison to cow’s milk for 4 weeks showed altered liver and skeletal muscle mitochondrial function and metabolism [12, 13]. Specifically, in isolated liver mitochondria, HM and DM-treated groups had increased succinate or palmitoylcarnitine-associated proton leak, increased states 3 and 4 respiration, and higher mitochondrial markers (e.g., carnitine palmitoyltransferase and citrate synthase activities) when compared to mitochondria from untreated control animals [12]. Mitochondria from animals fed cow’s milk shared some similarities to HM and DM, but the phenotype was complicated by significantly higher adiposity. In muscle mitochondria, leak and states 3 and 4 respiration were again higher in samples derived from HM or DM fed rats vs. untreated control rats, whereas these parameters were equivalent in cow’s milk fed and untreated control rats [13]. In addition, HM and DM increased liver and muscle levels of N-oleoylethanolamine (OEA), a regulator of lipid metabolism [13]. The authors speculated that this could contribute to burning of fat and protect the animals from developing certain obesity-associated metabolic and inflammatory sequelae. Furthermore, they speculated that diet-associated changes in microbiota and increased short chain fatty acid (butyrate) in HM- and DM-fed animals contributed to the differences in metabolism and mitochondrial function through as-yet unknown signaling pathways [12]. We have reported significant alterations in the bioregional gut microbiome when comparing HM- and MF-fed piglets [14], and several studies have indicated differences in gut microbiota comparing breastfed to formula-fed infants [15,16,17,18].
There is increasing appreciation for the impact of early-life diet on “programming” of physiological systems with potential metabolic consequences in childhood or adulthood. More important, there is growing consensus that nutritional “programming effects” persist and influence risk for allergies, asthma, obesity, diabetes, and cardiovascular disease later in life [1, 19,20,21,22,23,24]. We hypothesized that neonatal diet (HM and MF) would differentially affect mitochondrial respiration in the small intestine (ileum) and liver. To address our hypothesis, we used HM and MF-fed piglets under controlled experimental conditions, due to inherent variability associated with sow-fed piglets (e.g., farm housing, mother’s skin contact, and suckling diet). Tissues were collected from piglets fed HM and MF between postnatal day 2–21, to determine substrate- and ADP-driven respiration as well as mitochondrial DNA copy number. To our knowledge, this is the first study to characterize the impact of postnatal feeding paradigms on tissue mitochondrial function in a larger animal model.
Methods
Study design
Animals were maintained in accordance with the ethical guidelines for animal research established and approved by the Institutional Animal Care and Use Committee at the University of Arkansas for Medical Sciences. Animals were obtained from Metz Farm (Russellville, AR) that are approved for research. Two-day old White Dutch Landrace Duroc male piglets (Metz Farm, Russellville, AR) were transferred to individual housing at the vivarium in Arkansas Children’s Nutrition Center (ACNC). The study protocol and information on this cohort of animals has been published previously [14]. Piglets were obtained from 30 different sows and housed individually. Based on weights piglets were randomized using excel sheet formula to consume either human milk (HM, n = 26) or isocaloric dairy milk-based formula diet (MF; n = 26). HM was obtained from the Mothers’ Milk Bank of North Texas (Benbrook, TX). For the MF diet, Similac Advance powder was obtained from Ross Products (Abbott Laboratories, Columbus, OH). Both HM and MF diets were modified to meet the energy and nutrient recommendations of the National Research Council for growing pigs [25]. Diet composition, nutritional contents, and methods related to the feeding paradigm have been published previously [14]. Briefly, with the transition to ACNC, piglets were trained to drink from rubber nipples and were provided 1.047 MJ·kg− 1·d− 1 of either HM or MF. Piglets were fed with warmed diet every 2 h in the first week of the study, every 4 h in the second week of the study, and every 6 h in the third week of the study through day 21. At day 14, solid “starter pig food” (Teklad diet, TD 140608; Harlan) was slowly introduced until day 21 when 11 randomly selected piglets from each diet group were euthanized. Piglets were euthanized with a mask at an oxygen flow rate of 2.0 LPM and isofluorane mixture of 3–5% at and cardiovascular signs (pulse) were monitored to assure complete unconciousnees prior to exsanguination. The remaining animals were utilized for immunization challenge studies as previously published (n = 30) [14], and are therefore not included in the current paper. No significant difference in body weights was observed between HM and MF as previously published [14].
