Abstract
Background
Atrial fibrillation (AF) is the most common cardiac arrhythmia in the intensive care unit and is associated with increased morbidity and mortality. New-onset atrial fibrillation (NOAF) is often initially paroxysmal and fleeting, making it difficult to diagnose, and therefore difficult to understand the true burden of disease. Automated algorithms to detect AF in the ICU have been advocated as a means to better quantify its true burden.
Results
We used a publicly available 12-lead ECG dataset to train a deep learning model for the classification of AF. We then conducted an external independent validation of the model using continuous telemetry data from 984 critically ill patients collected in our institutional database. Performance metrics were stratified by signal quality, classified as either clean or noisy. The deep learning model was able to classify AF with an overall sensitivity of 84%, specificity of 89%, positive predictive value (PPV) of 55%, and negative predictive value of 97%. Performance was improved in clean data as compared to noisy data, most notably with respect to PPV and specificity.
Conclusions
This model demonstrates that computational detection of AF is currently feasible and effective. This approach stands to improve the efficiency of retrospective and prospective research into AF in the ICU by automating AF detection, and enabling precise quantification of overall AF burden.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Background
New-onset atrial fibrillation (NOAF) is the most common cardiac dysrhythmia in critically ill patients with a reported incidence as high as 46% [1]. It is most often described in patients with sepsis with an incidence of 10–40% [2], but is seen in a variety of illnesses such as acute respiratory distress syndrome [3], non-cardiac thoracic surgery [4, 5], and trauma [6]. Critical illness is an independent driver of NOAF due to arrhythmogenic triggers such as electrolyte disorders, vasoactive medications, fluid overload and hypoxia [7]. Acute events during critical illness (e.g., infection, ischemia) accelerate cardiac remodeling and fibrosis, begetting further arrhythmias [8]. NOAF is independently associated with prolonged duration of hospital stay [2, 9] and significant morbidity and mortality [10]. Patients with NOAF are at an increased risk for in-hospital ischemic stroke [11] and 18% of patients who develop NOAF are discharged from the intensive care unit (ICU) with ongoing atrial fibrillation (AF) [12]. Patients who develop NOAF in the ICU may be at increased risk of having recurrent AF in the future and suffer the consequences associated with it, such as stroke and heart failure [13].
Given the consequentiality of developing NOAF, prevention and treatment of this arrhythmia are of great potential benefit. Unfortunately, NOAF is difficult to study, as the ability to detect it in real time and quantify its burden is limited in most ICU settings. There is difficulty capturing the arrhythmia in patients who develop paroxysmal atrial fibrillation as fleeting arrhythmias are often missed on telemetry or ECG [14]. Most studies of NOAF are retrospective and are based on diagnostic codes in large databases which rely on the accuracy and completeness of those codes for identification of patients and their long-term outcomes.
Development of detection algorithms to identify patients with NOAF would allow for precise estimates of the incidence and prevalence and inform future studies of NOAF in a critical care setting. Automated monitoring algorithms for the detection of AF have previously been developed using heart rate variability techniques [15, 16], however their application to continuous, high-frequency ECG data collected in ICUs by bedside monitoring is limited. In this paper, we describe the development and validation of a deep learning model to classify AF in a large database of continuous telemetry data collected on critically ill patients.
Methods
Figure 1 shows a high-level overview of our experimental process.
Institutional dataset creation
The Queen’s University Health Sciences Research Ethics Board approved this study (File No: 6024689). The need for informed consent was waved because the data were collected as part of routine care and were stored in a de-identified format. Our main source of test data were collected from bedside monitors in the ICU at Kingston Health Sciences Centre (KHSC) in Kingston, Ontario, Canada, a 33-bed mixed-use medical-surgical, neurological, and trauma ICU. Both physiological waveforms and vital sign time series were collected from GE Solar monitors using Bedmaster software (Hill-Rom Holdings Inc, Chicago, Ill). These were stored in Critical Care Data Exchange Format [17] files for ease of querying and long-term archiving. Over 11TB of data were collected between 2015 and 2020, including oxygen saturation, arterial line, and ECG waveforms. ECG signals were sampled at 240 Hz and included leads I, II, III and V1.
