Abstract
Background
Genetic heterogeneity of the canine angiotensin converting enzyme (ACE) gene is functionally important because the degree of aldosterone breakthrough with ACE-inhibitor therapy is greater in variant positive dogs compared to variant negative dogs, but the prevalence of the variant is not known. The purpose of this study was to determine ACE gene variant-positive prevalence in a population of 497 dogs of different breeds.
Results
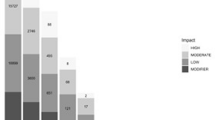
Overall variant-positive prevalence was 31%, with 20% of dogs heterozygous and 11% of dogs homozygous. The variant was overrepresented in Irish Wolfhounds (prevalence 95%; P < .001), Dachshunds (prevalence 90%; P < .001), Cavalier King Charles Spaniels (prevalence 85%; P < .001), Great Danes (prevalence 84%; P < .001), and Bull Mastiffs (prevalence 58%; P = .02). Irish Wolfhounds were more likely to be homozygous than heterozygous (P < .001).
Conclusions
Nearly one-third of dogs in this study were positive for a functionally important ACE gene variant, with wide prevalence variability between breeds. The clinical importance of high ACE gene variant-positive prevalence in some breeds requires further study because the highest prevalences were found in breeds that are predisposed to heart disease and therefore may be treated with ACE-inhibitors.
Plain English summary
Genetic differences in the angiotensin converting enzyme (ACE) have been shown to affect the way dogs respond to ACE-inhibitors, a class of medication which is used to treat dogs with heart disease. Dogs that have a genetic mutation of the ACE gene show less benefit with ACE-inhibitor medications than dogs without the mutation, but it is not known how common the mutation is in dogs. The purpose of this study was to determine the prevalence of this mutation (variant) in a large population of dogs of different breeds. The overall variant-positive prevalence in this population of 497 dogs was 31%. The variant was overrepresented in Irish Wolfhounds (prevalence 95%), Dachshunds (prevalence 90%), Cavalier King Charles Spaniels (prevalence 85%), Great Danes (prevalence 84%), and Bull Mastiffs (prevalence 58%). Irish Wolfhounds were more likely than other breeds to have 2 copies of the mutation (homozygous) than 1 copy of the mutation (heterozygous). The clinical importance of high ACE gene variant-positive prevalence in some breeds will require additional studies because some breeds are predisposed to heart disease, for which treatment with ACE-inhibitor medication might be recommended.
Similar content being viewed by others
Background
The angiotensin converting enzyme (ACE) is an important catalyst for the renin-angiotensin aldosterone system (RAAS) cascade by cleaving a carboxy-terminal dipeptide from angiotensin I to form angiotensin II [1]. When the RAAS is pathologically and chronically activated, angiotensin II, both directly and through stimulation of aldosterone synthesis, mediates maladaptive actions such as vasoconstriction, sodium retention, and pathological remodeling of cardiovascular and renal tissues [1]. Inhibitors of ACE are used clinically to reduce the formation of angiotensin II, thereby mitigating these effects which contribute to cardiovascular and renal disease progression, fluid retention, and systemic hypertension [1, 2]. Clinically effective RAAS suppression by ACE-inhibitors, however, is sometimes sub-optimal because of aldosterone breakthrough, non-ACE mediated angiotensin II formation, genetic variants affecting RAAS components, feedback mechanisms, and likely other unidentified factors [3,4,5,6,7]. The ACE gene intronic variant at canine chromosome 9:11507816:G > A has recently been shown to increase the magnitude of aldosterone breakthrough, despite adequate suppression of angiotensin II by ACE-inhibitors [4]. Genotype influence on non-angiotensin mediated aldosterone production holds clinical implications for dogs with advanced heart disease in both disease expression and response to treatments, but the prevalence of this variant in the canine population is unknown. This study sought to determine the prevalence of this ACE gene variant in a large number of dogs and to define breed predispositions. We hypothesized that this ACE gene variant would be common in dogs, but with variable breed distributions.
