Abstract
Background
Although, in general, cancer is considered a multifactorial disease, clustering of particular cancers in pedigrees suggests a genetic predisposition and could explain why some dog breeds appear to have an increased risk of certain cancers. To our knowledge, there have been no published reports of whole genome sequencing to investigate inherited canine mammary tumor (CMT) risk, and with little known about CMT genetic susceptibility, we carried out whole genome sequencing on 14 purebred dogs diagnosed with mammary tumors from four breed-specific pedigrees. Following sequencing, each dog’s data was processed through a bioinformatics pipeline. This initial report highlights variants in orthologs of human breast cancer susceptibility genes.
Results
The overall whole genome and exome coverage averages were 26.0X and 25.6X, respectively, with 96.1% of the genome and 96.7% of the exome covered at least 10X. Of the average 7.9 million variants per dog, initial analyses involved surveying variants in orthologs of human breast cancer susceptibility genes, BRCA1, BRCA2, CDH1, PTEN, STK11, and TP53, and identified 19 unique coding variants that were validated through PCR and Sanger sequencing. Statistical analyses identified variants in BRCA2 and STK11 that appear to be associated with CMT, and breed-specific analyses revealed the breeds at the highest risk. Several additional BRCA2 variants showed trends toward significance, but have conflicting interpretations of pathogenicity, and correspond to variants of unknown significance in humans, which require further investigation. Variants in other genes were noted but did not appear to be associated with disease.
Conclusions
Whole genome sequencing proves to be an effective method to elucidate risk of CMT. Risk variants in orthologs of human breast cancer susceptibility genes have been identified. Ultimately, these whole genome sequencing efforts have provided a plethora of data that can also be assessed for novel discovery and have the potential to lead to breakthroughs in canine and human research through comparative analyses.
Similar content being viewed by others
Plain ENGLISH summary
Despite the advances in sequencing technology and the success of previous canine whole genome sequencing research, we know of no other publications that report using whole genome sequencing to investigate a genetic risk (aka. a risk that can be passed down through generations) for canine mammary tumors in purebred dogs. This canine cancer type is comparable to human breast cancer, and as a result, genes that are known to influence inherited risk for breast cancer were investigated to determine if those same genes played a role in risk for dogs. We whole genome sequenced 14 purebred dogs from four different breeds; each of the dogs within a breed had been tied back to the same family tree (pedigree). From this study, we have identified mutations in genes BRCA2 and STK11 that could increase risk for those dogs with the mutations. These mutations seem to be present in some breeds more than others, thus affecting risk differently. Furthermore, the large dataset from this research allows for further exploration to find additional mutations that influence their risk for developing canine mammary tumors.
Background
The practice of breeding dogs for specific characteristics and traits has resulted in over 190 phenotypically diverse breeds, according to the American Kennel Club [1]. Defined as selective breeding, this practice has cultivated breed-specific gene pools that not only contribute towards each breed’s defining features but also disease susceptibility. To date, over 450 canine genetic diseases have been reported, many of which are monogenic and limited to a specific dog breed(s) [2,3,4]. Efforts to understand the genetic causes of such diseases began in the 1980s with the first canine genetic mutation identified in 1989 for hemophilia B, an X-linked disorder [5]. Since then, investigating hereditary diseases that segregate in purebred lines/pedigrees have fostered numerous genetic discoveries; over 130 canine hereditary diseases are now genetically explained [2,3,4]. Through these discoveries, it has been determined that there is much genetic overlap between canine and human disease. Importantly, the elucidation of certain hereditary canine diseases has even led to breakthroughs in human medicine, with diseases such as sleep disorders, Birt-Hogg-Dubé syndrome, and more [3, 6,7,8,9].