Model development
Building on our previous work [18], we used the publicly available Chapman 12-lead ECG dataset [19] for model development and training. This dataset consists of static 12-lead, 10 s ECG recordings from over 10,000 patients from Shaoxing People’s Hospital in Zhejiang, China. Leads I, II, III and V1 were extracted and the recorded segments were resampled from 500 to 240 Hz in order to match the dimensions of our institutional database at Kingston Health Sciences Center (KHSC). To translate between the KHSC classes and the Chapman diagnostic classes, we mapped the sinus bradycardia, sinus rhythm, sinus tachycardia and sinus irregularity classes to our sinus rhythm class, and the AF and atrial flutter classes to our AF class. We split the ECG segments with a 70/10/20% training/validation/testing split, stratified by rhythm type and patient gender.
Model architecture and training
A deep convolutional neural network [20] was chosen as our ECG classification model. This specific classifier [21] was derived from an existing architecture shown to have state-of-the-art performance for AF detection on single-lead, portable device ECG [22]. The base architecture was kept as-is, with the exception of changes to the input dimensions to accommodate our 4-lead KHSC data.
Once initialized, the model was trained on the Chapman training dataset. Hyperparameter tuning was conducted on the Chapman validation set to select a high-performing configuration, and the final model was evaluated on the Chapman test set to ensure performance was comparable to the results found in the study by Zheng et al. [19]. A full breakdown of the model architecture and training parameters is found in Additional file 1: Fig. S4. Model development was conducted using the PyTorch Python Library [23].
To create a retrospective patient cohort, we selected files from the KHSC institutional dataset that could be directly linked back to a known patient admission and stay. Because patients could be moved between different beds in the ICU over the course of their admission, there was a risk of a particular recording containing waveform data for multiple patients if monitors were not properly re-configured after a move. To that end, we excluded any stays that had a bed move to ensure a one-to-one correspondence between the patients and the data files. This strict matching also ensured that admission and discharge times could be aligned reliably. Further filtering was conducted to ensure a sufficient quantity of ECG signal was present in each patient’s recorded waveforms. We excluded any stays that did not have all four ECG leads recorded. We removed cases with less than 10 s of continuous, unbroken ECG data. For the remaining records, we randomly sampled a 10-s segment of continuous ECG signal for each patient to create our KHSC dataset.
To obtain ground truth labels for the segments, we defined a set of eight disjoint classes for annotation (Table 1). These included seven categories of rhythms and a catch-all “Abstain” class for records deemed too noisy or possessing insufficient clear signal to assign a label. Annotation was conducted by two critical care physicians experienced in ECG interpretation and identifying arrhythmias present in an ICU context. A third critical care physician acted as a tiebreaker in cases of disagreements, and if all three reviewers disagreed the third reviewer’s label was used. Because not all classes defined prior to annotation were present in the labeled dataset, only those with greater than ten samples were included for further analysis. A second round of annotation was conducted with a critical care physician to collect additional information on potentially noisy segments. This involved a simple binary label indicating whether each segment was "clean" or contained a significant amount of noise. Clean samples had no baseline wander, motion artifact or signal drop off. Samples labeled “noisy” could still be categorized if the rhythm was interpretable and were labeled “abstain” if the signal was too noisy to determine the underlying rhythm. Free text comments were also collected about specific types of noise present for each noisy sample.
Model evaluation
Due to the low expected prevalence of arrhythmias other than AF, we retained only segments labeled as sinus rhythm (SR) or AF from the KHSC dataset to test the pre-trained model. A standard set of binary classification metrics were employed: area under the receiver operator curve (AUROC), precision/positive predictive value (PPV), negative predicative value (NPV), specificity and recall/sensitivity. Segments with an AF label were considered as a positive test, while segments with a label of sinus rhythm were considered a negative test. In addition to evaluating overall performance on this subset of KHSC data, we derived independent performance measurements for segments that were identified as clean or noisy.
Results
Curation of local dataset
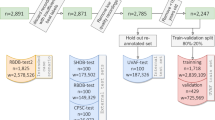
In the KHSC dataset, a total of 1043 patients were found to have an ICU stay with no bed moves. Of these stays, 59 did not contain all 4 leads or a minimum of 10 s of continuous ECG signal. The median recording duration of the remaining 984 patient waveform records before sampling was 11.9 h. 665 were annotated by two critical care physicians, with a kappa of 0.81. The reviewers disagreed on 47 segments, of which 17 were labeled “abstain” by at least one reviewer. All three reviewers disagreed on 9 segments of which 5 were labeled “abstain” by the third reviewer. Given the good agreement between physicians, an additional 319 samples were annotated by the first critical care physician. All 984 segments received noise annotations. A flowchart summary of the sampling and annotation process may be found in Fig. 2.
