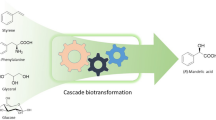
Abstract
Chiral phenylglycinol is a very important chemical in the pharmaceutical manufacturing. Current methods for synthesis of chiral phenylglycinol often suffered from unsatisfied selectivity, low product yield and using the non-renewable resourced substrates, then the synthesis of chiral phenylglycinol remain a grand challenge. Design and construction of synthetic microbial consortia is a promising strategy to convert bio-based materials into high value-added chiral compounds. In this study, we reported a six-step artificial cascade biocatalysis system for conversion of bio-based l-phenylalanine into chiral phenylglycinol. This system was designed using a microbial consortium including two engineered recombinant Escherichia coli cell modules, one recombinant E. coli cell module co-expressed six different enzymes (phenylalanine ammonia lyase/ferulic acid decarboxylase/phenylacrylic acid decarboxylase/styrene monooxygenase/epoxide hydrolase/alcohol dehydrogenase) for efficient conversion of l-phenylalanine into 2-hydroxyacetophenone. The second recombinant E. coli cell module expressed an (R)-ω-transaminase or co-expressed the (S)-ω-transaminase, alanine dehydrogenase and glucose dehydrogenase for conversion of 2-hydroxyacetophenone into (S)- or (R)-phenylglycinol, respectively. Combining the two engineered E. coli cell modules, after the optimization of bioconversion conditions (including pH, temperature, glucose concentration, amine donor concentration and cell ratio), l-phenylalanine could be easily converted into (R)-phenylglycinol and (S)-phenylglycinol with up to 99% conversion and > 99% ee. Preparative scale biotransformation was also conducted on 100-mL scale, (S)-phenylglycinol and (R)-phenylglycinol could be obtained in 71.0% and 80.5% yields, > 99% ee, and 5.19 g/L d and 4.42 g/L d productivity, respectively. The salient features of this biocatalytic cascade system are good yields, excellent ee, mild reaction condition and no need for additional cofactor (NADH/NAD+), provide a practical biocatalytic method for sustainable synthesis of (S)-phenylglycinol and (R)-phenylglycinol from bio-based L-phenylalanine.

Similar content being viewed by others
Introduction
Chiral vicinal amino alcohol moieties are important structural features of many pharmaceutically active molecules and natural products (Bergmeier et al. 2000; Gupta et al. 2018); besides, they can be used as chiral ligands or auxiliaries in asymmetric synthesis (Tan et al. 2017). In this context, enantiopure phenylglycinols are particularly interesting due to their potential use as a key building block for many pharmaceuticals synthesis, such as neurotrophic agent (Panek et al. 1998), h5-HT1D receptor agonists (Russell et al. 1999), antimitotic agents (Kim et al. 2004) and p21-activated kinases (PAK4) inhibitors (Guo et al. 2017).
The industrial synthesis of vicinal amino alcohols typically uses chemical methods. These methods mainly include reduction of amino acids (McKennon et al. 1993; Vandekerkhove et al. 2018), asymmetric ring-opening of epoxides (Overman et al. 1985; Jacobsen et al. 2000), addition of lithiated aziridines to boronic esters (Schmidt et al. 2009), addition of nucleophiles to aminocarbonyls (Reetz et al. 1991) and imines (Kobayashi et al. 1998), and asymmetric aminohydroxylation (AA) of styrene (Li et al. 1996; Sharpless et al. 1975). However, the conventional chemical methods for the synthesis of chiral vicinal amino alcohols are often affected by unsatisfied regioselectivity and enantioselectivity, low product yield, and the toxicity of the metal catalyst and N-protected amino alcohol product. Therefore, the chemical methods used for the industrial synthesis of chiral vicinal amino alcohols are considered to be non-viable for industrial application purposes. However, biocatalysis has been widely used in the synthesis of many useful chemicals, and it is a more sustainable method compared to the above-mentioned methods (You et al. 2021; Wang et al. 2020). In the past two decades, various biocatalytic alternatives have been reported for the synthesis of chiral vicinal amino alcohols, such as racemic amino alcohol kinetic resolution (Wu et al. 2017; Rouf et al. 2011), chiral amino ketones asymmetric reduction (Patel et al. 1993), and α-ketol asymmetric reduction amination (Zhang et al. 2019a; Chen et al. 2019). Recently, two types of biocatalytic cascades for the synthesis of chiral vicinal amino alcohols have been developed via asymmetric ring-opening of epoxides (Zhang et al. 2019b) and asymmetric aminohydroxylation of alkenes (Zhang et al. 2020). The results of these studies showed good conversion percentages (up to 99%) and ee values (> 99%) of amino alcohol products from the tested epoxides and alkenes. Although the substrates (i.e., epoxides and alkenes) used in these methods are not expensive and readily available, these substrates are mainly produced from non-renewable carbon resources. Consequently, these methods can be less sustainable compared to other techniques. Therefore, there is a need to find new biocatalytic strategies for synthesizing chiral vicinal amino alcohols from sustainable resources.
l-α-Amino acids are bio-based materials that can be produced from the fermentation of carbohydrates or waste protein sources (Pelckmans et al. 2017). In fact, the annual worldwide amino acids production is about four million tons (Becker et al. 2012). Therefore, the bio-based l-α-amino acids can make a significant contribution to the economic and ecological production of chemical products and materials. In the past decades, several methods have been developed for converting l-α-amino acids into other high added-value compounds (Fotheringham et al. 2006; Studte et al. 2008; Zhang et al. 2019c). For example, cascade biocatalysis processes have been used for conversion of l-phenylalanine into chiral styrene oxides, 1-phenylethane-1,2-diols, mandelic acids and phenylglycines (Zhou et al. 2016). Tan et al. (2021) described an in vivo four-enzyme cascade pathway for efficient conversion of l-tyrosine to d-p-hydroxyphenylglycine. For instance, these authors reported d-p-hydroxyphenylglycine conversion and ee percentages of 92.5% and > 99% from 50 g/L l-tyrosine, respectively (Tan et al. 2021). Moreover, a whole-cell cascade biocatalysis system was established for efficient production of glutarate (yield = 43.8 g/L) from l-lysine with a recombinant E. coli microbial consortium by applying a fed-batch strategy (Wang et al. 2019). Recently, Sekar et al. (2020) developed a one-pot cascade biotransformation process for conversion of l-phenylalanine to chiral phenylglycinol for the first time. About 42% conversion of (R)-phenylglycinol (99% ee) and 26% conversion of (S)-phenylglycinol (92% ee) were obtained from l-phenylalanine using engineered E. coli strains (Sekar et al. 2020). Unfortunately, the low substrate loadings, productivity and ee values significantly limit the industrial application of this process. Therefore, there is a need to develop more efficient biocatalysis systems for the synthesis of chiral phenylglycinol from l-phenylalanine.
In this study, a novel artificially designed cascade biocatalysis system was constructed for conversion of bio-based l-phenylalanine into chiral phenylglycinol. This system was designed using a microbial consortium including two recombinant E. coli cell modules (Scheme 1). One recombinant E. coli cell module co-expressed the following six enzymes: phenylalanine ammonia lyase (PAL), ferulic acid decarboxylase (Fdc1), phenylacrylic acid decarboxylase (Pad1), styrene monooxygenase (SMO), epoxide hydrolase (EH), and alcohol dehydrogenase (ADH). This module was used for achieving an efficient conversion of l-phenylalanine (L-PA, 1) into 2-hydroxyacetophenone (2-HAP, 6). The second recombinant E. coli cell module expressed the (R)-ω-transaminase or co-expressed the (S)-ω-transaminase, alanine dehydrogenase, and glucose dehydrogenase for conversion purposes of 2-HAP 6 into (S)- or (R)-phenylglycinol, respectively. Moreover, when combining these two recombinant E. coli modules, l-phenylalanine could be easily transformed to (R)-phenylglycinol and (S)-phenylglycinol with good conversion and excellent ee values.
Design and modularization of a de novo designed biocatalytic cascade for production of chiral phenylglycinol from L-phenylalanine. Cell module 1 involves PAL catalyzed deamination of L-phenylalanine 1, Fdc1 and Pad1 mediates the conversion of trans-cinnamic acid 2 to styrene 3, SMO catalyzed oxidation of styrene 3 to styrene oxide 4, SpEH catalyzed hydrolysis of styrene oxide 4 to diol 5 and GoSCR catalyzed oxidation of diol 5 to 2-hydroxyacetophenone 6. Cell module 2 involves MVTA catalyzed asymmetric amination of 2-hydroxyacetophenone 6 to (S)-phenylglycinol 7. Cell module 3 involves BMTA catalyzed asymmetric amination of 2-hydroxyacetophenone 6 to (R)-phenylglycinol 7, as well as AlaDH catalyzed recycling of L-alanine and GDH catalyzed regeneration of NADH cofactor
Materials and methods
Chemicals
l-Phenylalanine (l-PA), trans-cinnamic acid, styrene, (S)-1-phenyl-1, 2-ethanediol, styrene oxide, 2-hydroxyacetophenone (2-HAP), (±)-phenylglycinol, (R)-phenylglycinol, (S)-phenylglycinol, l-alanine (l-Ala), n-dodecane, pyridoxal-5′-phosphate (PLP) and (R)-α-methylbenzylamine (R-MBA) were from Energy Chemical and Titan Scientific (Shanghai, China). Yeast extract, tryptone and antibiotics (kanamycin, ampicillin and streptomycin) were from Sangon Biotech (Shanghai, China). T4 DNA ligase and restriction enzymes were from New England Biolabs (NEB, Beijing, China). Isopropyl β-d-1-thiogalactopyranoside (IPTG) and Taq plus DNA polymerase were purchased from Tsingke (Beijing, China). All other chemical reagents were obtained from commercial sources.