Tissue processing for mitochondrial functional analysis
Animals were fasted for 8 h prior to tissue collection. A subset of piglets were randomly chosen to assess mitochondrial function (HM, n = 8–11; MF, n = 8–11; see individual figures). We measured 50 cm from the distal end of small intestine, and at that mark tissue was cut as a 15 cm proximal sample. Ileal and liver samples for analyses of mitochondrial function were processed immediately after collection. A portion of ileum and liver (∼40 mg) was immediately submerged in ice-cold preservation buffer (BIOPS) containing 10 mM Ca-EGTA buffer, 20 mM imidazole, 20 mM taurine, 50 mM K-MES, 0.5 mM dithiothreitol, 6.56 mM MgCl2, 5.77 mM ATP, and 15 mM creatine phosphate (pH of 7.1) [26], for mitochondrial respiration analysis within 1–2 h of tissue collection.
Ileal and liver samples were minced (0.1–0.2 mm) using small and sharp forceps while on ice, and chemically permeabilized for 20 min in BIOPS buffer containing 50 μg/ml saponin at 4 °C [27,28,29,30]. Samples were transferred to 2 ml of MIR05 buffer (0.5 mM EGTA, 3 mM MgCl2, 60 mM K-lactobionate, 20 mM taurine, 10 mM KH2PO4, 20 mM HEPES, and 110 mM sucrose, and 1 mg/ml essential fatty acid-free bovine serum albumin (Sigma Aldrich, St. Louis, MO, USA; Lot SLBF5061V) followed by 10 min mixing on a shaker to wash away the remaining saponin. Permeabilized tissue explants were blotted on filter paper before being weighed on a precision microbalance. Less than 10 mg of wet weight ileum and about 3 mg of wet weight liver were transferred to an Oxygraph-2 k (O2k) respirometer chamber (Oroboros Instruments, Innsbruck, Austria) containing 2 ml of MIR05 buffer [28, 31, 32].
High-resolution respirometry (HRR)
Before each experiment, a background calibration was performed on each O2k polygraphic oxygen sensor (POS). This calibration was performed in MIR05 buffer at air saturation. Zero oxygen and instrumental background calibrations were performed at regular intervals throughout the data collection period (∼12 months) using dithionite titrations. This ensured acceptable POS instrumental background and stability over time [26]. Temperature was maintained at 37 ± 0.01 °C by an electronic Peltier during all high-resolution respirometry (HRR) experiments. O2 concentration within the MIR05 buffer was recorded at 2 to 4 s intervals from which O2 fluxes were calculated in the picomolar range (DatLab version 6; Oroboros Instruments, Innsbruck, Austria) [31]. Once tissue samples had been placed in the O2k chambers, a gas phase was created. About 1 ml of 99% O2 was injected into each O2k chamber, and equilibration of the gas phase and MIR05 O2 concentrations was monitored until an O2 concentration of ∼400 μM was achieved in the MIR05 buffer; O2 flux measurements were typically made when O2 concentrations were in the range of 200–400 μM to minimize any O2 dependency artefacts and to avoid potential limitations in oxygen diffusion in permeabilized tissue samples.
Mitochondrial respiratory capacity and function were evaluated by the sequential addition of substrates and inhibitors. A wide variety of substrate- inhibitor-titration protocols have been used for evaluation of mitochondrial respiration in different samples and under various experimental conditions [30, 33,34,35]. Here, we evaluated the contribution of complex I and II to mitochondrial respiration, as well as the impact of inhibition of complex V. A bioenergetics protocol was used that informs on specific respiratory pathways converging at the Q-junction of the electron transport chain (Table 1). Effects of NADH-linked substrates (N-pathway), including pyruvate (5 mM), malate (2 mM) and glutamate (PMG, 10 mM) were evaluated to determine non-ATP-linked respiration at Complex I [26]. The present protocol also evaluated oxygen consumption rate (pmol O2 · s− 1 · mg wet weight tissue− 1) to determine the oxidative phosphorylation capacity (“OXPHOS capacity” (P)) after adenosine diphosphate (ADP, 5 mM) addition [27, 28, 32]. Subsequently, the “S-pathway” was evaluated using succinate as substrate (10 mM) to determine respiration at Complex II. Also, proton leak respiration at complex I and II was determined by adding oligomycin (OMY; 0.2 μg/ml) after substrates and ADP were provided. The respiratory oxidative capacity results were corrected by the residual oxygen respiration (ROX) obtained after antimycin A (AMA; 12.5 μM) inhibition of electron transport chain Complex III (a.k.a. UQ-cytochrome c oxidoreductase) at the end of the experiment.