Table 1 shows a breakdown of class counts and identified noise levels for the KGH data. Of the ECG segments annotated, the majority class was sinus rhythm and the majority arrhythmia was AF at 12.5%.
The sinus rhythm class contains all sinus rhythms regardless of rate, including sinus tachycardia and sinus bradycardia. No segments were labeled as ventricular tachycardia/fibrillation or other types of tachy/bradycardia.
Twenty-seven percent of records were found to contain some amount of noise, whereas segments in the non-diagnostic class were found to be almost completely noisy. Upon review of the segments in this class that were not labeled as noisy, we found that the discrepancy could be explained by disagreements during overreading. The AF class had the next highest noise level at 36%, more than twice that of sinus rhythm. Comments left for noisy segments fell into one of three categories: baseline signal artifact, baseline wander, and other unspecified artifacts. Representative examples for clean and each category of noise and counts of each type of noise may be found in Additional file 1: Table S3 and Additional file 1: Fig. S5.
After removing 115 segments not labeled as either SR or AF, we were left with a final testing dataset of 869 segments. The prevalence of positive AF samples in this subset was 14.1%, while 19.7% of samples were identified as noisy.
Model performance
In the test set portion of the Chapman dataset, the deep learning model for binary classification of AF (positive) versus SR (negative) achieved an AUROC of 0.985, PPV of 0.828, sensitivity of 0.950 and specificity of 0.940 (Additional file 1: Fig. S6). The prevalence of the AF class in this test set was 23.5%, compared to 36.5% across the entire Chapman dataset. Model performance in the KHSC dataset is summarized in Table 2. Overall, the classifier demonstrated high sensitivity and specificity in identifying AF, with better performance seen among the clean segments compared to the noisy segments. Full precision–recall curves of KHSC data may be found in Fig. 3.
Discussion
In this study, we applied a deep learning model for the classification of AF adapted from static 12-lead ECGs, to continuous telemetry data collected from an ICU. Despite training on a different type of ECG data, the model was highly sensitive and specific, with performance in clean data being better than that in noisy data.
Prior studies on AF detection from ICU telemetry data have been developed using continuous ECG telemetry waveform data from the Medical Information Mart for Intensive Care (MIMIC III) data set, and have primarily focused on engineered features derived from waveform characteristics [16, 24, 25]. In the study by Walkey et al. telemetry data from three cohorts of 50 patients was analyzed for interpretable signal using automated signal and noise detection. Two-minute segments were then assessed using an algorithm that detects R-waves using variable-frequency complex demodulation-based ECG reconstruction that evaluates the sample entropy, which would be expected to be higher in AF samples than in normal sinus rhythm. To differentiate premature atrial and ventricular beats, Poincaré plots and p-wave detection algorithms were used. When only signal and noise detection and R–R interval indices were used the algorithm had a sensitivity of 91%, a specificity of 71%, a PPV of 48%, a NPV of 96% and an accuracy of 76%. This improved with the addition of Poincaré plots and P-wave indices to a sensitivity of 100%, a specificity of 95%, a PPV of 85%, a NPV of 100% and an accuracy of 96% [25]. Similarly, Bashar et al. analyzed 198 septic subjects from the MIMIC III database using a heart rate variability algorithm using statistical parameters such as the root mean square of successive differences, Shannon entropy, and sample entropy, followed by Poincaré plots for premature atrial contraction/premature ventricular contraction (PAC/PVC) detection. The samples included 50 patients in a training set and 49 patients in a validation set, of which 11 were in AF. The sensitivity, specificity, accuracy, PPV and NPV were 100%, 94.74%, 95.92%, 84.62%, and 100%, respectively. The algorithm was tested on an additional 49 patients whose ECGs did not contain any PAC/PVC and a second test set of 50 patients where 25 patients had AF and 25 had NSR with PAC/PVCs. The combined test sets of 99 patients had a sensitivity, specificity, accuracy, PPV and NPV of 100%, 98%, 98.99%, 98%, and 100% [16].
In contrast to these studies, we have applied our model to a new data set from our institution that is more recent than the MIMIC III dataset. We have used a larger sample size than previous studies and have the added benefit of deploying the algorithm on raw data without any computationally expensive pre-processing. Our prevalence-dependent performance metrics (PPV) are more realistic given the class imbalance in our data (14.1% AF), which is in keeping with estimated prevalence of atrial fibrillation in ICUs, compared to the 50% AF prevalence in test sets in other studies [16, 24].