Interestingly, despite the fact that some dog breeds appear to have an increased risk of certain cancer types, little is known about the etiology. Although, in general, cancer is considered a multifactorial disease, clustering of particular cancers in pedigrees suggests a genetic predisposition [10]. In humans, the study of cancer families has revealed genetic mutations that severely increase lifetime risk of developing particular cancers; for instance, high-risk mutations in BRCA1, BRCA2, CDH1, PTEN, STK11, and TP53 all result in hereditary cancer syndromes (such as hereditary breast cancer syndrome, Li Fraumeni syndrome and Cowden Syndrome) associated with an increased risk of breast cancer as well as other cancer types [11]. Therefore, breed or kennel/pedigree-based studies should be a beneficial approach to determine cancer genetic risk in dogs. This approach was successful in identifying the susceptibility locus for multifocal renal cystadenocarcinoma and nodular dermatofibrosis (RCND) in a German Shepherd pedigree [9]. RCND, an inherited cancer syndrome, is an autosomal dominantly inherited trait that is caused by a mutation in the Birt-Hogg-Dubé (BHD) gene, which is named after the equivalent human cancer syndrome [12,13,14]. Similar to how the BHD mutation in German Shepherds predisposes them to RCND, there are likely yet-to-be-discovered mutations that explain particular cancer incidences in other breeds.
With little known about canine mammary tumor (CMT) genetic susceptibility [15], we decided to carry out whole genome sequencing (WGS) on 14 purebred dogs diagnosed with CMT from four different breeds (Golden Retriever, Siberian Husky, Dalmatian, and Standard Schnauzer). The CMT-affected dogs from each breed were linked back to a common ancestor through pedigree analysis. Even though it is highly debated as to which dog breeds have the greatest CMT susceptibility or prevalence, we hypothesized that a cluster of CMT in these pedigrees is indicative of a genetic predisposition. Previous attempts to study CMT genetics either focused on small cohorts of multiple breeds or English Springer Spaniels (ESS) [15]. Multiple studies have indicated that the ESS from Sweden is a high-risk breed; however, it is worth noting that dogs in Sweden are rarely spayed - a procedure known to greatly reduce the risk of CMT [16,17,18]. Nevertheless, studying ESSs has revealed apparent CMT-associated SNVs, including ones in BRCA1 and BRCA2, but the causative alleles have yet to be identified [19,20,21]. To our knowledge, there have been no published reports of WGS to investigate CMT inherited-genetic risk. Furthermore, outside of ESS, there have been no breed-specific analyses. Considering that different WGS efforts in dogs have recently proven to be advantageous in elucidating genetic susceptibility to disease [22,23,24,25,26,27], differences in body types [28], as well as adaptions against parasites [29], we have compiled and processed WGS data to begin the exploration of breed-specific CMT-risk alleles and, in this initial report, to specifically reveal the coding variants detected in orthologs of the high-risk human breast cancer susceptibility genes.
Materials and methods
CMT sample acquisition
DNA samples from 82 purebred CMT-affected dogs, representing 32 different American Kennel Club (AKC) recognized breeds (Table 1), were obtained from the Canine Health Information Center (CHIC) DNA Repository, which is a part of the Orthopedic Foundation for Animals (OFA; https://www.ofa.org/about/dna-repository). Briefly, this repository stores canine DNA samples and corresponding genealogic and phenotypic information to facilitate genetics research. Dog owners submit either blood or buccal samples to the repository along with their pets’ health history. Ultimately, researchers request access to samples pertaining to a disease of interest along with any additional information submitted. An unfortunate limitation of this resource is the lack of collected data. Being reliant on the owner’s knowledge and willingness to share, along with a generic survey used for all collected samples/phenotypes, information such as CMT pathology/histology, age of onset, and spay/neuter status were not provided to the research team.
Of the 82 acquired samples, both blood-extracted DNA and buccal swabs were obtained. DNA was purified from the provided buccal swabs using the QIAamp DNA Mini Kit (Cat No./ID: 51304). Of the 32 represented breeds, 15 had multiple samples per breed (Table 1); thus, pedigree analyses were performed to identify breed-specific common ancestors and determine the level of relationship. Specifically, a dog’s registration and breeding information were entered into online (and mainly breed-specific) databases to build pedigrees. From this, 12 different pedigrees were generated.
Sequencing and bioinformatics
Fourteen DNA samples from four pedigrees were chosen for WGS. This included five Golden Retriever samples (three female, two male), three Siberian Husky females, three Standard Schnauzer females, and three Dalmatian females (Fig. 1). The selected dogs from each breed were AKC-registered and located within the same pedigree. Also, utilizing the CHIC database, offspring information of each dog was recorded to attempt to determine intact status as hormone exposure can affect the likelihood of development of CMT (Additional File 1).