Results
The variant was present in 32 breeds and absent in 22 breeds in the 497 dogs screened (Table 1). Allele frequency of the chromosome 9:11507816:G > A variant within the population was 21%. Overall variant-positive prevalence within the study population was 31%, with 20% of dogs heterozygous, and 11% of dogs homozygous.
Twenty-one breeds were represented by ≥10 dogs and were evaluated for breed predisposition for the variant (Table 2). The variant was overrepresented in Irish Wolfhounds (prevalence 95%; P < .001), Dachshunds (prevalence 90%; P < .001), Cavalier King Charles Spaniels (prevalence 85%; P < .001), Great Danes (prevalence 84%; P < .001), and Bull Mastiffs (prevalence 58%; P = .02). The variant was underrepresented in Golden Retrievers (prevalence 10%; P = .001), Newfoundlands (prevalence (7%; P = .046), Rottweilers (prevalence 6%; P = .03), Boxers (prevalence 5%; P = .01), Miniature Schnauzers (prevalence 9%; P = .001), French Bulldogs (prevalence 0%; P = .001), and Pomeranians (prevalence 0%; P = .01). Of the 11 breeds with ≥5 variant-positive dogs, only Irish Wolfhounds were more likely to be homozygous than heterozygous (P < .001; Table 3).
Discussion
The ACE gene variant at chromosome 9:11507816:G > A was found in nearly one-third of dogs, with unequal breed distribution. The breeds with the highest prevalences include some that are predisposed to heart disease, which could have clinical importance if RAAS suppression is indicated for treatment [4]. Two of the breeds with high variant-positive prevalence (≥84%) are predisposed to dilated cardiomyopathy (Irish Wolfhound and Great Dane) and 2 other breeds with high variant-positive prevalence (≥85%) are predisposed to degenerative mitral valve disease (Cavalier King Charles Spaniel and Dachshund) [8,9,10,11]. Both of these cardiac diseases are commonly treated with ACE-inhibitors. Our study also showed that Irish Wolfhounds were more likely to be homozygous for the variant than heterozygous, but the clinical importance of this finding is unknown. Conversely, 7 other breeds were less likely to have the ACE gene variant and some of these breeds are also predisposed to cardiac disease (e.g. Boxer, Pomeranian, Miniature Schnauzer).
The efficacy of ACE-inhibitors in delaying disease progression and prolonging survival in dogs with both preclinical and clinical mitral valve disease is debated because published study results are contradictory [12,13,14,15]. The relatively high prevalence of a functionally important ACE gene variant in some breeds but not others and the effect of this variant on aldosterone breakthrough, could explain discordant findings depending on the enrolled population of dogs [4]. Future studies investigating the efficacy of ACE-inhibitors in dogs with naturally occurring disease should consider genotype in study planning.
This study has limitations that impact interpretation. Although the database was relatively large, the number of dogs in each breed varied and was low for some breeds. Therefore, breed-specific assessments may have been underpowered and findings could be different with a greater number of dogs. Additionally, we did not analyze for breed predisposition if the number of dogs in the database was < 10 for a breed, and not all breeds were represented in the database. Therefore, other breed predispositions may be present but not uncovered by this study. Although the database did not include large families, the degree of relatedness between dogs was not explored in this study. Specific geographic origins within North America were not available and so the potential for geographic genotypic differences in the ACE gene variant remains unexplored. It is also possible that variant-positive prevalence was linked to the disease for which the dogs were sequenced, and therefore prevalence within larger or different populations could be different.
Conclusions
Nearly one-third of dogs in this study were positive for a functionally important ACE gene variant, and for some breeds predisposed to naturally occurring heart disease, the prevalence was much higher. Genotyping for this variant may advance personalized canine medicine and permit targeted clinical trials in the future.