Several traditional learning models for the detection of atrial fibrillation have been published and are generally based on the mathematical characteristics of waveform analysis and typically require manual feature selection, extraction and classification. These extracted features may perform well in the data set in which they are developed, but require assumptions that may limit their reliability and their generalizability is uncertain. The advantage of a deep learning model is twofold: the selection and extraction features are built into the model, thus eliminating the need for separate steps, and the model is able to “learn” as more data are presented to it, with potential for model performance to be continuously improved [26]. Although deep learning approaches have been explored for static ECG [27], end-to-end deep learning approaches have seen very limited uptake for streaming data from ICU bedside monitors [28]. Transfer learning (training a model on one dataset and fine-tuning on a much smaller dataset before evaluating) has also been evaluated as a method for developing AF classification models when only limited annotated ECG data are available locally [29]. Building on this, we demonstrate that a deep learning model trained on static ECGs can identify AF in telemetry data, and expect that the performance of this model will improve with continued training.
Diagnosis of NOAF and inclusion into studies of this arrhythmia has traditionally relied on manual interpretation of ECGs, recordings of observed heart rhythms by bedside nurses, and billing codes (such as ICD-9 codes). Study of NOAF is further limited by a lack of common definition of the condition, with studies reporting different heart rates, durations of NOAF, and criteria for “new-onset” [30]. Use of automated algorithms could revolutionize the study of NOAF in critically ill patients. While heart rate variability algorithms require clean ECG samples of sufficient duration to reliably calculate statistical parameters, our machine learning approach allows the use of 10-s segments of telemetry data which are far less cumbersome to manually annotate and have the additional benefit of granularity that allows capture of very brief periods of NOAF that may be otherwise missed given the paroxysmal nature of the arrhythmia. This would aid in quantifying the burden of NOAF that is clinically important and contribute to a uniform definition of NOAF to be used in future studies. Continuous monitoring with computational classification may enable prediction of the development of atrial fibrillation [31] and enable the stratification of patients with varied duration of NOAF, potentially allowing predictive enrichment in clinical trials. Clinically, enhanced detection of these arrhythmias has the potential to markedly enhance our understanding of the true burden of disease and management of this arrhythmia in critically ill patients. Real-time identification of NOAF would enable immediate notification of practitioners and rapid administration of treatment if needed. This study provides the ground work for development of predictive models, allowing for identification of patients at higher risk of developing the arrhythmia to optimize prophylaxis and risk mitigation for a precision medicine approach.
Our study has some limitations. This model was trained on the Chapman training set which is not composed exclusively of critically ill patients. Despite this, the performance of the model on telemetry data of exclusively critically ill patients was very good. Model performance degraded slightly in the presence of background noise or artifact of the type that is sometimes seen in telemetry data due to patient movement, lead displacement, or bedside procedures. However, it still showed good performance when noisy data were included. Our use of a binary classifier resulted in the removal of 115 (12%) samples with non-AF and non-sinus rhythms, of which 87 were classified as non-diagnostic. While this is less than the > 25% loss using other noise detection and pre-processing methods, it does limit its use in a “real-time” clinical setting where the model would have to evaluate all rhythm strips. While 665 ECG samples were annotated by two reviewers, an additional 319 samples were annotated by a single reviewer. The agreement between reviewers was excellent and this was unlikely to contribute uncertainty to the model. Although it is possible to reduce the false positive rate by tuning the model decision threshold, doing so comes with the trade-off of decreased sensitivity. A future validation study to evaluate model performance in a prospective and continuous fashion is warranted. Further work is also needed to ‘clean’ raw data by removing artifacts.