Purebred dog pedigrees and selected samples for WGS. Offspring of WGS samples are not depicted here, see Additional File 1 for offspring information
Samples were prepared for Illumina platform WGS at HudsonAlpha Institute for Biotechnology’s Genome Sequencing Laboratory and the sequencing was carried out on Illumina HiSeq X. Paired FASTQ files were obtained from HudsonAlpha with sequencing data for each sample; the quality of the raw FASTQ files was determined using FASTQC. After assuring quality files, this sequencing data was carried through an in-house bioinformatics canine pipeline that was adapted from the Genome Analysis Toolkit (GATK) best practices bioinformatics pipeline [30] (Fig. 2). In brief, each sample file had Illumina adapters trimmed using the program Trimmomatic [31]. Samples were then aligned to the canine genome CanFam3.1 [32] using BWA mem [33]. Duplicate reads were marked using a Picard tool from version 1.79 (http://broadinstitute.github.io/picard/); then indels were realigned and base quality scores were recalibrated referencing the CanFam3.1 dbsnp data using Base Quality Score Recalibrator (BQSR) as part of the GATK v.3.4.46 [34]. Additionally, using GATK, coverage was calculated using the Depth of Coverage tool, and genomic variant calling format (GVCF) files were generated using Haplotype Caller and then merged through genotyping GVCF files. ANNOVAR [35] was used to annotate the VCF files using gene prediction from Ensembl build version 75. Variants were filtered by a Quality by Depth threshold of at least 12.
Coding variants within orthologs of human breast cancer susceptibility genes were isolated using the following coordinates: BRCA1 (ENSCAFT00000023190.4): chr9:19960910–20,024,390, BRCA2 (ENSCAFT00000010309.3): chr25:7734450–7,797,815, CDH1 (ENSCAFT00000032333.3): chr5:80759112–80,834,940, PTEN (ENSCAFT00000024821.3): chr26:37853135–37,913,097, STK11 (ENSCAFT00000031055.3): chr20:57556289–57,625,288, and TP53 (ENSCAFT00000026465.3): chr5:32560598–32,574,109. All coding variants identified through WGS were validated through PCR and Sanger sequencing. Once the variant list was finalized, protein sequences for the orthologous human genes (BRCA1 (NP_009231), BRCA2 (NP_000050), CDH1 (NP_004351), PTEN (NP_000305), STK11 (NP_000446), and TP53 (NP_000537)) were compared to the canine protein sequences (that corresponded to the above canine gene accession numbers) through EMBOSS Water alignment (https://www.ebi.ac.uk/Tools/psa/emboss_water/). These alignments were used to determine the corresponding human amino acid of each coding variant. The ClinVar database was then checked to see if a human mutation was identified in that position [36].
Controls
Control data were obtained through Ensembl by accessing each canine gene’s variant table [37], which reports population genetic information from the European Variation Archive (EVA; https://www.ebi.ac.uk/eva/?eva-study=PRJEB24066). EVA provides data from the “High quality variant calls from multiple dog genome project – Run 1” representing WGS data of over 200 dogs from multiple breeds. Variants were filtered based on GATK’s best practices filtering guidelines, and the resulting variants and corresponding frequencies are accessible on the web through Ensembl’s database. Exact breed and sex information of these control dogs was unknown. This EVA control dataset is similar to the use of publically available databases that present general population control data for human disease genetic studies [38,39,40,41,42].
Statistical analyses
For all the BRCA1, BRCA2, CDH1, PTEN, STK11, and TP53 coding variants validated in the 14 CMT cases, allele frequencies were calculated in both cases and controls. Major and minor alleles were defined based on EVA control data. Subsequently, the Fisher Exact test was carried out to determine any statistically significant allele frequency differences between the EVA controls and the overall CMT cohort, as well as each specific breed. The Fisher Exact test, a test of contingency tables that calculates statistical significance based on a probability scale, is typically used as a statistical test for allele frequency [38, 43, 44]. This statistical analysis method has been considered a solution for analysis with small cell counts, which is why this analysis method was chosen for our analyses [45]. P-values were calculated using Fisher Exact test in R (v 3.5.1), which were not adjusted for multiple testing.