Mitochondrial DNA copy number
DNA was extracted from ~ 25 mg of frozen ileal tissue using QIAamp Fast DNA Tissue Kit (QIAGEN, Germantown, MD) following the manufacturer’s protocol. Approximately 500 mg of the liver was homogenized in PBS (Gibco, Thermo Fisher). DNA was extracted from the equivalent of ~ 25 mg of liver using QIAamp Fast DNA Tissue Kit. DNA standards for each target were generated using primers listed in Table 2 to amplify the mitochondrial genes ND1 (519 base pair (bp) amplicon) and Cox1 (477 bp amplicon), and the nuclear gene β-actin (496 bp amplicon) using GoTaq Green Master Mix (Promega, Madison, WI). PCR products were separated on a 1% agarose gel and amplicon bands were excised and extracted using QIAquick Gel Extraction Kit (QIAGEN, Germantown, MD). Concentrations were determined using a NanoDrop 1000 Spectrophotometer (NanoDrop, Wilmington, DE) and a Qubit 2.0 fluorometer using the dsDNA HS Assay Kit (Invitrogen). Copy number per μl of DNA was determined using the equation:
number of copies/μl = (amount (ng/μl) * 6.022 × 1023 number /mole) / (length (bp) * 1 × 109 ng/g * 650 g/mole of bp).
Standards were serially diluted 10-fold from 1 X 108 copies/μl to 1 X 103 copies/μl to construct a 6-point standard curve. For real-time PCR, new primers located within the amplicon for ND1, Cox1 and β-actin (Table 2) were designed using Integrated DNA Technology’s (IDT) PrimerQuest Tool (www.idtdna.com/Primerquest/Home/Index). DNA (200 pg) was used in a 10 μl PCR reaction using Fast SYBR Green Master Mix (Applied Biosystems) on a ViiA 7 Real-Time PCR System (Applied Biosystems). mtDNA copy number was calculated using standard curves and normalized using the abundance of the β-actin nuclear gene. All available piglet samples were used to determine mitochondrial DNA copy number (HM, n = 11; MF, n = 11 per tissue).
Gene expression
RNA was extracted from ~ 30 mg of frozen ileum tissue using RNeasy Plus Mini Kit (Qiagen) according to manufacturer’s protocol. For liver RNA, ~ 100 mg of frozen livers were homogenized using miRNeasy Mini Kit (Qiagen) and homogenate equivalent to ~ 30 mg of liver from each sample was used for RNA extraction. Concentration was determined using UV absorbance. One μg of total RNA was used to create cDNA using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA) according to manufacturer’s protocol. cDNA was diluted and 10 ng of cDNA was used in a PCR reaction using Fast SYBR Green Master Mix (Applied Biosystems) on a ViiA 7 Real-Time PCR System (Applied Biosystems). A standard curve was generated by pooling undiluted cDNA from different treatments to create a master pool. The master cDNA mix was serially diluted 5-fold a total of four times, for an arbitrary five-point standard curve. RNA expression was normalized for the ileum using the geometric mean of three (Rps16, Rpl27, and 18S) reference RNAs and for the liver using the geometric mean of two (Rps16 and 18S) reference RNAs that were not altered by treatment. All available piglet samples were used to determine mitochondrial gene expression (HM, n = 11; MF, n = 11). One MF fed piglet liver sample was removed due to poor RNA quality (RIN ~ 6.5). Primers were designed using IDT’s PrimerQuest and are listed in Table 3.
Statistical analyses
All data are presented as means ± SEM. The data were analyzed by two-tailed t-test and p < 0.05 was considered to be statistically significant between the groups.
Results
Mitochondrial respiration phenotypes
A trace of oxygen flux using permeabilized ileum and liver samples derived from a 21-day old piglet fed with HM is shown in Fig. 1. This protocol provides assessment of “respiratory leak” (L) supported primarily by electron flow through complex I of the respiratory chain after addition of pyruvate, malate and glutamate (PMG-L). The protocol also provides the OXPHOS capacity (P) after ADP is added following the pyruvate, malate and glutamate (PMG-P); P represents maximal Complex I-linked ADP-linked OXPHOS capacity through the N-pathway, electron transfer occurs by Complex I. Subsequently, succinate (SUC) is added to induce complex II-linked respiration. We determined the contribution of electron flow from both complex I and II respiration, representing the maximal ADP-linked OXPHOS capacity that was measured in ileum. Next, OMY was added to assess leak respiration when substrates and ADP were provided, but ATP synthase inhibited, measuring proton leak from complex I and II.
We assessed the impact of neonatal diets on mitochondrial respiration in permeabilized ileum on day 21 (Fig. 2a). No significant differences were observed in the permeabilized ileum explants for all parameters tested: PMG respiratory leak, Complex I-linked OXPHOS capacity, measured after adding ADP, Complex I- and II-linked OXPHOS capacity (CI + II P) measured after addition of succinate (SUC), and Complex I and II-linked leak (OMY). In the liver (Fig. 2b), higher Complex I-linked OXPHOS capacity and higher Complex I- and II-linked OXPHOS capacity were observed in MF-fed piglets compared to HM-fed piglets.