Conclusion
We adapted a deep learning model trained on static ECG data, to continuous telemetry data from an ICU in order to detect AF in critically ill patients. The model showed a sensitivity of 84%, a specificity of 89%, a PPV of 55%, and a NPV of 97%, allowing enhanced detection of AF beyond the 14% baseline prevalence of AF in the dataset. This model provides the foundation for enhanced detection and predictive model development to improve our understanding of the true burden of AF and its clinical consequences, and enhance classification and stratification of patients with NOAF for further studies.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Seguin P, Launey Y (2010) Atrial fibrillation is not just an artefact in the ICU. Crit Care 14(4):182
Klein Klouwenberg PM, Frencken JF, Kuipers S, Ong DS, Peelen LM, van Vught LA, Schultz MJ, van der Poll T, Bonten MJ, Cremer OL et al (2017) Incidence, predictors, and outcomes of new-onset atrial fibrillation in critically ill patients with sepsis. A cohort study. Am J Respir Crit Care Med 195(2):205–211
Ambrus DB, Benjamin EJ, Bajwa EK, Hibbert KA, Walkey AJ (2015) Risk factors and outcomes associated with new-onset atrial fibrillation during acute respiratory distress syndrome. J Crit Care 30(5):994–997
Christians KK, Wu B, Quebbeman EJ, Brasel KJ (2001) Postoperative atrial fibrillation in noncardiothoracic surgical patients. Am J Surg 182(6):713–715
Passman RS, Gingold DS, Amar D, Lloyd-Jones D, Bennett CL, Zhang H, Rusch VW (2005) Prediction rule for atrial fibrillation after major noncardiac thoracic surgery. Ann Thorac Surg 79(5):1698–1703
Seguin P, Laviolle B, Maurice A, Leclercq C, Malledant Y (2006) Atrial fibrillation in trauma patients requiring intensive care. Intensive Care Med 32(3):398–404
Sibley S, Muscedere J (2015) New-onset atrial fibrillation in critically ill patients. Can Respir J 22(3):179–182
Bosch NA, Cimini J, Walkey AJ (2018) Atrial Fibrillation in the ICU. Chest 154(6):1424–1434
Bedford JP, Gerry S, Hatch RA, Rechner I, Young JD, Watkinson PJ (2020) Hospital outcomes associated with new-onset atrial fibrillation during ICU admission: a multicentre competing risks analysis. J Crit Care 60:72–78
Chen AY, Sokol SS, Kress JP, Lat I (2015) New-onset atrial fibrillation is an independent predictor of mortality in medical intensive care unit patients. Ann Pharmacother. https://doi.org/10.1177/1060028015574726
Walkey AJ, Wiener RS, Ghobrial JM, Curtis LH, Benjamin EJ (2011) Incident stroke and mortality associated with new-onset atrial fibrillation in patients hospitalized with severe sepsis. JAMA 306(20):2248–2254
Kanji S, Williamson DR, Yaghchi BM, Albert M, McIntyre L, Canadian Critical Care Trials G (2012) Epidemiology and management of atrial fibrillation in medical and noncardiac surgical adult intensive care unit patients. J Crit Care 27(3):326.e321-328
Walkey AJ, Hammill BG, Curtis LH, Benjamin EJ (2014) Long-term outcomes following development of new-onset atrial fibrillation during sepsis. Chest 146(5):1187–1195
Moss TJ, Calland JF, Enfield KB, Gomez-Manjarres DC, Ruminski C, DiMarco JP, Lake DE, Moorman JR (2017) New-onset atrial fibrillation in the critically ill. Crit Care Med 45(5):790–797
Zhang K, Aleexenko V, Jeevaratnam K (2020) Computational approaches for detection of cardiac rhythm abnormalities: are we there yet? J Electrocardiol 59:28–34
Bashar SK, Hossain MB, Ding E, Walkey AJ, McManus DD, Chon KH (2020) Atrial fibrillation detection during sepsis: study on MIMIC III ICU data. IEEE J Biomed Health Inform 24(11):3124–3135
Laird P, Wertz A, Welter G, Maslove D, Hamilton A, Heung Yoon J, Lake DE, Zimmet AE, Bobko R, Randall Moorman J et al (2021) The critical care data exchange format: a proposed flexible data standard for combining clinical and high-frequency physiologic data in critical care. Physiol Meas. https://doi.org/10.1088/1361-6579/abfc9b
Chen B, Javadi G, Jamzad A, Hamilton A, Sibley S, Abolmaesumi P, Maslove D, Mousavi P: detecting atrial fibrillation in ICU telemetry data with weak labels. In: Ken J, Serena Y, Mark S, Michael S, Rajesh R, eds., Proceedings of the 6th Machine Learning for Healthcare Conference, vol. 149. Proceedings of Machine Learning Research: PMLR; 2021; pp. 176--195.
Zheng J, Zhang J, Danioko S, Yao H, Guo H, Rakovski C (2020) A 12-lead electrocardiogram database for arrhythmia research covering more than 10,000 patients. Sci Data 7(1):48
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444
Goodfellow SD, Goodwin A, Greer R, Laussen PC, Mazwi M, Eytan D: Towards understanding ECG rhythm classification using convolutional neural networks and attention mappings. In: Finale D-V, Jim F, Ken J, David K, Rajesh R, Byron W, Jenna W, Eds., Proceedings of the 3rd Machine Learning for Healthcare Conference, vol. 85. Proceedings of Machine Learning Research: PMLR; 2018; pp. 83--101.