Results
Sequencing and annotation
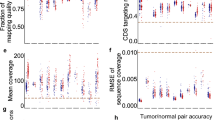
WGS of the 14 dogs yielded an average sequencing depth of 26.0X (Table 2). On average, 99.13% of the reads aligned to the reference, resulting in 99.7, 99.1, 96.1 and 75.6% of the genome being covered at least 1X, 5X, 10X, and 20X, respectively (Table 2). An average of 7,909,896 variants were called for each dog, the majority of which were non-coding, with an average of 40,965 coding variants per dog. The overall average sequencing depth of the exome, according to Ensembl build version 75, was 25.6X; 99.8, 99.4, 96.7, and 76.0% of the exome was covered at least 1X, 5X, 10X, and 20X, respectively (Additional File 2).
Variant analyses
A total of 19 coding variants, 13 nonsynonymous and six synonymous, were detected in BRCA1, BRCA2, CDH1, PTEN, STK11, and TP53 (Table 3; Additional File 3). The nonsynonymous variants included ten missense variants (only one of which was considered possibly damaging based on Polyphen analysis), two non-frameshifting deletions, and one non-frameshifting indel (Table 3; Additional File 3). Of the 19 total variants, 11 had been previously reported in CMT canine cohorts (Table 3). Three STK11 missense variants were identified (Table 3), one of which was detected in a single breed (Additional File 3). These three STK11 variants have yet to be reported, not only in CMT studies, but also in the EVA control dataset (Table 3). Consequently, they appear to be associated with an increased risk of CMT, and each variant may affect breeds differently (Tables 3 and 4). Additionally, significant P-values were generated for BRCA2 variants (Tables 3 and 4). Variants in other genes were noted but did not appear to be associated with disease.
Discussion
In an effort to study CMT heritability, our group acquired germline DNA from CMT-affected purebred dogs whose samples were submitted to the CHIC repository by the owner. Based on the hypothesis that dogs from the same breed/lineage share ancestral CMT-genetic risk factors, WGS was carried out on 14 samples from four generated pedigrees, including Golden Retriever, Siberian Husky, Standard Schnauzer, and Dalmatian. However, it is important to note that even if our hypothesis holds true in future studies that validate our findings or through novel CMT-gene discovery efforts, some cases within each pedigree could be phenocopies, representing sporadic cases not due to a familial genetic variant. This has to be kept in mind since ages of onset were not available through CHIC, and early ages of onset are associated with hereditary risk.
Our CMT-affected cohort represents dogs from the United States and did not include any ESS, which is the only breed to date that has had breed-specific CMT-genetic analyses [19,20,21]. To our knowledge, there have been no published reports of WGS to investigate the inherited risk of CMT. However, a compilation of next-generation sequencing efforts was used to compare human breast tumors to CMTs, and somatic mutations were identified [18]. Additionally, a limited number of studies have investigated germline CMT risk, and only a few risk variants have been identified with significance [15]. On our initial quest to find inherited breed-specific CMT-risk alleles, it is important to note that all CMT-affected dogs chosen for WGS were female except two closely related Golden Retrievers males. In addition to family history and early age of onset, male breast cancer is a hallmark of hereditary breast cancer in humans [11]; in fact, genetic predisposition significantly elevates the risk of male breast cancer, which is otherwise rare in the general population [49]. Therefore, assuming CMT genetic risk is similar to human genetic susceptibility, these two CMT-affected males suggest genetic factors are playing a role and were selected to enhance the prospects of discovery.