Respiratory fluxes of piglet ileum on day 21, from animals consuming human milk (HM) or cow milk-based formula (MF). a Mass-specific respirometry results of permeabilized ileum tissue of 21-day old piglets fed either HM or MF (n = 11/diet group). b Mass-specific respirometry results of permeabilized liver of 21-day old piglets fed either HM (n = 8) or MF (n = 8) for a period of 21 days starting on postnatal day 2. PMG: pyruvate, malate and glutamate; ADP: adenosine diphosphate; SUC: succinate; OMY: oligomycin; The data are presented as mean ± SEM. The data were analyzed by 2-tailed t-test and *p < 0.05, **p < 0.01 was considered significant between the diet groups
Mitochondrial DNA
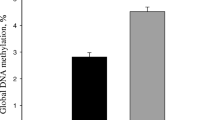
Mitochondrial DNA copy number, normalized to nuclear DNA copy number, did not significantly differ on day 21 between the HM and MF diet groups in ileum or liver (Fig. 3). Notably, mtDNA was ~ 2–3 times higher in liver compared to ileum.
Mitochondrial DNA (mtDNA) copy number in 21 day old piglet ileum and liver tissues. a Day 21 mtDNA (n = 11/ diet group) of ileum and b day 21 mtDNA (n = 11/diet group) of liver. mtDNA values are normalized to the nuclear DNA target gene beta-actin. The data are presented as mean ± SEM. The data were analyzed by 2-tailed t-test to determine the significance between the diet groups. HM - Human milk; MF - Cow milk-based formula
Expression of select genes relevant to metabolism and mitochondrial bioenergetics
We measured gene expression for select transcripts involved in mitochondrial function and metabolism from ileum and liver. In ileum, no significant differences were noted between the diet groups with respect to Ubiquinol-Cytochrome C Reductase (Uqcr10), Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1-Alpha (Pgc1α), Cytochrome C (Cycs), Heat Shock Protein Family A, Member 9 (Hspa9), p53, Nuclear Respiratory Factor 1 (Nrf1), TNF Receptor Associated Protein 1 (Trap1), and Transcription Factor A mitochondria (Tfam) transcript abundances at day 21 (Fig. 4a). Interestingly, in ileum Peroxisome Proliferator-Activated Receptor Beta (Pparβ) expression was significantly lower in MF group at day 21 relative to the HM group, while Cytochrome C Oxidase Assembly Protein (Sco2) expression was significantly higher (Fig. 4a). In liver, p53, Trap1 and Pparβ transcript abundances were higher in MF-fed relative to HM-fed piglets (Fig. 4b).
Gene expression analysis of piglet ileum and liver at day 21, from animals consuming human milk (HM) or cow milk-based formula (MF). a lleum (n = 11/ diet group) and b liver (n = 10–11/diet group) samples expression of Uqcr10, Pgc1α, Hspa9, p53, Nrf1, Trap1, Tfam, Cycs, Pparβ and Sco2. Normalized arbitrary units were calculated using geometric mean of 2–3 reference genes and standard curves generated for each target (see methods section). The data are presented as mean ± SEM. The data were analyzed by 2-tailed t-test, *p < 0.05, **p < 0.01 was considered significant between the diet groups
Discussion
The current proof-of-principle study focusing on piglet ileal and liver tissue uncovered novel findings with respect to infant diet and tissue mitochondrial function, and suggest that select aspects of liver bioenergetics are altered by neonatal diet.
The area of developmental programming of bioenergetics, which encompasses systems involving the mitochondrial management of fuels and metabolites to ultimately support efficient energy conversion to ATP and regulation of reactive oxygen species generation, remains largely unexplored. It may be expected that one’s diet patterns and macronutrient availability would influence mitochondrial function and adaptations that would ensure proper maintenance and development of bioenergetics systems. This may be especially profound in tissues that are proximal to gut-derived signals. Signals from the gut microbiome, or that are changed coincident with microbiome shifts, reduce liver or muscle expression of some metabolism genes (e.g., PGC-1α, SIRT1, and AMPK) and regulate mitochondrial biogenesis and function (reviewed in [36]). The most obvious example is butyrate, which can mediate PGC-1α induction in rodent muscle. Administration of the probiotic Lactobacillus rhamnosus CNCMI-4317 to adult animals appeared to activate PPAR-α pathways coincident with higher oxidative phosphorylation capacity. Several clinical and animal studies have shown gut microbiota differences in breast-fed versus formula-fed neonates [14, 37,38,39,40,41], and other studies have demonstrated that gut microbiota may increase fatty acid uptake and oxidation through secondary bile acids [42,43,44]. The gut microbes Bacteroides, Eubacterium, and Clostridium genera degrade the primary bile acids to secondary bile acids [45]. Interestingly, from the same piglets studied herein, we have reported higher abundance of Bacteroides in HM-fed piglets relative to MF-fed piglets [14]. In addition, rats fed with HM also showed increased Bacteroides colonization compared to control animals along with enhanced mitochondrial activity/proton leakage [12]. Since the GI tract is in direct contact with gut microbes, and other splanchnic tissues such as liver are immediately downstream, mitochondria and mitochondrial regulators in splanchnic tissue cells may be particularly sensitive to microbe-derived signals. Such a hypothesis awaits testing in future studies.