Clifford GD, Liu C, Moody B, Lehman LH, Silva I, Li Q, Johnson AE, Mark RG (2010) AF classification from a short single lead ECG recording: the physionet/computing in cardiology challenge 2017. Comput Cardiol 2017:44
Adam Paszke SG, Francisco Massa, Adam Lerer, James Bradbury, Gregory Chanan, Trevor Killeen, Zeming Lin, Natalia Gimelshein, Luca Antiga, Alban Desmaison, Andreas Kopf, Edward Yang, Zachary DeVito, Martin Raison, Alykhan Tejani, Sasank Chilamkurthy, Benoit Steiner, Lu Fang, Junjie Bai, Soumith Chintala: PyTorch: an imperative style, high-performance deep learning library. Advances in Neural Information Processing Systems Curran Associates, Inc 2019; pp. 8026–8037.
Bashar SK, Ding E, Albuquerque D, Winter M, Binici S, Walkey AJ, McManus DD, Chon KH (2019) Atrial fibrillation detection in ICU patients: a pilot study on MIMIC III data(.). Annu Int Conf IEEE Eng Med Biol Soc 2019:298–301
Walkey AJ, Bashar SK, Hossain MB, Ding E, Albuquerque D, Winter M, Chon KH, McManus DD (2021) Development and validation of an automated algorithm to detect atrial fibrillation within stored intensive care unit continuous electrocardiographic data: observational study. JMIR Cardio 5(1):e18840
Wang J (2020) A deep learning approach for atrial fibrillation signals classification based on convolutional and modified Elman neural network. Futur Gener Comput Syst 2020(102):670–679
Hong S, Zhou Y, Shang J, Xiao C, Sun J (2020) Opportunities and challenges of deep learning methods for electrocardiogram data: a systematic review. Comput Biol Med 122:103801
Yoon D, Lim HS, Jung K, Kim TY, Lee S (2019) Deep learning-based electrocardiogram signal noise detection and screening model. Healthc Inform Res 25(3):201–211
Weimann K, Conrad TOF (2021) Transfer learning for ECG classification. Sci Rep 11(1):5251
Drikite L, Bedford JP, O’Bryan L, Petrinic T, Rajappan K, Doidge J, Harrison DA, Rowan KM, Mouncey PR, Young D et al (2021) Treatment strategies for new onset atrial fibrillation in patients treated on an intensive care unit: a systematic scoping review. Crit Care 25(1):257
Bashar SK, Ding EY, Walkey AJ, McManus DD, Chon KH (2021) Atrial fibrillation prediction from critically ill sepsis patients. Biosensors (Basel). https://doi.org/10.3390/bios11080269
Acknowledgements
Not applicable.
All work was performed at Kingston Health Sciences Centre and Queen’s University.
Funding
UHKF Women’s Giving Circle Fund, Queen’s Catalyst Fund.
Author information
Authors and Affiliations
Contributions
DM, SS and PM conceived and planned the experiment. BC, AH, and PL developed the algorithm and performed the computations and were supervised by PM. DM, SS and PL performed all annotations. SS, DM, PM, BC, and JC prepared the final manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by Queen’s University Health Sciences Research Ethics Board—Reference number 6024689. Consent in this study was waived because the data were collected as part of routine care and were stored in a de-identified format.
Consent for publication
Not applicable.
Competing interests
The authors have no competing interests to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Figure S4. Diagram of the neural network. Table S3. Specific categories of noise observed in the labeled KGH dataset during the noise annotation process. Segments not provided with a noise label are listed here as "No comment". Figure S5. Example segments for each noise category in the labeled KGH dataset. Only lead I is shown on these 10-s samples. a Clean atrial fibrillation b atrial fibrillation with baseline wander c normal sinus rhythm with baseline artifacts d non-diagnostic sample (annotator abstained). Figure S6. ROC curves for the deep learning model on clean, noisy, and all labeled sinus and AF samples from the KGH dataset
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, B., Maslove, D.M., Curran, J.D. et al. A deep learning model for the classification of atrial fibrillation in critically ill patients. ICMx 11, 2 (2023). https://doi.org/10.1186/s40635-022-00490-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40635-022-00490-3