Unlike human disease gene discovery efforts, which have capitalized on whole-exome sequencing (WES) to facilitate discovery upon the introduction of next-generation sequencing [50], WGS has been the methodology of choice for identifying the genetic factors associated with inherited canine diseases. WGS and WES involve the re-sequencing of a genome or exome, respectively, which was made possible for canines once the first reference genome was published in 2005 [51]. In 2013, the first WGS [52,53,54] and WES [55] studies identified mutations associated with inherited canine disorders. Since that time, despite improvements to canine exome designs [56], the use of WES lagged behind. A possible reason for this is the cost. From our experience, when determining which of the two sequencing approaches to take for this study, the cost of WES was surprisingly expensive. WES baits alone were ~ $1000 per sample, which was the total cost per sample for WGS (to yield an average sequencing depth of at least 15X). Additional benefits to WGS include, (a) avoiding technical enrichment biases associated with WES capture, (b) more uniformity regarding sequencing-quality parameters, (c) the ability to explore both coding and non-coding regions, (d) the ability to better detect variants in coding regions (including regions targeted by a WES kit), and (e) the continued usefulness of the data as the annotation of the canine reference genome improves and gaps are filled [22, 32, 57,58,59].
Upon WGS of the 14 CMT-affected dogs, individual average sequencing depths ranged from 12.1 to 36.6X and overall averaged 26.0X. Aiming to achieve an average sequencing depth of, at least, 15X, all but one dog yielded such results (Table 2). Ultimately, the overall average was comparable to other canine WGS studies using Illumina technology [23,24,25,26]. On average, 99.7% of the genome was covered at least 1X, which is comparable to the Illumina-generated data in Gilliam et al. [23]. Noteworthy, it was higher than Sayyab et al. who used Ion Proton technology and reported an average sequencing depth of 9.2X and that 96% of the genome was covered at least 1X [22]. Viluma et al., who carried out another Ion Proton study, determined that 80% of the genome was covered at least 4X [60]; this is vastly different from our data, which covered 99.1% of the genome at 5X or greater. Similar to the two Ion Proton studies, our group also sought to determine the coverage of the canine exome through our WGS efforts. Not only did our study produce greater coverage for the canine genome, we additionally determined higher coverage results for the canine exome. Previously, Sayyab et al. reported that 91% of the exome was covered at least 1X, and Viluma et al. reported 77% of the exome was covered at least 4X. Contrarily, we obtained 99.8 and 99.4% of the exome at 1X and 5X, respectively (Additional File 2). In fact, these results far surpass the 5X coverage noted by Broeckx et al. regarding their improved canine exome design; they stated that just over 90% of the targeted base pairs were covered at least 5X [56]. Furthermore, Broeckx et al. had an average sequencing depth of 68.3X, which emphasizes the issue of lack of uniformity regarding targeted captures.
On average, each of the 14 dogs had 7.9 million variants called. Overall, this is comparable to the number of variants reported in the WGS studies that had similar sequencing depths [23,24,25,26]. The majority of the variants were non-coding, which, in the future, provides data for exploration. However, for this study, we focused on coding variants, specifically in orthologs of high-risk human breast cancer susceptibility genes, BRCA1, BRCA2, CDH1, PTEN, STK11, and TP53 [11], as an initial gene exclusion approach, acknowledging that this dataset will be subsequently analyzed to investigate risk in other coding and non-coding regions of the genome. Through our initial analysis, 19 different coding variants were identified through WGS and confirmed through PCR and Sanger sequencing (Table 3). Interestingly, this list of variants gave insight regarding the complications of next-generation sequencing in dogs. Using a reference sequence derived from a Boxer for the alignment and, similarly, gene transcripts derived from the latest genome assembly for the annotation, we noted instances when the data could have easily been misconstrued. Firstly, four BRCA2 variants were homozygous in all 14 CMT-affected dogs. This observation hinted that each alternate allele could in fact be the true wild-type (major) allele for the species since the four reference alleles appear to be unique to the Boxer. This was confirmed when we determined that all EVA control dogs were also homozygous for the four alternate alleles, as well as when we compared the Boxer reference protein sequence to the BRCA2 protein sequence for the Basenji (Basenji-breed-1.1) and the dingo dog (ASM325472v1). The reference genome is of an unaffected female Boxer, but that is the difficulty when studying a disease with age-related risk. These four BRCA2 variants, with alleles that appear to be extremely rare in the species according to the control data, need to be further investigated to determine if they contribute toward disease risk in the Boxer. Unfortunately, we did not sequence any Boxers in this study, but their assessment would require a careful analysis of controls to properly interpret the data, which stresses that analyzing controls from multiple breeds can have extreme benefits.