Methods
A database of 497 canine whole genome sequences from 54 breeds sequenced for various disease-specific studies from 10/1/2014 to 12/31/2020 was utilized to genotype dogs at chromosome 9:11507816. All sequenced dogs were from North America but the specific geographic origins were not available. The reasons for sequencing in this group of dogs were: cardiac (57.1%), neurologic (8.5%), renal (8.2%), immunologic (5.8%), musculoskeletal (6.2%), respiratory (2.6%), ophthalmologic (2.6%), healthy (3.4%), unspecified (1.8%), hepatobiliary (1.6%), metabolic (1.6%), and dermatologic (0.4%).
Samples were classified as wild type, heterozygous, or homozygous for the ACE gene variant at chromosome 9:11507816:G > A. Allele frequency and overall variant-positive prevalence (heterozygous and homozygous) within our canine study population were calculated.
Breed predisposition was evaluated for breeds with ≥10 dogs in the dataset by using Fisher’s exact test to compare the variant-positive prevalence of each breed to that of all the remaining dogs in the sample population. Effect size was determined by calculating the odds ratio for each breed (the odds of being variant-positive for each of these breeds was divided by the odds of being variant-positive for all the other dogs in the sample population except those of that particular breed). Breeds were considered overrepresented if the odds ratio (and 95% CI) of having the variant was > 1.0 and underrepresented if < 1.0.
Differences in homozygous and heterozygous status were evaluated for breeds with ≥5 variant-positive dogs using Fisher’s exact test to compare the prevalence of homozygosity for each of these breeds to the prevalence of homozygosity for variant-positive dogs from the other breeds combined. Breeds without variant-positive dogs (i.e. breeds with only wildtype dogs) were not used for this analysis. Effect size was determined by calculating the odds ratio for each breed (the odds of being homozygous for each of these breeds divided by the odds of being homozygous for the variant-positive dogs from all the other breeds combined. Homozygosity was considered overrepresented in a breed if the odds ratio (and 95% CI) of being homozygous was > 1.0 and underrepresented if < 1.0. Statistical analysis was performed using commercially available software (GraphPad Prism 8, San Diego CA, USA). Significance was set at P < .05.
Availability of data and materials
Datasets are presented within the manuscript. Data for individual dog genotyping are available from the corresponding author upon reasonable request.
Abbreviations
- ACE:
-
Angiotensin converting enzyme
- RAAS:
-
Renin-angiotensin-aldosterone system
References
Ames MK, Atkins CE, Pitt B. The renin-angiotensin-aldosterone system and its suppression. J Vet Intern Med. 2019;33(2):363–82.
Keene BW, Atkins CE, Bonagura JD, Fox PR, Fuentes VL, Uechi M, et al. ACVIM consensus guidelines for the diagnosis and treatment of myxomatous mitral valve disease in dogs. J Vet Intern Med. 2019;33(3);1–14.
Ames MK, Atkins CE, Eriksson A, Hess AM. Aldosterone breakthrough in dogs with naturally occurring myxomatous mitral valve disease. J Vet Cardiol. 2017;19:218–27.
Adin D, Atkins C, Domenig O, DeFrancesco T, Keene B, Tou S, et al. Renin-angiotensin aldosterone profile before and after angiotensin-converting enzyme-inhibitor administration in dogs with angiotensin-converting enzyme gene polymorphism. J Vet Intern Med. 2020;34(2):600.
Zakrzewski-Jakubiak M, De Denus S, Dubé MP, Bélanger F, White M, Turgeon J. Ten renin-angiotensin system-related gene polymorphisms in maximally treated Canadian Caucasian patients with heart failure. Br J Clin Pharmacol. 2008;65(5):742–51.
Uehara Y, Miura S, Yahiro E, Saku K. Non-ACE pathway-induced angiotensin II production. Curr Pharm Des. 2013;19:3054–9.
Lefebvre H, Duparc C, Naccache A, Lopez AG, Castanet M, Louiset E. Paracrine regulation of aldosterone secretion in physiological and pathophysiological conditions. Vitam Horm. 2019;109:303–39.