Plasmids, microorganisms and media
The expression plasmids (pET28a (+), pETduet-1, pRSFduet-1 and pCDFduet-1) were from Novagen (Madison, WI, USA). Previously constructed recombinant plasmids (pET28a-SMO, pETduet-SpEH, pET28a-GoSCR, pET28a-MVTA, pET28a-BMTA and pETduet-GDH) (Zhang et al. 2020) were stored in our lab. The host strain E. coli T7 supercompetent cells were purchased from NEB (Beijing, China). The E. coli strains were grown at 37 °C in Luria–Bertani (LB) medium or Terrific Broth (TB) medium. Antibiotics ampicillin (0.1 mg/mL), kanamycin (0.05 mg/mL) and streptomycin (0.1 mg/mL) were utilized for the selection of recombinant E. coli cells. Bacillus subtilis sp. 168 stored in our lab was maintained on MRS agar slants and grown in MRS medium at 30 °C.
Construction of the recombinant E. coli strains
All the primers used in this study were synthesized by Tsingke (Beijing, China) and listed in Additional file 1: Table S1. All the constructed plasmids and recombinant E. coli cells are listed in Additional file 1: Table S2. E. coli (GoSCR), E. coli (GDH) (Cui et al. 2017), E. coli (MVTA) (Zhang et al. 2019a), E. coli (SpEH) (Zhang et al. 2019b), E. coli (SMO) and E. coli (BMTA) (Zhang et al. 2020) were constructed as described previously.
Regarding the recombinants E. coli (PAL), E. coli (Fdc1) and E. coli (Pad1), the PAL gene from Arabidopsis thaliana (Cochrane et al. 2004) and the Fdc1 and Pad1 genes from Aspergillus niger (Payne et al. 2015) were synthesized by Tsingke (Beijing, China). The DNA fragments of PAL, Fdc1, and Pad1 were then amplified by polymerase chain reaction (PCR) using the primers listed in Additional file 1: Table S1. After, the PCR products were isolated, double digested with the restriction endonucleases (NdeI and XhoI), and ligated into pET28a (+), which were further digested with the same restriction enzymes. The resulting recombinant plasmids (pET28a-PAL, pET28a-Fdc1 and pET28a-Pad1) were separately transformed into the competent E. coli T7 to form E. coli (PAL), E. coli (Fdc1), and E. coli (Pad1).
For the design of E. coli (AlaDH), the genome DNA was extracted from Bacillus subtilis sp. 168 by using a Bacterial DNA extraction kit supplied by Sangon Biotech (China). After, the DNA fragment of the AlaDH (alanine dehydrogenase) gene was amplified from the genomic DNA of Bacillus subtilis sp. 168 with PCR using the primers shown in Additional file 1: Table S1. The PCR product was isolated and double digested with BamHI and PstI. After, it was ligated into the expression vector pETduet-1 at the BamHI/PstI sites to form the recombinant pETduet-AlaDH. The constructed recombinant plasmid pETduet-AlaDH was then transformed into the competent E. coli T7 to form the recombinant E. coli (pETduet-AlaDH). In addition, the recombinant E. coli (pCDFduet-AlaDH) was constructed using a similar procedure.
Moreover, the recombinant E. coli (Fdc1-Pad1) was designed by amplifying the DNA fragments of Fdc1 and Pad1 with PCR using the primers listed in Additional file 1: Table S1. The Fdc1 gene was double-digested with the BamHI and NotI and ligated into the pRSFduet-1 at the BamHI/NotI sites to form the recombinant pRSFduet-Fdc1. In addition, the Pad1 gene was double-digested with the restriction endonucleases NdeI and XhoI and ligated into pRSFduet-Fdc1 at the BamHI/XhoI sites to form the pRSFduet-Fdc1-Pad1 (named as RFP). Furthermore, the recombinant plasmid pRSFduet-Fdc1-Pad1 was then transformed into the competent E. coli T7 to form the recombinant E. coli (pRSFduet-Fdc1-Pad1) cells, designated as E. coli (RFP). Furthermore, the recombinant E. coli (pETduet-Fdc1-Pad1) (designated as E. coli (DFP)) and E. coli (pCDFduet-Fdc1-Pad1) (designated as E. coli (CFP)) were constructed in a similar way.
In the case of E. coli (pETduet-SpEH-PAL), the DNA fragments of both epoxide hydrolase (SpEH) and PAL enzymes were amplified with PCR using the primers shown in Additional file 1: Table S1. The SpEH gene was double-digested with the restriction endonucleases BamHI and HindIII and ligated into the pETduet-1 at the BamHI/HindIII sites to form the recombinant pETduet-SpEH. Also, the PAL gene was double-digested with NdeI and XhoI and ligated into the pETduet-SpEH at the NdeI/XhoI sites to form the recombinant pETduet-SpEH-PAL (known as DEA). In addition, the recombinant plasmid pETduet-SpEH-PAL was transformed into the competent E. coli T7 to form E. coli (pETduet-SpEH-PAL) which was named E. coli (DEA).
Moreover, the following recombinants were constructed in a similar way: E. coli (pCDFduet-PAL-SMO) (designated as E. coli (CAS)), E. coli (pRSFduet-SpEH-GoSCR) (named as E. coli (REG)), E. coli (pCDFduet-GoSCR-SMO) (known as E. coli (CGS)), E. coli (pETduet-SpEH-SMO) (designated as E. coli (DES)), and E. coli (pRSFduet-GoSCR-PAL) (named as E. coli (RGA)). The enzyme GoSCR referred to NAD+-dependent alcohol dehydrogenase.
Regarding the recombinant E. coli (PAL-Fdc1-Pad1-SMO-SpEH-GoSCR), the constructed recombinant plasmids pRSFduet-Fdc1-Pad1, pETduet-SpEH-PAL, and pCDFduet-GoSCR-SMO had different antibiotic resistance genes. These recombinants were simultaneously transformed into the competent E. coli T7 to form the recombinant E. coli (PAL-Fdc1-Pad1-SMO-SpEH-GoSCR) cells, which were designated as E. coli (RFP-DEA-CGS). Furthermore, the recombinant E. coli (DFP-CAS-REG) and E. coli (CFP-DES-RGA) cells were similarly constructed with different recombinant plasmid combinations. These recombinants co-expressed PAL, Fdc1, Pad1, SMO, SpEH, and GoSCR.
For E. coli (pETduet-GDH-AlaDH), the GDH and AlaDH genes were amplified by PCR using the primers shown in Additional file 1: Table S1. After, the GDH (glucose dehydrogenase) gene was double-digested with BamHI and HindIII and ligated into the pETduet-1 at the BamHI/HindIII sites to form the recombinant pETduet-GDH. Then, the AlaDH gene was double-digested with the restriction endonucleases KpnI and XhoI and further ligated into the pETduet-GDH at the KpnI/XhoI sites to obtain a new recombinant plasmid pETduet-GDH-AlaDH (known as DGA). This recombinant plasmid was then transformed into the competent E. coli T7 to form the recombinant E. coli (pETduet-GDH-AlaDH), designated as E. coli (DGA).
For E. coli (BMTA-GDH-AlaDH), the constructed recombinant plasmids pET28a-BMTA and pETduet-GDH-AlaDH were simultaneously transformed into the competent E. coli T7 to form the recombinant E. coli (BMTA-GDH-AlaDH), designated as E. coli (EB-DGA). These two recombinant plasmids had different antibiotic resistance genes. Similarly, the recombinant plasmids pET28a-BMTA, pETduet-GDH, and pCDFduet-AlaDH were simultaneously transformed into the competent E. coli T7 to form the recombinant E. coli (BMTA-GDH-AlaDH), named E. coli (EB-DG-CA). The enzyme BMTA refers to (S)-selective ω-transaminase.
The expression of all genes from the above-constructed recombinant E. coli strains was confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and testing the activity of corresponding enzymes.