In addition to microbial signals, there is the possibility that food components themselves influence mitochondrial function. Rat models (post-weaned, adult animals) indicate that mitochondrial function of liver and skeletal muscle can be modulated by dietary components [13, 46,47,48,49,50,51]. However, nothing is known about the effect of postnatal diet on mitochondrial function, especially in the splanchnic tissues that would be exposed to dietary components and microbial-derived factors that might regulate metabolic pathways. For instance, human milk and donkey milk improved glucose and lipid metabolism, and modified mitochondria in adult rat skeletal muscle when compared to untreated control animals [13], although it could not be ascertained if these differences were driven by food components or other factors emanating from the gut microbes or host tissues. Furthermore, diet components have been shown to impact mitochondrial bioenergetics in animal models and cell culture (reviewed in [52]). In liver and muscle there is evidence that exposure to high levels of saturated fatty acids promotes mitochondrial fission-like events, decreased proton leak, and higher ROS, whereas ω3 polyunsaturated fatty acids have the opposite effect in animal models. Whether or not fats and other components of the neonatal diet influence mitochondrial function or programming of mitochondrial bioenergetics remains to be evaluated experimentally. Regardless of specific mechanisms, the current study indicates that HM and MF feeding can elicit different bioenergetics phenotypes in the liver: piglets fed MF displayed significantly higher ADP-linked respiration. The increased permeabilized liver respiration was not due to differences in the number of mitochondria (since copy number per genomic beta-actin was not different between the diet groups), and was not due to gross differences in proton leak. From these observations, it appears that higher ATP turnover in the MF liver drove higher O2 consumption. On the other hand, neonatal feeding paradigm did not appear to alter ileum bioenergetics compared to HM fed piglets, highlighting a potential tissue specificity in the diet-splanchnic tissue bioenergetics interplay. The current study design does not allow us to determine the specific mechanisms involved, but one speculation is that in the case of ileum there is more proximal exposure to diet- and microbe-derived tissue regulators, whereas metabolic regulation in the liver is an aggregate of gut-derived and systemic factors. Future studies are needed to validate these findings in other models, and to determine if additional regions of the gut display changes in mitochondrial function in response to neonatal feeding paradigms.
Conclusion
Our data demonstrate that neonatal diet impacts liver mitochondrial bioenergetics phenotypes. In addition, in the presence of ADP, mitochondrial respiration was increased in MF piglets relative to HM-fed piglets, suggesting that formula feeding led to a higher innate hepatic ATP turnover. Future studies are needed to understand how diet-associated differences in gut tissue mitochondrial respiration come about, and if there are potential long-term ramifications with respect to energy homeostasis, oxidative stress, and other outcomes. Clearly, more work needs to be done to evaluate this working model of neonatal diet effects on mitochondrial function. The specific diet- or microbe-derived signals, and/or host signals, that regulate mitochondrial activities (and their temporal relationship to the neonatal period), as well as specific gut regions impacted, remain to be identified. Should infant diet effects on bioenergetics described herein recapitulate in human tissues, it would have profound implications in terms of understanding fundamental molecular events that differentiate physiological responses to formula-feeding and breastfeeding.
Availability of data and materials
All generated or analyzed data are included in the article. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ACNC:
-
Arkansas Children’s Nutrition Center
- ADP:
-
Adenosine diphosphate
- AMA:
-
Antimycin A
- DM:
-
Donkey milk
- HM:
-
Human milk
- HRR:
-
High-resolution respirometry
- OEA:
-
Oleoylethanolamine
- OMY:
-
Oligomycin
- PMG-L:
-
Pyruvate, malate and glutamate
- SUC:
-
Succinate
References
Owen CG, Martin RM, Whincup PH, Smith GD, Cook DG. Effect of infant feeding on the risk of obesity across the life course: a quantitative review of published evidence. Pediatrics. 2005;115(5):1367–77.