Vollmar C, Vollmar A, Keene BW, Fox PR, Reese S, Kohn B. Dilated cardiomyopathy in 151 Irish wolfhounds: characteristic clinical findings, life expectancy and causes of death. Vet J. 2019;245:15–21. https://doi.org/10.1016/j.tvjl.2018.12.018.
Oyama MA, Chittur SV, Reynolds CA. Decreased triadin and increased calstabin2 expression in great danes with dilated cardiomyopathy. J Vet Intern Med. 2009;23:1014–9.
Garncarz M, Parzeniecka-Jaworska M, Hulanicka M, Jank M, Szaluś-Jordanow O, Kurek A. Mitral regurgitation in dachshund dogs without heart murmurs. J Vet Res. 2017;61(3):363–6.
Markby GR, Macrae VE, Corcoran BM, Summers KM. Comparative transcriptomic profiling of myxomatous mitral valve disease in the cavalier king Charles spaniel. BMC Vet Res. 2020;16(1):1–14.
Kvart C, Häggström J, Pedersen HD, Hansson K, Eriksson A, Järvinen AK, et al. Efficacy of enalapril for prevention of congestive heart failure in dogs with myxomatous valve disease and asymptomatic mitral regurgitation. J Vet Intern Med. 2002;16(1):80–8.
Atkins CE, Keene BW, Brown WA, Coats JR, Crawford MA, DeFrancesco TC, et al. Results of the veterinary enalapril trial to prove reduction in onset of heart failure in dogs chronically treated with enalapril alone for compensated, naturally occurring mitral valve insufficiency. J Am Vet Med Assoc. 2007;231(7):1061–9.
Wess G, Kresken JG, Wendt R, Gaugele J, Killich M, Keller L, et al. Efficacy of adding ramipril (VAsotop) to the combination of furosemide (Lasix) and pimobendan (VEtmedin) in dogs with mitral valve degeneration: the VALVE trial. J Vet Intern Med. 2020;34(6):2232–41.
BENCH (BENazepril in Canine Heart disease) Study Group. The effect of benazepril on survival times and clinical signs of dogs with congestive heart failure: Results of a multicenter, prospective, randomized, double-blinded, placebo-controlled, long-term clinical trial. J Vet Cardiol. 1999;1(1):7–18 [cited 2016 Jun 14]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19081317.
Acknowledgements
Not applicable.
Funding
This work was supported by the Morris Animal Foundation (Grant #D14CA-810) and a Mark L. Morris Jr. Investigator Award, (D16CA509).
Author information
Authors and Affiliations
Contributions
Drs Meurs, Stern, and Friedenberg contributed cases to the genetic database. Dr. Adin conceived the study design and performed data compilation, analysis and primary manuscript preparation. Dr. Atkins provided data interpretation and subject expertise. All authors contributed intellectually to data interpretation and manuscript writing and editing. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Institutional Animal Care and Use Committees at North Carolina State University (14-142-0; 17-168-0), University of Minnesota (1907-37204A, 1907-37205A, 1606-33916A) and University of California, Davis (21857) approved sample collections from these dogs, and client consent was obtained at the time of sampling.
Consent for publication
Not applicable.
Competing interests
Dr. Adin has received funding from CEVA Animal Health and has provided consultation and sponsored lectures for Ceva Animal Health and Boehringer Ingelheim. Dr. Atkins has received funding from and has consulted for CEVA Animal Health, Boehringer-Ingelheim, and Vetoquinol. Drs. Meurs, Stern and Friedenberg declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Adin, D.B., Atkins, C.E., Friedenberg, S.G. et al. Prevalence of an angiotensin-converting enzyme gene variant in dogs. Canine Genet Epidemiol 8, 6 (2021). https://doi.org/10.1186/s40575-021-00105-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40575-021-00105-2