Enzyme expression and activity analysis
The constructed recombinant E. coli cells were cultivated at 37 °C for 12 h in 5 mL LB medium (10 g/L peptone, 5 g/L yeast extract and 10 g/L NaCl, pH7.0) containing appropriate antibiotics. Two milliliter of seed culture was then inoculated into 100 mL of Terrific Broth (TB) medium (12 g/L tryptone, 24 g/L yeast extract, 4 g/L glycerol, 17 mM KH2PO4, 72 mM K2HPO4) supplemented with the appropriate antibiotics, and cultivated at 37 °C for 2–3 h. When OD600 value of the recombinant E. coli cells reached 0.6–0.8, IPTG (0.5 mM) was then added to the medium to induce the protein expression. After incubation for 12–20 h at 20 °C and 200 rpm, cells were harvested by centrifugation (8000 rpm) at 4 °C for 10 min, washed twice with cold saline and resuspended in sodium phosphate buffer (100 mM, pH 7.5). The cell pellets were put on ice and lysed by ultra-sonication for 90 times at 400 W for 4 s with 6 s of interval. The cell debris were removed by centrifugation (12,000 rpm, 30 min) at 4 °C, and the soluble portion of cell lysate was stored at − 80 °C for further use.
The activity of phenylalanine ammonia lyase (PAL) was measured as described previously (Cochrane et al. 2004). The reaction mixture consisted of 100 mM sodium phosphate buffer (pH 8.5), 5 mM l-phenylalanine substrates, and an appropriate amount of enzyme in a total volume of 0.5 mL. The reaction mixture was incubated at 30 °C for 5 min. The reaction samples (100 μL) were mixed with acetonitrile (900 μL) containing 2 mM acetophenone (internal standard), centrifuged at 12,000 rpm for 10 min. The concentration of the trans-cinnamic acid was determined by HPLC analysis. 1 unit of activity refers to the amount of catalyst that catalyzed the conversion of 1 μmol l-phenylalanine to trans-cinnamic acid per min.
The activity of decarboxylases (Fdc1 and Pad1) was measured as described previously (Payne et al. 2015). The reaction mixture consisted of 100 mM sodium phosphate buffer (pH 7.0), 5 mM trans-cinnamic acid, and an appropriate amount of enzyme in a total volume of 0.5 mL. The reaction mixture was incubated at 30 °C for 5 min, the reaction samples (100 μL) were mixed with acetonitrile (900 μL) containing 2 mM acetophenone (internal standard), centrifuged at 12,000 rpm for 10 min. The concentration of the styrene was determined by HPLC analysis. 1 unit of activity refers to the amount of catalyst that catalyzed the conversion of 1 μmol trans-cinnamic acid to styrene per minute.
The activity of styrene monooxygenase (SMO) was measured as described previously (Xu et al. 2009). The reaction mixture consisted of 100 mM sodium phosphate buffer (pH 7.0), 5 mM styrene substrate, 1 mM NADH, and an appropriate amount of enzyme in a total volume of 0.5 mL. After incubation at 30 °C for 5 min, the reaction samples (300 µL) were saturated with sodium chloride, and extracted with ethyl acetate (EtOAc) containing 5 mM of n-dodecane (internal standard). The organic phase was dried over anhydrous Na2SO4, and the concentration of styrene oxide was determined by GC analysis. 1 unit of activity refers to the amount of catalyst that catalyzed the conversion of 1 μmol styrene to styrene oxide per minute.
The activity of epoxide hydrolase (SpEH) was measured as described previously (Wu et al. 2013). The reaction mixture consisted of 100 mM sodium phosphate buffer (pH 7.0), 10 mM styrene oxide substrate, and an appropriate amount of enzyme in a total volume of 0.5 mL. After incubation at 30 °C for 5 min, the reaction samples (300 µL) were saturated with sodium chloride, and extracted with EtOAc containing 5 mM of n-dodecane (internal standard). The organic phase was dried over anhydrous Na2SO4, and the concentration of the 1-phenyl-1,2-ethanediol was determined by GC analysis. 1 unit of activity refers to the amount of catalyst that catalyzed the conversion of 1 μmol styrene oxide to 1-phenyl-1,2-ethanediol per minute.
The oxidation activity of alcohol dehydrogenase was measured as described previously (Zhang et al. 2020). The reaction mixture consisted of 100 mM sodium phosphate buffer (pH 7.5), 0.2 mM NAD+, 10 mM 1-phenyl-1,2-ethanediol and an appropriate amount of enzyme in a total volume of 1.0 mL, the reaction was performed at 30 °C for 1 min. 1 unit of activity refers to the amount of catalyst that catalyzed the conversion of 1 μmol NAD+ to NADH per minute.
The activity of alanine dehydrogenase (AlaDH) was measured as described previously (Lerchner et al. 2016). The reaction mixture consisted of 100 mM sodium phosphate buffer (pH 7.5), 0.2 mM NADH, 5 mM pyruvate, 200 mM NH4Ac and an appropriate amount of enzyme in a total volume of 1.0 mL. The reaction was performed at 30 °C for 1 min. 1 unit of activity refers to the amount of catalyst that catalyzed the conversion of 1 μmol NADH to NAD+ per minute.
The activity of transaminase was assayed as described in our previous study (Zhang et al. 2017). The activity of glucose dehydrogenase (GDH) was measured as described in our previous study (Cui et al. 2017).
The activity of recombinant E. coli (DFP-CAS-REG) cells was measured by testing the formation of 2-HAP 6. The reaction mixture consisted of 100 mM sodium phosphate buffer (pH 7.5), 10 mM L-PA 1, 10 mM glucose and 5 g cell dry weight (cdw)/L recombinant E. coli cells in a total volume of 1.0 mL, the reaction was performed at 25 °C and 200 rpm for 10 min. The concentration of 2-HAP was determined by GC analysis. 1 unit of activity refers to the amount of catalyst that catalyzed the conversion of 1 μmol L-PA to 2-HAP per min.
The protein concentration was determined by the Bradford method (Bradford 1975).
In vitro conversion of L-PA 1 into 2-HAP 6 and chiral phenylglycinol 7
The freshly prepared recombinant E. coli cells (E. coli (PAL), E. coli (Fdc1), E. coli (Pad1), E. coli (SMO), E. coli (SpEH), E. coli (GoSCR), E. coli (BMTA) and E. coli (MVTA)) were washed twice with sterile deionized water, and resuspended in sterile deionized water to an OD600 of 30, respectively. The cell pellets were put on ice and lysed by ultra-sonication for 90 times at 400 W for 4 s with 6 s of interval. After centrifugation for 30 min at 4 °C and 12,000 rpm, the soluble portion of cell lysate was frozen at − 80 °C overnight. Then the frozen cell-free extracts were lyophilized for 48 h using a vacuum freeze dryer (SCIENTA-10, China) at a temperature of − 50 °C and a vacuum of 0.055 mbar. For conversion of L-PA 1 to 2-HAP 6, the reaction mixture consisted of 100 mM sodium phosphate buffer (pH 7.5), 10–20 mM L-PA, 0.5 mM NADH, 15 mg/mL PAL, 15 mg/mL Fdc1, 15 mg/mL Pad1, 20 mg/mL SMO, 10 mg/mL SpEH, 20 mg/mL GoSCR in a total volume of 5 mL. For conversion of L-PA 1 to phenylglycinol 7, the reaction mixture consisted of 100 mM sodium phosphate buffer (pH 7.5), 10–20 mM L-PA 1, 0.5 mM NADH, 15 mg/mL PAL, 15 mg/mL Fdc1, 15 mg/mL Pad1, 20 mg/mL SMO, 10 mg/mL SpEH, 20 mg/mL GoSCR, 15 mg/mL MVTA or 15 mg/mL BMTA, 0.1 mM PLP, and 15–25 mM (R)-MBA or 200 mM L-Ala in a total volume of 5 mL. The reactions were conducted at 30 °C and 200 rpm. At appropriate intervals, samples were taken for GC analysis.
In vivo conversion of L-PA 1 into 2-HAP 6 with the resting cells of recombinant E. coli cells
The freshly prepared recombinant E. coli (RFP-DEA-CGS), E. coli (DFP-CAS-REG) and E. coli (CFP-DES-RGA) cells were washed twice with sterile deionized water and frozen at − 80 °C overnight. Then the frozen cells were lyophilized for 48 h using the vacuum freeze dryer at a temperature of − 50 °C and a vacuum of 0.055 mbar. The reaction mixture consisted of 100 mM sodium phosphate buffer (pH 7.5), 10 g cdw/L lyophilized recombinant E. coli cells, 10 mM L-PA 1 and 10 mM glucose in a total volume of 5 mL. The reactions were performed at 25 °C and 200 rpm. At appropriate intervals, samples were taken for GC analysis.
In vivo conversion of 2-HAP 6 into (S)- or (R)-phenylglycinol 7
The freshly prepared recombinant E. coli (MVTA) and E. coli (EB-DGA) cells were washed twice with sterile deionized water and frozen at − 80 °C overnight and lyophilized for 48 h using the vacuum freeze dryer at a temperature of − 50 °C and a vacuum of 0.055 mbar. The reaction mixture consisted of 100 mM sodium phosphate buffer (pH 8.0), 10 g cdw/L lyophilized recombinant E. coli cells, 10 mM 2-HAP 6, 0.1 mM PLP, 10% DMSO, 20 mM R-MBA or 400 mM L-Ala (including 150 mM NH3/NH4Cl) in a total volume of 5 mL. The reactions were performed at 30 °C and 200 rpm. At appropriate intervals, samples were taken for GC analysis.