Owen CG, Martin RM, Whincup PH, Davey-Smith G, Gillman MW, Cook DG. The effect of breastfeeding on mean body mass index throughout life: a quantitative review of published and unpublished observational evidence. Am J Clin Nutr. 2005;82(6):1298–307.
Feldman-Winter L, Burnham L, Grossman X, Matlak S, Chen N, Merewood A: Weight gain in the first week of life predicts overweight at 2 years: A prospective cohort study. Matern Child Nutr 2018, 14(1)..
Gale C, Logan KM, Santhakumaran S, Parkinson JR, Hyde MJ, Modi N. Effect of breastfeeding compared with formula feeding on infant body composition: a systematic review and meta-analysis. Am J Clin Nutr. 2012;95(3):656–69.
Savino F, Liguori SA, Benetti S, Sorrenti M, Fissore MF. Cordero di Montezemolo L: high serum leptin levels in infancy can potentially predict obesity in childhood, especially in formula-fed infants. Acta Paediatr. 2013;102(10):e455–9.
Liu Z, Neuringer M, Erdman JW, Jr., Kuchan MJ, Renner L, Johnson EE, Wang X, Kroenke CD: The effects of breastfeeding versus formula-feeding on cerebral cortex maturation in infant rhesus macaques. Neuroimage. 2018;184:372-85.
Mercer KE, Bhattacharyya S, Diaz-Rubio ME, Piccolo BD, Pack LM, Sharma N, Chaudhury M, Cleves MA, Chintapalli SV, Shankar K, et al. Infant formula feeding increases hepatic cholesterol 7alpha hydroxylase (CYP7A1) expression and fecal bile acid loss in neonatal piglets. J Nutr. 2018;148(5):702–11.
Lonnerdal B. Infant formula and infant nutrition: bioactive proteins of human milk and implications for composition of infant formulas. Am J Clin Nutr. 2014;99(3):712S–7S.
Visentin S, Vicentin D, Magrini G, Santandreu F, Disalvo L, Sala M, Fasano V, Gonzalez HF. Red blood cell membrane fatty acid composition in infants fed formulas with different lipid profiles. Early Hum Dev. 2016;100:11–5.
Gianni ML, Roggero P, Baudry C, Fressange-Mazda C, Galli C, Agostoni C, le Ruyet P, Mosca F. An infant formula containing dairy lipids increased red blood cell membrane omega 3 fatty acids in 4 month-old healthy newborns: a randomized controlled trial. BMC Pediatr. 2018;18(1):53.
Wu X, Jackson RT, Khan SA, Ahuja J, Pehrsson PR. Human Milk Nutrient Composition in the United States: Current Knowledge, Challenges, and Research Needs. Curr Dev Nutr. 2018;2(7):nzy025.
Trinchese G, Cavaliere G, Canani RB, Matamoros S, Bergamo P, De Filippo C, Aceto S, Gaita M, Cerino P, Negri R, et al. Human, donkey and cow milk differently affects energy efficiency and inflammatory state by modulating mitochondrial function and gut microbiota. J Nutr Biochem. 2015;26(11):1136–46.
Trinchese G, Cavaliere G, De Filippo C, Aceto S, Prisco M, Chun JT, Penna E, Negri R, Muredda L, Demurtas A, et al. Human Milk and donkey Milk, compared to cow Milk, reduce inflammatory mediators and modulate glucose and lipid metabolism, Acting on Mitochondrial Function and Oleylethanolamide Levels in Rat Skeletal Muscle. Front Physiol. 2018;9:32.
Miklavcic JJ, Badger TM, Bowlin AK, Matazel KS, Cleves MA, LeRoith T, Saraf MK, Chintapalli SV, Piccolo BD, Shankar K, Yeruva L. Human Breast-Milk Feeding Enhances the Humoral and Cell-Mediated Immune Response in Neonatal Piglets. J Nutr. 2018;148(11):1860-70. https://doi.org/10.1093/jn/nxy170.
Balmer SE, Wharton BA. Diet and faecal flora in the newborn: breast milk and infant formula. Arch Dis Child. 1989;64(12):1672–7.
Bezirtzoglou E, Tsiotsias A, Welling GW. Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe. 2011;17(6):478–82.
Davis EC, Wang M, Donovan SM. The role of early life nutrition in the establishment of gastrointestinal microbial composition and function. Gut Microbes. 2017;8(2):143–71.
Fanaro S. Vigi V: [infant formulas supplemented with prebiotics: intestinal microbiota and immune responses]. Minerva Pediatr. 2008;60(3):327–35.
Thompson AL. Developmental origins of obesity: early feeding environments, infant growth, and the intestinal microbiome. Am J Hum Biol. 2012;24(3):350–60.