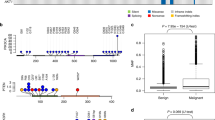
Similar to the example above, there were other instances where the alternate allele in the Boxer was in fact the major allele in controls. This was the case for two BRCA2 variants that appear to be associated with CMT risk, particularly in the Siberian Huskies. According to the Boxer reference sequence and annotation using transcript ID ENSCAFT00000010309.3, these two variants were named BRCA2:c.T1158G (p.C386W) and BRCA2:c.9995_9996insAAA (p.M3332delinsIK), which were previously reported in CMT heritability studies [46,47,48, 61]. Thus, the Boxer had a cysteine at amino acid 386 and a methionine at 3332. However, the major allele in the EVA control dogs translated to most dogs having a tryptophan at amino acid 386, and isoleucine-lysine at position 3332, which also resembles that of the references for Basenji dog breed and the dingo dog and, most interestingly, corresponds to the conserved human residues. Comparing allele frequencies between the CMT cases and EVA controls revealed that cysteine at amino acid 386 and a methionine at 3332 were associated with an increased risk of CMT. In fact, these alleles appear to be most strongly associated with CMT risk in Siberian Huskies (Table 4). These associations will need to be validated by studying larger cohorts. Boxers should also be studied to determine the true allele frequencies in that breed. If a cysteine at position 386 and a methionine at 3332 are actually more common in Boxers, they could be at an elevated disease risk. Noteworthy, the human BRCA2 residue W395 corresponds to W386 in these dogs (Fig. 3), and while a cysteine mutation at W395 has not been found in human hereditary breast cancer cases, two pathogenic truncation mutations have been reported at that position (ClinVar Variation IDs: 266612 and 265053), along with the missense variant, W395G, which is considered a variant of unknown significance (VUS; ClinVar Variation ID: 51078) [36]. Similarly, human BRCA2 residues I3312 and K3313 correspond to the conserved isoleucine-lysine in dogs at 3332 (Fig. 3), and BRCA2 p.I3312M has been reported as another VUS (ClinVar Variation ID: 52921) [36]. VUS are genetic variants of unknown clinical significance, and it has been reported that as many as 15% of people who undergo BRCA1 and BRCA2 genetic screening are informed of a detected VUS, which are inconclusive results [62].
In addition to BRCA2 c.T1158G (p.C386W), we identified three other BRCA2 missense variants that had been previously reported in CMT studies assessing inherited risk; this included BRCA2:c.A428G (p.H143R), BRCA2:c.A2401C (p.K801Q), and BRCA2:c.A4304G (p.K1435R; Table 3) [15, 46, 47]. Even though neither of these variants generated a significant P-value when investigating the overall CMT cohort, those P-values appeared to be trending towards significance. Nonetheless, breed-specific analyses suggested that BRCA2 p.H143R is associated with CMT-risk in Golden Retrievers (Table 4). This variant was previously described as possibly damaging by Borge et al. [46, 63], but PolyPhen2 analysis predicts it to be benign [64]. Similarly, contradictory pathogenicity predictions were noted for BRCA2 p.K801Q. It was predicted to be possibly damaging using PolyPhen2 but was initially reported by Borge et al. in 2011 and predicted to be benign [46, 63]. Moreover, the Polyphen2-suggested benign variant, p.K1435R, was reported by Yoshikawa et al. in 2008 as possibly deleterious upon blood and CMT analyses, including loss-of-heterozygosity studies [47]. Altogether, knowing that current computational prediction methods misclassify a significant percentage of clinically valid missense variants [65], and that the P-values generated for those variants were, at least, trending towards significance, larger genotyping and functional studies will be required for true classification. Additionally, all three missense variants are conserved in humans (Fig. 3), and, most interestingly, the equivalent mutations of canine p.H143R and p.K1435R have been identified in humans as p.H150R and p. K1440R, respectively (ClinVar Variation IDs: 51657 and 51632) [36]. These variants are classified as VUS, similar to the other BRCA2 VUS mentioned above. Overall, VUS include missense variants as well as in-frame insertions and deletions, both of which were detected in this study; this overlap with human and dogs offers another avenue for exploration since the reclassification of VUS is a current hot topic [66, 67].