Conversion of L-PA 1 into (S)-phenylglycinol 7 with the mixture of resting cells of E. coli (RFP-DEA-CGS) and E. coli (MVTA)
The reaction mixture consisted of 100 mM sodium phosphate buffer (pH 8.0), 20 g cdw/L lyophilized recombinant E. coli (RFP-DEA-CGS) cells, 15 g cdw/L lyophilized recombinant E. coli (MVTA) cells, 10–50 mM L-PA 1, 5–40 mM glucose, 0.1 mM PLP, 25–40 mM (R)-MBA in a total volume of 5 mL. The reactions were performed at 25 °C and 200 rpm. At appropriate intervals, samples were taken for GC analysis.
Conversion of L-PA 1 into (R)-phenylglycinol 7 with the mixture of resting cells of E. coli (RFP-DEA-CGS) and E. coli (EB-DGA)
The reaction mixture consisted of 100 mM sodium phosphate buffer (pH 8.0), 15 g cdw/L lyophilized recombinant E. coli (RFP-DEA-CGS) cells, 20 g cdw/L lyophilized recombinant E. coli (EB-DGA) cells, 10–50 mM L-PA 1, 5–40 mM glucose, 0.1 mM PLP, 100–600 mM L-Ala and 0–200 mM NH3/NH4Cl in a total volume of 5 mL. The reactions were performed at 25 °C and 200 rpm. At appropriate intervals, samples were taken for GC analysis.
Preparation experiment
For preparation of (S)-phenylglycinol 7, the reaction was conducted in 100 mL sodium phosphate buffer (100 mM, pH 7.5) containing 20 mM (330.4 mg) L-PA 1, 20 g cdw/L E. coli (RFP-DEA-CGS), 15 g cdw/L E. coli (MVTA), 10 mM glucose, 0.1 mM PLP and 30 mM (R)-MBA. For preparation of (R)-phenylglycinol 7, the reaction was conducted in 100 mL sodium phosphate buffer (100 mM, pH 7.5) containing 20 mM (330.4 mg) L-PA 1, 15 g cdw/L E. coli (RFP-DEA-CGS), 20 g cdw/L E. coli (EB-DGA), 20 mM glucose, 0.1 mM PLP, 400 mM L-Ala and 150 mM NH3/NH4Cl. The reactions were performed at 25 °C and 200 rpm for 12 h. After the reactions were completed, the reaction mixtures were basified (pH > 10) by adding NaOH (10 N), and extracted with EtOAc for three times (3 × 50 mL). The combined organic phases were dried over anhydrous Na2SO4. The solvent was removed by evaporation and the residue was purified using a silica gel column with EtOAc/methanol (10:1) as eluent.
(S)-7 was obtained as a white solid in 71.0% yield (194.8 mg) and > 99% ee. 1H NMR (400 MHz, 298 K, CDCl3) δH 7.34–7.26 (5H, m, Ph), 4.02 (1H, dd, 3JHH = 4.0 Hz, 8.5 Hz, CH), 3.73–3.70 (1H, dd, 2J HH = 11.0 Hz, 3J HH = 4.0 Hz, CH2), 3.56–3.52 (1H, dd, 2J HH = 11.0 Hz, 3J HH = 8.5 Hz, CH2), 2.45 (br, 3H).
(R)-7 was obtained as a yellow oil in 80.5% yield (220.9 mg) and > 99% ee. 1H NMR (400 MHz, 298 K, CDCl3) δH 7.37–7.26 (5H, m, Ph), 4.07–4.03 (1H, dd, 3JHH = 4.0 Hz, 8.5 Hz, CH), 3.76–3.72 (1H, dd, 2J HH = 11.0 Hz, 3J HH = 4.0 Hz, CH2), 3.58–3.54 (1H, dd, 2J HH = 11.0 Hz, 3J HH = 8.5 Hz, CH2), 2.40 (br, 3H).
Assay methods
For GC analysis of styrene 3, (S)-1-phenyl-1, 2-ethanediol 5 and 2-HAP 6, the reaction samples (300 µL) were saturated with sodium chloride, mixed with 300 µL of EtOAc containing 20 mM of n-dodecane (internal standard), after centrifugation (12,000 rpm) at 4 °C for 5 min, the organic phase was separated and dried over anhydrous Na2SO4. The concentration of styrene, (S)-1-phenyl-1, 2-ethanediol and 2-HAP was determined by GC analysis. For GC analysis of phenylglycinol 7, the reaction samples (300 µL) were basified (pH > 10) by adding NaOH (10 N), saturated with sodium chloride, and extracted with EtOAc (300 µL) containing 20 mM of n-dodecane (internal standard). The organic phase was dried over anhydrous Na2SO4, the concentration of the phenylglycinol 7 was determined by GC analysis. GC analysis was conducted using Shimadzu GC-14C gas chromatography system with a flame ionization detector. The column was an Agilent HP-5 (30 m × 0.32 mm × 0.25 mm). Parameter: injector temperature, 250 °C; detector temperature, 275 °C; column temperature: 120 °C. The enantiomeric excess of (S)-phenylglycinol or (R)-phenylglycinol was determined by chiral GC as described previously (Zhang et al. 2020).
Results and discussion
Design and in vitro construction of the artificial biocatalytic cascade for conversion of L-PA 1 into chiral phenylglycinol 7
A sequential cascade biocatalysis was designed by retrosynthetic strategy to convert bio-based L-PA 1 into chiral phenylglycinol 7 (Scheme 1). First, L-PA 1 was deaminized to trans-cinnamic acid 2 by the PAL enzyme, followed by oxidative decarboxylation of trans-cinnamic acid 2 to styrene 3 by PAD. Moreover, the styrene 3 compound was subsequently oxidized to styrene oxide 4 by SMO, and styrene oxide 4 was then hydrolyzed to 1-phenyl-1,2-ethanediol 5 by EH. This 1-phenyl-1,2-ethanediol 5 compound was oxidized to 2-HAP 6 by ADH. Finally, an asymmetric reduction amination converted the 2-HAP 6 to chiral phenylglycinol 7 by an ω-transaminase (ωTA).
Moreover, PAL from Arabidopsis thaliana (Cochrane et al. 2004), which plays an important role in the process of ammonia removal, was selected for converting L-PA 1 into trans-cinnamic acid 2 for in vitro reconstruction of the biocatalysis cascade. In the second decarboxylation step, Fdc1 and Pad1 from Aspergillus niger were selected for conversion of trans-cinnamic acid 2 into styrene 3. In fact, both enzymes can work together more efficiently in the decarboxylation process of the former compound (Payne et al. 2015). In the third step, the NADH-dependent SMO from Pseudomonas sp. VLB120 was used for converting styrene 3 into (S)-styrene epoxide 4 (Xu et al. 2009). Furthermore, in the fourth step, SpEH from Sphingomonas sp.HXN-200 was used for selective hydrolysis of 4 to (S)-diol 5 (Wu et al. 2013). After, in the fifth step, the GoSCR enzyme from Gluconobacter oxydans 621H (Cui et al. 2017) was selected for oxidation of (S)-diol 5 into 2-HAP 6. In the final amination step, an (R)-selective ω-transaminase (MVTA) from Mycobacterium vanbaalenii (Zhang et al. 2019) and the BMTA enzyme from Bacillus megaterium SC6394 (Zhang et al. 2020) were then employed to convert 2-HAP 6 to (S)-7 and (R)-7, respectively. The NAD+ and NADH recycling system was generated in the third and fifth step of the catalytic process. All the enzymes used in this study were lyophilized cell-free extracts.
In addition, the activities of these lyophilized enzymes were tested considering their corresponding substrates (Additional file 1: Table S3). After, the L-PA (10–20 mM) and NADH (0.5 mM) compounds were added into the reaction system including a set of six different enzymes (PAL, Fdc1, Pad1, SMO, SpEH, and GoSCR). Interestingly, 2-HAP 6 was obtained with a 99% conversion percentage from 10–20 mM L-PA without optimizing the reaction conditions (Table 1). In addition, the (R)-7 and (S)-7 compounds were obtained with an ee value of > 99% and between a range of conversion percentages from 66.8 to 87.2% from 10 to 20 mM L-PA with the mixture of seven enzymes (PAL, Fdc1, Pad1, SMO, SpEH, GoSCR, and MVTA or BMTA) (Table 1). These results showed that the designed cascade biocatalysis complex was successfully in vitro constructed for conversion of L-PA 1 into chiral 7. However, the in vitro cascade reactions needed the application of multiple enzymes. In addition, the cell-free enzymes were easily affected by a rapid deactivation, and the redox reaction required an expensive cofactor (NAD+/NADH), which can increase the reaction costs. Consequently, it was decided to construct the recombinant E. coli whole cells that were able to co-express the necessary enzymes for conversion of L-PA 1 into chiral phenylglycinol 7.