Fewtrell MS. Breast-feeding and later risk of CVD and obesity: evidence from randomised trials. Proc Nutr Soc. 2011;70(4):472–7.
Owen CG, Whincup PH, Cook DG. Breast-feeding and cardiovascular risk factors and outcomes in later life: evidence from epidemiological studies. Proc Nutr Soc. 2011;70(4):478–84.
Yan J, Liu L, Zhu Y, Huang G, Wang PP. The association between breastfeeding and childhood obesity: a meta-analysis. BMC Public Health. 2014;14:1267.
Miliku K, Robertson B, Sharma AK, Subbarao P, Becker AB, Mandhane PJ, Turvey SE, Lefebvre DL, Sears MR, Investigators CS, et al. Human milk oligosaccharide profiles and food sensitization among infants in the CHILD study. Allergy. 2018;73(10):2070–3.
Moossavi S, Miliku K, Sepehri S, Khafipour E, Azad MB. The prebiotic and probiotic properties of human Milk: implications for infant immune development and pediatric asthma. Front Pediatr. 2018;6:197.
Council NR. Nutrient requirements of swine: eleventh revised edition. Washington, DC: The National Academies Press; 2012.
Chicco AJ, Le CH, Schlater A, Nguyen A, Kaye S, Beals JW, Scalzo RL, Bell C, Gnaiger E, Costa DP, et al. High fatty acid oxidation capacity and phosphorylation control despite elevated leak and reduced respiratory capacity in northern elephant seal muscle mitochondria. J Exp Biol. 2014;217(Pt 16):2947–55.
Chu MJ, Phillips AR, Hosking AW, MacDonald JR, Bartlett AS, Hickey AJ. Hepatic mitochondrial function analysis using needle liver biopsy samples. PLoS One. 2013;8(10):e79097.
Grattagliano I, Portincasa P, Cocco T, Moschetta A, Di Paola M, Palmieri VO, Palasciano G. Effect of dietary restriction and N-acetylcysteine supplementation on intestinal mucosa and liver mitochondrial redox status and function in aged rats. Exp Gerontol. 2004;39(9):1323–32.
Kuznetsov AV, Strobl D, Ruttmann E, Konigsrainer A, Margreiter R, Gnaiger E. Evaluation of mitochondrial respiratory function in small biopsies of liver. Anal Biochem. 2002;305(2):186–94.
Porter C, Hurren NM, Cotter MV, Bhattarai N, Reidy PT, Dillon EL, Durham WJ, Tuvdendorj D, Sheffield-Moore M, Volpi E, et al. Mitochondrial respiratory capacity and coupling control decline with age in human skeletal muscle. Am J Physiol Endocrinol Metab. 2015;309(3):E224–32.
Pesta D, Gnaiger E. High-resolution respirometry: OXPHOS protocols for human cells and permeabilized fibers from small biopsies of human muscle. Methods Mol Biol. 2012;810:25–58.
Koliaki C, Szendroedi J, Kaul K, Jelenik T, Nowotny P, Jankowiak F, Herder C, Carstensen M, Krausch M, Knoefel WT, et al. Adaptation of hepatic mitochondrial function in humans with non-alcoholic fatty liver is lost in steatohepatitis. Cell Metab. 2015;21(5):739–46.
Krumschnabel G, Fontana-Ayoub M, Sumbalova Z, Heidler J, Gauper K, Fasching M, Gnaiger E. Simultaneous high-resolution measurement of mitochondrial respiration and hydrogen peroxide production. Methods Mol Biol. 2015;1264:245–61.
Porter C, Herndon DN, Borsheim E, Chao T, Reidy PT, Borack MS, Rasmussen BB, Chondronikola M, Saraf MK, Sidossis LS. Uncoupled skeletal muscle mitochondria contribute to hypermetabolism in severely burned adults. Am J Physiol Endocrinol Metab. 2014;307(5):E462–7.
Ost M, Doerrier C, Gama-Perez P, Moreno-Gomez S. Analysis of mitochondrial respiratory function in tissue biopsies and blood cells. Curr Opin Clin Nutr Metab Care. 2018;21(5):336–42.
Clark A, Mach N. The crosstalk between the gut microbiota and mitochondria during exercise. Front Physiol. 2017;8:319.
Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, Sears MR, Becker AB, Scott JA, Kozyrskyj AL, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ. 2013;185(5):385–94.
Poroyko V, White JR, Wang M, Donovan S, Alverdy J, Liu DC, Morowitz MJ. Gut microbial gene expression in mother-fed and formula-fed piglets. PLoS One. 2010;5(8):e12459.
Korpela K, de Vos WM. Early life colonization of the human gut: microbes matter everywhere. Curr Opin Microbiol. 2018;44:70–8.