Construction of the recombinant E. coli cell modules for conversion of L-PA into 2-HAP
Co-expression of multi-enzymes in one microbial cell will usually result in serious protein expression burden and redox constraints, but these negative factors can be reduced by distributing the biocatalytic pathway among different cell modules (Wang et al. 2020). Therefore, a synthetic microbial consortium was designed and constructed to convert L-PA 1 into chiral phenylglycinol 7. This microbial consortium included two different recombinant E. coli cell modules (Scheme 1). Therefore, one recombinant E. coli cell module co-expressed the PAL/Fdc1/Pad1/SMO/EH/GoSCR enzymatic set for converting L-PA 1 into 2-HAP 6. The second recombinant E. coli cell module expressed the MVTA enzyme or co-expressed the BMTA/AlaDH/GDH enzymes set for converting 2-HAP into (S)- or (R)-phenylglycinol 7, respectively.
Additionally, a recombinant E. coli cells module 1 that co-expressed six enzymes (PAL, Fdc1, Pad1, SMO, SpEH, and GoSCR) was constructed for converting L-PA 1 into 2-HAP 6. Also, three expression plasmids (pETDuet-1, pRSFDuet-1, and pCDFDuet-1) were used for co-expression purposes of two different enzymes with various combinations (Additional file 1: Figure S1–S3). Theoretically, there are 15 different plasmid combinations, eight of them were randomly selected and constructed to co-express six enzymes. Specifically, three recombinant E. coli cells, E. coli (RFP-DEA-CGS), E. coli (DFP-CAS-REG), E. coli (CFP-DES-RGA) with three different plasmids combinations were successfully constructed (Fig. 1A). Furthermore, a SDS-PAGE analysis of the crude lysate present in the constructed recombinant E. coli cells showed that the six enzymes were successfully expressed in the E. coli cells (Fig. 1B). The performances of the constructed recombinant E. coli cells in the conversion of L-PA 1 into 2-HAP 6 were tested using 10 mM L-PA. The results showed that the recombinant E. coli (RFP-DEA-CGS) cells had the highest activity (5.6 U/g cdw) with the amino acid L-PA 1. By using the recombinant E. coli (RFP-DEA-CGS) cells, the conversion percentage of 2-HAP 6 raised to 93% within a 7-h period (Fig. 1C). Then, a recombinant E. coli (RFP-DEA-CGS) was selected as the best candidate to convert L-PA 1 into 2-HAP 6. The cell growth and specific activity of the recombinant E. coli (RFP-DEA-CGS) were further analyzed (Fig. 1D). Moreover, the highest activity value of E. coli (RFP-DEA-CGS) was 5.8 U/g cdw at OD600 (3.0) after a 14-h induction period.
Construction of recombinant E. coli cells co-expression of six enzymes for conversion of L-PA 1 to 2-HAP 6. A Construction of recombinant E. coli cells co-expression of six enzymes based on different plasmid configurations. B SDS-PAGE analysis of recombinant E. coli cells. M: marker; lane 1: cell-free extract of E. coli (RFP-DEA-CGS), lane 2: cell-free extract of E. coli (CFP-DES-RGA); lane 3: cell-free extract of E. coli (DFP-CAS-REG). C Activities of three different recombinant E. coli cells for conversion of L-PA 1 to 2-HAP 6. D Cell growth and specific activity of E. coli (RFP-DEA-CGS) for conversion of L-PA 1 to 2-HAP 6
Conversion of L-PA 1 into 2-HAP 6 by E. coli (RFP-DEA-CGS)
After the recombinant E. coli (RFP-DEA-CGS) was selected for converting L-PA into 2-HAP, the reaction conditions were optimized with 20 mM L-PA. The highest conversion percentage of 2-HAP was 92% at pH 7.5 and 25 °C (Fig. 2A-B). Moreover, the addition of 10 mM glucose increased the conversion percentage of 2-HAP 6 to 95% (Fig. 2C). For 10 mM L-PA 1, 15 g cdw/L of E. coli (RFP-DEA-CGS) was enough to convert L-PA 1 into 2-HAP 6 with a 99% conversion percentage. However, 25 g cdw/L of E. coli (RFP-DEA-CGS) was needed for conversion of 20 mM L-PA 1 into 2-HAP 6. In the case of 30 mM L-PA, the conversion percentage of 2-HAP 6 was 91.6% using 25 g cdw/L of E. coli (RFP-DEA-CGS). However, the conversion of 2-HAP 6 for 50 mM L-PA decreased to 46.2% using 25 g cdw/L of E. coli (RFP-DEA-CGS) (Fig. 2D and Additional file 1: Figure S4). In fact, the 2-HAP 6 compound is a very important structural moiety in several synthetic intermediates used in organic and medicinal chemistry. For example, 2-HAP can be used as a building block in the synthesis of oxazolone carboxamides, which belong to a novel class of acid ceramidase inhibitors. Also, 2-HAP can act as a useful pharmacological tool in treating sphingolipid-mediated disorders (Caputo et al. 2020) and as a potent inhibitor of urease (Tanaka et al. 2004).
Reaction condition optimization for conversion of L-PA 1 to 2-HAP 6. A Effect of different pH on the conversion of 2-HAP 6; B effect of different temperature on the conversion of 2-HAP 6; C effect of glucose concentration on the conversion of 2-HAP 6; D effect of different cell density of E. coli (RFP-DEA-CGS) on the conversion of different concentration of L-PA 6, blue: 10 mM of L-PA; yellow: 20 mM of L-PA; green: 30 mM of L-PA; red: 50 mM of L-PA
Construction of recombinant E. coli cell modules for conversion of 2-HAP into chiral phenylglycinol
The MVTA enzyme was selected to catalyze the asymmetric amination of 2-HAP 6 into (S)-7, and R-MBA was used as an optimum amine donor in the conversion process to acetophenone (AP) (Zhang et al. 2019a). Therefore, the recombinant E. coli (MVTA) was then constructed for conversion of 2-HAP 6 into (S)-phenylglycinol 7. In this case, the whole-cell specific activity of the recombinant E. coli (MVTA) cells with 2-HAP 6 was 22.2 U/g cdw. Regarding the bioconversion of 2-HAP 6 into (R)-7, BMTA was used for catalyzing the amination of the 2-HAP 6 into (R)-7. In addition, L-Ala serves as an optimum amine donor that can lead to pyruvate. Thus, pyruvate can then be recycled to alanine by employing the AlaDH enzyme, which consumes ammonia and NADH. Also, NADH can be recycled using GDH. Consequently, some recombinant E. coli cells were constructed to co-express BMTA, AlaDH, and GDH (Fig. 3A). In fact, an SDS-PAGE showed that all the enzymes were successfully expressed (Fig. 3B), and the activities of the recombinant E. coli cells were tested with 10 mM 2-HAP 6. Therefore, the recombinant E. coli (EB-DGA) cells showed a highest activity of 8.3 U/g cdw with 2-HAP 6 and a 95% conversion percentage of (R)-7 during a 12-h reaction period (Fig. 3C).
Construction of recombinant E. coli cells for conversion of 2-HAP 6 to (R)-phenylglycinol 7. A Construction of recombinant E. coli cells co-expression of three enzymes (BMTA, AlaDH and GDH) based on different plasmid configurations. B SDS-PAGE analysis of recombinant E. coli cells. M: marker; lane 1: cell-free extract of E. coli (EB-DGA), lane 2: cell-free extract of E. coli (EB-DG-CA). C Activities of two different recombinant E. coli cells for conversion of 2-HAP 6 to (R)-phenylglycinol 7
Conversion of L-PA 1 into (S)-7 by the designed E. coli (RFP-DEA-CGS) and the E. coli (MVTA) consortium
After the biocatalytic modules were constructed, the lyophilized E. coli (RFP-DEA-CGS) and E. coli (MVTA) cells were combined to convert L-PA 1 into (S)-7. The reaction conditions were first optimized with 20 mM of L-PA 1. The results showed that the highest conversion percentage of (S)-7 (78%) was obtained at a pH of 7.5 and 25 °C. In this case, 10 mM glucose and 30 mM R-MBA were used as substrates, and E. coli (RFP-DEA-CGS) and E. coli (MVTA) were combined at a 1:1 ratio with 15 g cdw/L of each recombinant (Fig. 4A–E). Moreover, the combination of two recombinant E. coli cells at a 2:1.5 ratio (20 g cdw/L of E. coli (RFP-DEA-CGS) and 15 g cdw/L of E. coli (MVTA)) resulted in the highest conversion of (S)-7 (80%) and > 99% ee. Following the above optimized reaction conditions, different L-PA 1 concentrations (10–50 mM) were tested in a period of 7 h (Fig. 4F), and about 98% of conversion of (S)-7 was obtained with 10 mM L-PA 1. Moreover, (S)-7 was obtained with only a 34.4% conversion percentage for 30 mM L-PA 1. Surprisingly, a further increase in the substrate concentration to 50 mM decreased the conversion percentage of (S)-7 to 12.9%. In fact, the remaining intermediate product was mainly diol 5 (85%), and almost no 2-HAP 6 was detected in the reaction mixture. This finding implied that the reactivity of GoSCR with respect to the diol intermediate compound in this cascade biocatalysis system could be severely inhibited by R-MBA. Therefore, the effect of different concentrations of R-MBA (0–60 mM) on the conversion of 2-HAP 6 by E. coli (RFP-DEA-CGS) was examined with 20 mM L-PA 1. The results showed that the conversion of 2-HAP 6 was significantly decreased with the increasing concentration of R-MBA (Additional file 1: Figure S5).