Fanaro S, Chierici R, Guerrini P, Vigi V. Intestinal microflora in early infancy: composition and development. Acta Paediatr Suppl. 2003;91(441):48–55.
Hesla HM, Stenius F, Jaderlund L, Nelson R, Engstrand L, Alm J, Dicksved J. Impact of lifestyle on the gut microbiota of healthy infants and their mothers-the ALADDIN birth cohort. FEMS Microbiol Ecol. 2014;90(3):791–801.
Neis EP, Dejong CH, Rensen SS. The role of microbial amino acid metabolism in host metabolism. Nutrients. 2015;7(4):2930–46.
Kuipers F, Bloks VW, Groen AK. Beyond intestinal soap--bile acids in metabolic control. Nat Rev Endocrinol. 2014;10(8):488–98.
Joyce SA, Gahan CG. Bile acid modifications at the microbe-host Interface: potential for Nutraceutical and pharmaceutical interventions in host health. Annu Rev Food Sci Technol. 2016;7:313–33.
Gerard P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens. 2013;3(1):14–24.
Leduc-Gaudet JP, Reynaud O, Chabot F, Mercier J, Andrich DE, St-Pierre DH, Gouspillou G: The impact of a short-term high-fat diet on mitochondrial respiration, reactive oxygen species production, and dynamics in oxidative and glycolytic skeletal muscles of young rats. Physiol Rep 2018, 6(4)..
Coudray C, Fouret G, Lambert K, Ferreri C, Rieusset J, Blachnio-Zabielska A, Lecomte J, Ebabe Elle R, Badia E, Murphy MP, et al. A mitochondrial-targeted ubiquinone modulates muscle lipid profile and improves mitochondrial respiration in obesogenic diet-fed rats. Br J Nutr. 2016;115(7):1155–66.
Cooper MA, McCoin C, Pei D, Thyfault JP, Koestler D, Wright DE. Reduced mitochondrial reactive oxygen species production in peripheral nerves of mice fed a ketogenic diet. Exp Physiol. 2018;103(9):1206–12.
Cavaliere G, Trinchese G, Bergamo P, De Filippo C, Mattace Raso G, Gifuni G, Putti R, Moni BH, Canani RB, Meli R, et al. Polyunsaturated fatty acids attenuate diet induced obesity and insulin resistance, modulating mitochondrial respiratory uncoupling in rat skeletal muscle. PLoS One. 2016;11(2):e0149033.
Oriquat GA, Ali MA, Mahmoud SA, Eid RM, Hassan R, Kamel MA: Improving hepatic mitochondrial biogenesis as a postulated mechanism for the antidiabetic effect of Spirulina platensis in comparison with metformin. Appl Physiol Nutr Metab. 2019;44(4):357-64.
Cardoso AR, Cabral-Costa JV, Kowaltowski AJ. Effects of a high fat diet on liver mitochondria: increased ATP-sensitive K+ channel activity and reactive oxygen species generation. J Bioenerg Biomembr. 2010;42(3):245–53.
Putti R, Sica R, Migliaccio V, Lionetti L. Diet impact on mitochondrial bioenergetics and dynamics. Front Physiol. 2015;6:109.
Acknowledgements
We thank Matt Ferguson, Bobby Fay, and Trae Pittman for vivarium help with the piglets.
Funding
This work was funded by USDA-ARS Projects 6026–51000-010-05S and 6026–51000-010-06S. EC, MC, and EB are also partly supported by the Arkansas Biosciences Institute, the major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000 and by the NIH (P20GM109096). EC is also supported by COMPETE 2020: POCI-01-0145-FEDER-007440. In addition, LY is also supported by the NIH P20GM121293 and NIH R21AI146521. The funding body was not involved in study design, data collection, analysis, and interpretation and writing of the manuscript.
Author information
Authors and Affiliations
Contributions
The authors’ responsibilities were as follows - EC, SHA, EB, LY: designed and/or interpreted the research; EC, SHA, EB, and LY: wrote the manuscript; EC, AKB, MC, MLB and KOM: conducted technical aspects of the research; EC, SHA, EB, and LY: had primary responsibility for the final content and for editing the manuscript; and all authors: read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Animals were maintained in accordance with the ethical guidelines for animal research established and approved by the Institutional Animal Care and Use Committee at the University of Arkansas for Medical Sciences. All animals used were obtained from Metz Farm that approved the use of animals for research study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Carvalho, E., Adams, S.H., Børsheim, E. et al. Neonatal diet impacts liver mitochondrial bioenergetics in piglets fed formula or human milk. BMC Nutr 6, 13 (2020). https://doi.org/10.1186/s40795-020-00338-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40795-020-00338-7