Reaction condition optimization for conversion of L-PA 1 (20 mM) to (S)-7 by the designed E. coli (RFP-DEA-CGS) and E. coli (MVTA) consortium. A Effect of different pH on the conversion of (S)-7; B effect of different temperature on the conversion of (S)-7; C effect of glucose concentration on the conversion of (S)-7; D effect of R-MBA concentration on the conversion of (S)-7; E effect of different cell density of E. coli (RFP-DEA-CGS) and E. coli (MVTA) on the conversion of (S)-7; F effect of different L-PA 1 concentration on the conversion of (S)-7
The time course in the bioconversion of 10–30 mM L-PA 1 into (S)-7 is shown in Fig. 5. In fact, (S)-7 could be obtained with 99% and 84% conversion percentages (> 99% ee) from 10 and 20 mM L-PA at 12 h, respectively (Additional file 1: Figures S6 and S8). For 30 mM L-PA, the highest conversion percentage of (S)-7 was only 35% (Fig. 5A). Furthermore, the analysis of the intermediate products produced from L-PA (20 mM) showed that the (S)-7 (35%), diol 5 (35%), styrene 3 (8%), and 2-HAP 6 (5%) were the main remaining products in the reaction system at 1 h. This finding implied that 83% of L-PA 1 was converted to styrene 3 due to the high activity of PAL, Fdc1, and Pad1. After 1 h, the concentration of diol 5 began to decrease, while the concentration of (S)-7 increased linearly. At 7 h, a trace amount of diol 5 (< 1%) and 2-HAP 6 (< 1%) were detected in the reaction mixture, and the conversion of (S)-7 reached 75%. Finally, the conversion of (S)-7 was continually increased to 84% at 12 h (Fig. 5B).
Time course of the cascade reaction for the synthesis of (S)-7 from L-PA 1. A Time course of the cascade reaction for conversion of 10–30 mM L-PA 1 to (S)-7; B time course of the cascade reaction for intermediate produced from L-PA 1. Reaction conditions (5 mL): 100 mM phosphate buffer (pH 7.5), 10–30 mM L-PA, 10 mM glucose, 20 g cdw/L E. coli (RFP-DEA-CGS), 15 g cdw/L E. coli (MVTA), 30 mM (R)-MBA and 0.1 mM PLP, at 25 °C. All biotransformations were performed in duplicate and conversion was determined by GC, error limit: < 2% of the state values
Conversion of L-PA 1 into (R)-7 by the designed E. coli (RFP-DEA-CGS) and the E. coli (EB-DGA) consortium
The lyophilized E. coli (RFP-DEA-CGS) and E. coli (EB-DGA) cells were combined to create another E. coli microbial consortium for conversion purposes from L-PA 1 to (R)-7. The reaction conditions were optimized with 20 mM of L-PA 1. Figure 6A–F shows that the combination of E. coli (RFP-DEA-CGS) and E. coli (EB-DGA) at a 1.5:2 ratio (15 g cdw/L of E. coli (RFP-DEA-CGS) and 20 g cdw/L of E. coli (EB-DGA) led to the highest conversion percentage of (R)-7 (85%) at pH 7.5 and 25 °C with 20 mM glucose and 400 mM L-Ala. In addition, a reaction mixture with 150 mM NH3/NH4Cl (regeneration of L-Ala by AlaDH) resulted in a 95% conversion percentage of (R)-7. Further attempts to decrease the usage of L-Ala showed that the (R)-7 yield was not evidently improved (< 85%) when the L-Ala concentration was below 400 mM (data not shown). In fact, Koszelewski et al. (2010) reported that although only a catalytic amount of alanine is technically required in the L-Ala regeneration system, the non-favored reaction equilibrium of the amination step can delay the reaction. However, the excess of alanine allows a reasonable overall reaction rate. The time course for the conversion of 10–30 mM L-PA 1 into (R)-7 is shown in Fig. 7. In fact, (R)-7 was obtained with conversion percentages of 99% and 95% (> 99% ee) from 10 and 20 mM L-PA 1 at 12 h, respectively (Additional file 1: Figures S7–S8). However, (R)-7 was obtained with a 72.8% conversion percentage and ee values higher than 99% using a 30-mM L-PA substrate after a 16-h reaction period (Fig. 7A). Specifically, an analysis of the intermediate products from the L-PA 1 (20 mM) reaction showed that 2-HAP 6 (38.6%), (R)-7 (26.2%), diol 5 (10.0%), and styrene 3 (7%) were the main products obtained after 1 h. Therefore, L-PA 1 (> 84%) was converted into styrene 3 due to the high activity of PAL, Fdc1, and Pad1. In fact, this event is consistent with the conversion reaction of L-PA 1 into (S)-7. However, 2-HAP 6 (38.6%) and diol 5 (10.0%) remained in this reaction system, which implies that most of the diol 5 compound was converted into 2-HAP 6. Also, the activity of GoSCR was not affected in this system. After a 1-h reaction period, the concentration of 2-HAP began to decrease, while the concentration of (S)-7 increased linearly. In fact, (R)-7 was produced in about a 95% conversion percentage and > 99% ee at 12 h. Moreover, a trace of 2-HAP 6 (1.5%) was detected in the system, and the diol 5 and styrene 3 could not be detected at this reaction time (Fig. 7B).
Reaction condition optimization for conversion of L-PA 1 (20 mM) to (R)-7 by the designed E. coli (RFP-DEA-CGS) and E. coli (EB-DGA) consortium. A Effect of different pH on the conversion of (R)-7; B effect of different temperature on the conversion of (R)-7; C effect of glucose concentration on the conversion of (R)-7; D effect of different cell density of E. coli (RFP-DEA-CGS) and E. coli (MVTA) on the conversion of (R)-7; E effect of L-Ala concentration on the conversion of (R)-7; F: effect of NH3/NH4Cl concentration on the conversion of (R)-7
Time course of the cascade reaction for the synthesis of (R)-7 from L-PA 1. A Time course of the cascade reaction for conversion of 10–30 mM L-PA to (R)-7; B time course of the cascade reaction for intermediate produced from L-PA 1. Reaction conditions (5 mL): 100 mM phosphate buffer (pH 7.5), 10–30 mM L-PA, 20 mM glucose, 15 g cdw/L E. coli (RFP-DEA-CGS), 20 g cdw/L E. coli (EB-DGA), 400 mM L-ala, 150 mM NH3/NH4Cl, 0.1 mM PLP, at 25 °C. All biotransformations were performed in duplicate and conversion was determined by GC, error limit: < 2% of the state values
Preparation of (S)-7 and (R)-7 from L-PA
To examine the scalability of this cascade biocatalysis, preparative experiments were conducted on a 100-mL scale via the constructed recombinant E. coli microbial consortium. After the reactions were completed (12 h), the reaction solution was extracted with ethyl acetate and flash chromatography, pure (S)-7 and (R)-7 were prepared with 71.0% and 80.5% isolated yields, > 99% ee, and 5.19 g/L.d and 4.42 g/L.d volumetric productivity, respectively (Table 2, Additional file 1: Figures S9-S10).
Conclusions
In general, an artificial biocatalytic cascade was successfully constructed in this study by using a recombinant E. coli microbial consortium for converting bio-based l-phenylalanine into chiral phenylglycinol. Seven kinds of enzymes (PAL, Pad1, Fdc1, SMO, SpEH, GoSCR, and MVTA or BMTA) were first examined for the in vitro conversion of these two compounds. Both (R)-phenylglycinol and (S)-phenylglycinol were obtained within a conversion percentage range of 66.8–87.2% (> 99% ee) from 10–20 mM l-phenylalanine. Additionally, a recombinant E. coli microbial consortium, that included two recombinant E. coli cell modules, was constructed for the efficient conversion of l-phenylalanine into chiral phenylglycinol (up to 99% and > 99% ee). Moreover, the synthetic potential of this cascade biocatalysis was further confirmed by the constructed recombinant E. coli microbial consortium [E. coli (RFP-DEA-CGS), E. coli (MVTA) or E. coli (RFP-DEA-CGS), E. coli (EB-DGA)]. In this case, (S)-phenylglycinol and (R)-phenylglycinol were obtained with good yields (71.0% and 80.5%, respectively) and ee values higher than 99%. Finally, the advantages of this method are its mild reaction conditions, good yields and excellent ee values, and not requiring additional cofactors. Therefore, this system can enhance the sustainable synthesis of (S)-phenylglycinol or (R)-phenylglycinol from bio-based L-phenylalanine.
Availability of data and materials
All data generated or analyzed during this study are included in this article.
Abbreviations
- L-PA:
-
l-Phenylalanine
- 2-HAP:
-
2-Hydroxyacetophenone
- PAK4:
-
P21-activated kinases
- HDAC:
-
Histone deacetylase
- PDK1:
-
3-Phosphoinositide-dependent protein kinase-1
- AA:
-
Asymmetric aminohydroxylation
- PAL:
-
Phenylalanine ammonia lyase
- PAD:
-
Phenylacrylic acid decarboxylase
- SMO:
-
Styrene monooxygenase
- EH:
-
Epoxide hydrolase
- ADH:
-
Alcohol dehydrogenase
- MVTA:
-
(R)-Selective ω-transaminase from Mycobacterium vanbaalenii
- BMTA:
-
(S)-Selective ω-transaminase from Bacillus megaterium SC6394
- AlaDH:
-
Alanine dehydrogenase
- GDH:
-
Glucose dehydrogenase
- R-MBA:
-
(R)-α-Methylbenzylamine
- PLP:
-
Pyridoxal-5′-phosphate
- IPTG:
-
Isopropyl β-d-1-thiogalactopyranoside
- LB:
-
Luria–Bertani
- TB:
-
Terrific Broth
- Fdc1:
-
Ferulic acid decarboxylase
- Pad1:
-
Phenylacrylic acid decarboxylase
- FID:
-
Flame ionization detector
- GoSCR:
-
Polyol dehydrogenase from Gluconobacter oxydans 621H
- EtOAc:
-
Ethyl acetate
- cdw:
-
Cell dry weight
- L-Ala:
-
l-Alanine
References
Becker J, Wittmann C (2012) Systems and synthetic metabolic engineering for amino acid production–the heartbeat of industrial strain development. Curr Opin Biotechnol 23:718–726
Bergmeier SC (2000) The synthesis of vicinal amino alcohols. Tetrahedron 56(17):2561–2576
Bradford MM (1975) Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal Biochem 72:248–254
Caputo S, Di Martino S, Cilibrasi V, Tardia P, Mazzonna M, Russo D, Penna I, Summa M, Bertozzi SM, Realini N, Margaroli N, Migliore M, Ottonello G, Liu M, Lansbury P, Armirotti A, Bertorelli R, Ray SS, Skerlj R, Scarpelli R (2020) Design, synthesis, and biological evaluation of a series of oxazolone carboxamides as a novel class of acid ceramidase inhibitors. J Med Chem 63(24):15821–15851
Chen FF, Cosgrove SC, Birmingham WR, Mangas-Sanchez J, Citoler J, Thompson MP, Zheng GW, Xu JH, Turner NJ (2019) Enantioselective synthesis of chiral vicinal amino alcohols using amine dehydrogenases. ACS Catal 9(12):11813–11818
Cochrane FC, Davin LB, Lewis NG (2004) The Arabidopsis phenylalanine ammonia lyase gene family: kinetic characterization of the four PAL isoforms. Phytochemistry 65:1557–1564
Cui ZM, Zhang JD, Fan XJ, Zheng GW, Chang HH, Wei WL (2017) Highly efficient bioreduction of 2-hydroxyacetophenone to (S)- and (R)-1-phenyl-1,2-ethanediol by two substrate tolerance carbonyl reductases with cofactor regeneration. J Biotechnol 243:1–9
Fotheringham I, Archer I, Carr R, Speight R, Turner NJ (2006) Preparative deracemization of unnatural amino acids. Biochem Soc Trans 34(2):287–290
Guo J, Zhu M, Wu TX, Hao CZ, Wang K, Yan ZZ, Huang WX, Wang J, Zhao DM, Cheng MS (2017) Discovery of indolin-2-one derivatives as potent PAK4 inhibitors: structure-activity relationship analysis, biological evaluation and molecular docking study. Bioorg Med Chem 25(13):3500–3511
Gupta P, Mahajan N (2018) Biocatalytic approaches towards the stereoselective synthesis of vicinal amino alcohols. New J Chem 42(15):12296–12327
Jacobsen EN (2000) Asymmetric catalysis of epoxide ring-opening reactions. Acc Chem Res 33(6):421–431
Kim S, Park JH, Koo SY, Kim JI, Kim MH, Kim JE, Jo K, Choi HG, Lee SB, Jung SH (2004) Novel diarylsulfonylurea derivatives as potent antimitotic agents. Bioorg Med Chem Lett 14(24):6075–6078
Kobayashi S, Ishitani H, Ueno M (1998) Catalytic asymmetric synthesis of both syn- and anti-β-amino alcohols. J Am Chem Soc 120(2):431–432
Koszelewski D, Tauber K, Faber K, Kroutil W (2010) ω-Transaminases for the synthesis of non-racemic α-chiral primary amines. Trends Biotechnol 28(6):324–332
Lerchner A, Jarasch A, Skerra A (2016) Engineering of alanine dehydrogenase from Bacillus subtilis for novel cofactor specificity. Biotechnol Appl Biochem 63(5):616–624
Li G, Chang HT, Sharpless KB (1996) Catalytic asymmetric aminohydroxylation (AA) of olefins. Angew Chem Int Ed 35(4):451–453
McKennon MJ, Meyers AI (1993) A convenient reduction of amino acids and their derivatives. J Org Chem 58(13):3568–3571
Overman LE, Sugai S (1985) A convenient method for obtaining trans-2-aminocyclohexanol and trans-2-aminocyclopentanol in enantiomerically pure form. J Org Chem 50(21):4154–4155
Panek JS, Masse CE (1998) An improved synthesis of (4S, 5S)-2-phenyl-4-(methoxycarbonyl)-5-isopropyloxazoline from (S)-phenylglycinol. J Org Chem 63(7):2382–2384
Patel RN, Banerjee A, Howell JM, McNamee CG, Brozozowski D, Mirfakhrae D, Nanduri V, Thottathil JK, Szarka LJ (1993) Microbial synthesis of (2R,3S)-(−)-N-benzoyl-3-phenyl isoserine ethyl ester-a taxol side-chain synthon. Tetrahedron: Asymmetry 4 (9): 2069–2084.
Payne KA, White MD, Fisher K, Khara B, Bailey SS, Parker D, Rattray NJ, Trivedi DK, Goodacre R, Beveridge R, Barran P, Rigby SE, Scrutton NS, Hay S, Leys D (2015) New cofactor supports α, β-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition. Nature 522:497
Pelckmans M, Renders T, Van de Vyver S, Sels BF (2017) Bio-based amines through sustainable heterogeneous catalysis. Green Chem 19(22):5303–5331
Reetz MT (1991) New approaches to the use of amino acids as chiral building blocks in organic synthesis [new synthetic methods (85)]. Angew Chem Int Ed Engl 30(12):1531–1546
Rouf A, Gupta P, Aga MA, Kumar B, Parshad R, Taneja SC (2011) Cyclic trans-β-amino alcohols: preparation and enzymatic kinetic resolution. Tetrahedron Asymmetry 22(24):2134–2143
Russell MGN, Matassa VG, Pengilley RR, van Niel MB, Sohal B, Watt AP, Hitzel L, Beer MS, Stanton JA, Broughton HB, Castro JL (1999) 3-[3-(Piperidin-1-yl)propyl]indoles as highly selective h5-HT1D receptor agonists. J Med Chem 42(24):4981–5001
Schmidt F, Keller F, Vedrenne E, Aggarwal VK (2009) Stereocontrolled synthesis of β-amino alcohols from lithiated aziridines and boronic esters. Angew Chem Int Ed 48(6):1149–1152
Sekar BS, Mao J, Lukito BR, Wang Z, Li Z (2020) Bioproduction of enantiopure (R)- and (S)-2-phenylglycinols from styrenes and renewable feedstocks. Adv Synth Catal 363(7):1892–1903
Sharpless KB, Patrick DW, Truesdale LK, Biller SA (1975) New reaction. Stereospecific vicinal oxyamination of olefins by alkyl imido osmium compounds. J Am Chem Soc 97(8):2305–2307
Studte C, Breit B (2008) Zinc-catalyzed enantiospecific sp3–sp3 cross-coupling of α-hydroxy ester triflates with Grignard reagents. Angew Chem Int Ed 47(29):5451–5455
Tan Q, Wang X, Xiong Y, Zhao Z, Li L, Tang P, Zhang M (2017) Chiral amino alcohol accelerated and stereocontrolled allylboration of iminoisatins: highly efficient construction of adjacent quaternary stereogenic centers. Angew Chem Int Ed 56(17):4829–4833
Tan X, Zhang S, Song W, Liu J, Gao C, Chen X, Liu L, Wu J (2021) A multi-enzyme cascade for efficient production of d-p-hydroxyphenylglycine from l-tyrosine. Bioresour Bioprocess 8:41
Tanaka T, Kawase M, Tani S (2004) α-Hydroxy ketones as inhibitors of urease. Bioorg Med Chem 12(2):501–505
Vandekerkhove A, Claes L, De Schouwer F, Van Goethem C, Vankelecom IFJ, Lagrain B, De Vos DE (2018) Rh-catalyzed hydrogenation of amino acids to biobased amino alcohols: tackling challenging substrates and application to protein hydrolysates. ACS Sustain Chem Eng 6(7):9218–9228
Wang X, Su R, Chen K, Xu S, Feng J, Ouyang P (2019) Engineering a microbial consortium based whole-cell system for efficient production of glutarate from l-lysine. Front Microbiol 10:341
Wang F, Zhao J, Li Q, Yang J, Li R, Min J, Yu X, Zheng GW, Yu HL, Zhai C, Acevedo-Rocha CG, Ma L, Li A (2020) One-pot biocatalytic route from cycloalkanes to α, ω-dicarboxylic acids by designed Escherichia coli consortia. Nat Commun 11(1):1–10
Wu S, Li A, Chin YS, Li Z (2013) Enantioselective hydrolysis of racemic and meso-epoxides with recombinant Escherichia coli expressing epoxide hydrolase from Sphingomonas sp. HXN-200: preparation of epoxides and vicinal diols in high ee and high concentration. ACS Catal 3(4):752–759
Wu HL, Zhang JD, Zhang CF, Fan XJ, Chang HH, Wei WL (2017) Characterization of four new distinct ω-transaminases from Pseudomonas putida NBRC 14164 for kinetic resolution of racemic amines and amino alcohols. Appl Biochem Biotechnol 181(3):972–985
Xu Y, Jia X, Panke S, Li Z (2009) Asymmetric dihydroxylation of aryl olefins by sequential enantioselective epoxidation and regioselective hydrolysis with tandem biocatalysts. Chem Commun. https://doi.org/10.1039/b820889a
You ZN, Zhou K, Han Y, Yang BY, Chen Q, Pan J, Qian XL, Li CX, Xu JH (2021) Design of a self-sufficient hydride-shuttling cascade for concurrent bioproduction of 7, 12-dioxolithocholate and l-tert-leucine. Green Chem 23(11):4125–4133
Zhang JD, Wu HL, Meng T, Zhang CF, Fan XJ, Chang HH, Wei WL (2017) A high-throughput microtiter plate assay for the discovery of active and enantioselective amino alcohol-specific transaminases. Anal Biochem 518:94–101
Zhang JD, Zhao JW, Gao LL, Chang HH, Wei WL, Xu JH (2019a) Enantioselective synthesis of enantiopure β-amino alcohols via kinetic resolution and asymmetric reductive amination by a robust transaminase from Mycobacterium vanbaalenii. J Biotechnol 290:24–32
Zhang JD, Yang XX, Jia Q, Zhao JW, Gao LL, Gao WC, Chang HH, Wei WL, Xu JH (2019b) Asymmetric ring opening of racemic epoxides for enantioselective synthesis of (S)-β-amino alcohols by a cofactor self-sufficient cascade biocatalysis system. Catal Sci Technol 9(1):70–74
Zhang JD, Zhao JW, Gao LL, Zhao J, Chang HH, Wei WL (2019c) One-pot three-step consecutive transformation of l-α-amino acids to (R)- and (S)-vicinal 1,2-diols via combined chemical and biocatalytic process. ChemCatChem 11(20):5032–5037
Zhang JD, Yang XX, Dong R, Gao LL, Li J, Li X, Huang SP, Zhang CF, Chang HH (2020) Cascade biocatalysis for regio- and stereoselective aminohydroxylation of styrenyl olefins to enantiopure arylglycinols. ACS Sustain Chem Eng 8(49):18277–18285
Zhou Y, Wu S, Li Z (2016) Cascade biocatalysis for sustainable asymmetric synthesis: from biobased l-phenylalanine to high-value chiral chemicals. Angew Chem Int Ed 128(38):11819–11822
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 21772141), the Shanxi Province Science Foundation for Youths (Grant No. 201701D221042), the Key Research and Development (R&D) Project of Shanxi Province (201803D31050).
Author information
Authors and Affiliations
Contributions
JDZ conceived the idea and plan of this project. NQ performed most of the experiments. LLG performed part of the experiments in construction of dual-expression vectors. JL, FCZ and HHC performed part of the experiments in the product analysis. JDZ supervised the whole research and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors approved the consent for publishing the manuscript to Bioresources and Bioprocessing.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Primers used in this study. Table S2. Recombinant strains constructed in this study. Table S3. The specific activity of enzymes used in this study. Figure S1. Recombinant plasmid constructed. Figure S2. Recombinant E. coli cells co-expression of multiple enzymes. Figure S3. SDS-PAGE of the cell-free extracts of recombinant E. coli cells co-expression of multiple enzymes. A: lane M:marker, lane 1, 2: E. coli (DEA), lane 3, 4: E. coli (CGS); B: laneM:marker, lane 1, 2: E. coli (REG); C: laneM:marker, lane 1, 2: E. coli (DFP); D: laneM:marker, lane1: E. coli (CFP); E: laneM:marker, lane 1, 2: E. coli (RFP), lane 3: E. coli (DES); F: lane M:marker, lane 1, 2: E. coli (RGA); G: lane M:marker, lane 1: E. coli (CAS). Figure S4. Achiral GC chromatograms of 2-HAP. A: 2-HAP standard. B: 2-HAP produced by conversion of L-phenylalanine (10 mM) with resting cells of E. coli (RFP-DEA-CGS) (15 g cdw/L) at 3 h. C: 2-HAP produced by conversion of L-phenylalanine (20 mM) with resting cells of E. coli (RFP-DEA-CGS) (25 g cdw/L) at 9 h. D: 2-HAP produced by conversion of L-phenylalanine (50 mM) with resting cells of E. coli (RFP-DEA-CGS) (25 g cdw/L) at 12 h. IS: Internal standard (n-dodecane), 4.05 min; 2-HAP: 2-hydroxyacetophenone, 4.91 min. Figure S5. Effect of different concentration of RMBA on E. coli (RFP-DEA-CGS) cells for conversion of L-PA to 2-HAP. Figure S6. Achiral GC chromatograms of (S)-phenylglycinol. A: (S)-phenylglycinol standard. B: (S)-phenylglycinol produced by conversion of L-phenylalanine (10 mM) with resting cells of E. coli (RFP-DEA-CGS) (20 g cdw/L) and E. coli (MVTA) (15 g cdw/L) at 12 h. C: (S)-phenylglycinol produced by conversion of L-phenylalanine (20 mM) with resting cells of E. coli (RFPDEA-CGS) (20 g cdw/L) and E. coli (MVTA) (15 g cdw/L) at 12 h. IS: Internal standard (n-dodecane), 4.05 min; RMBA: (R)-(+)-1-phenylethylamine, 2.62 min; AP: acetophenone, 2.78 min; Product (S)-phenylglycinol, 6.05 min. Figure S7. Achiral GC chromatograms of (R)-phenylglycinol. A: (R)-phenylglycinol standard. B: (R)-phenylglycinol produced by conversion of L-phenylalanine (10 mM) with resting cells of E. coli (RFP-DEA-CGS) (15 g cdw/L) and E. coli (EB-DGA) (20 g cdw/L) at 9 h. C: (R)-phenylglycinol produced by conversion of L-phenylalanine (20 mM) with resting cells of E. coli (RFP-DEA-CGS) (15 g cdw/L) and E. coli (EB-DGA) (20 g cdw/L) at 12 h. IS: Internal standard (n-dodecane), 4.05 min; (R)-phenylglycinol, 6.05 min. Figure S8. Chiral GC chromatograms of phenylglycinol. A: (±)- phenylglycinol standard. B: (R)-phenylglycinol standard. C: (S)-phenylglycinol standard. D: (R)-phenylglycinol produced by conversion of L-phenylalanine (20 mM) with resting cells of E . coli (RFP-DEA-CGS) (15 g cdw/L) and E. coli (EB-DGA) (20 g cdw/L) at 12 h. E: (S)-phenylglycinol produced by conversion of L-phenylalanine (20 mM) with resting cells of E. coli (RFP-DEA-CGS) (20 g cdw/L) and E. coli (MVTA) (15 g cdw/L) at 9 h. (R)-phenylglycinol, 28.7 min; (S)-phenylglycinol, 29.3 min. Figure S9. Chiral GC chromatograms of phenylglycinol. A: (±)- phenylglycinol standard. B: (R)-phenylglycinol standard. C: (S)-phenylglycinol standard. D: Preparation of (R)-phenylglycinol from L-phenylalanine. E: Preparation of (S)-phenylglycinol from L-phenylalanine. (R)-phenylglycinol, 28.7 min; (S)-phenylglycinol, 29.3 min. Figure S10. 1H NMR spectra analysis of 7.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, J., Qi, N., Gao, L. et al. One-pot synthesis of (R)- and (S)-phenylglycinol from bio-based l-phenylalanine by an artificial biocatalytic cascade. Bioresour. Bioprocess. 8, 97 (2021). https://doi.org/10.1186/s40643-021-00448-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40643-021-00448-5