Regarding the other assessed genes, STK11 displayed the most interesting results. Three missense variants were identified, STK11 c.C109T (p.P37S), STK11 c.A286G (p.M96V), and STK11 c.T293C (p.F98S), all of which appear to play a role in CMT risk. Our findings suggest that STK11 is a CMT susceptibility gene, corroborating a similar claim in a recent publication by Canadas et al. [68]. Canadas and colleagues suggested that the minor allele (T) of rs22928814, which lies within an intron of STK11, was associated with an increased risk of CMT. Interestingly, this allele, which the authors reported to have a frequency of 25.7 and 14.9% in cases and controls, respectively [68], has a frequency of 26.6% in EVA controls according to Ensembl [37], which is more similar to the frequency reported in the CMT cases and stresses the need for validation studies. Of note, this variant was not detected in any of the CMT-affected dogs sequenced in this study. However, the three missense variants identified in this study appear to be extremely rare alleles since they were not reported in EVA controls. Regarding STK11 p.M96V and p.F98S, breed-specific P-values of 0.0002 and 0.0148 were generated for the Siberian Huskies and Standard Schnauzers, respectively (Table 4). Additionally, STK11 p.P37S was only detected in one Dalmatian, and breed-specific analyses suggest that this variant possibly increases risk of CMT in that breed. Overall, these findings mimic the phenomena in humans that rare STK11 variants increase risk of disease [11]. However, it is worth noting that these variants are not in a conserved region with human STK11 protein sequence. How these STK11 variants, along with the identified BRCA2 variants, specifically contribute towards risk needs to be further studied. Firstly, variants in both STK11 and BRCA2 appear to be tightly linked, thus determining the true risk alleles in both BRCA2 and STK11 is important. Also, polygenic risk assessment in humans is another hot topic [69], and demonstrating the same concept in dogs would further validate their usefulness as a model of hereditary breast cancer [15].
Conclusions
To our knowledge, we carried out the first study to assess inherited CMT risk through WGS data analysis, and we investigated risk through both multiple breed and breed-specific analyses. This manuscript reports the variants detected in six orthologs of high-risk human breast cancer susceptibility genes as an initial gene exclusion approach, acknowledging that this WGS dataset will be subsequently analyzed to investigate risk in other coding and non-coding regions of the genome. Through our initial efforts, we identified variants in BRCA2 and STK11 that appear to be associated with CMT risk. These variants could alter risk in many breeds but appear to be more prevalent in some breeds compared to others. Additionally, we identified several BRCA2 variants that correspond to VUS in humans. Indeed, these results need to be validated; the identified variants now require further investigation to determine the role they play in risk in both humans and dogs, which we plan to promptly address. For instance, noting the limitation of using a control dataset of multiple unknown breeds, we plan to acquire control samples to determine breed-specific allele frequencies. Furthermore, functional studies are pertinent to determine pathogenicity. Ultimately, in addition to this initial gene exclusion effort, this dataset provides the opportunity for novel discovery and has the potential to lead to further breakthroughs in canine and human breast cancer research through comparative analyses. Overall, in the era of personalized medicine, identifying risk variants not only provides better risk assessment and opportunities to selectively breed out a pathogenic mutation, it also can provide insight towards disease mechanism and aid in the development of targeted therapies [10, 70].
Availability of data and materials
The dataset supporting the conclusions of this article is available upon request through communication with the corresponding author. The DNA samples that were sequenced during this project are available through the CHIC repository (https://www.ofa.org/about/dna-repository). Sample selection can be shared through communication with the corresponding author.
Abbreviations
- RCND:
-
Renal cystadenocarcinoma and nodular dermatofibrosis
- BHD :
-
Birt-Hogg-Dubé
- CMT:
-
Canine mammary tumor
- WGS:
-
Whole genome sequencing
- ESS:
-
English Springer Spaniels
- AKC:
-
American Kennel Club
- CHIC:
-
Canine Health Information Center
- OFA:
-
Orthopedic Foundation for Animals
- GATK:
-
Genome Analysis Toolkit
- GVCF:
-
Genomic variant calling format
- EVA:
-
European Variation Archive
- SNV:
-
Single nucleotide variant
- WES:
-
Whole exome sequencing
- VUS:
-
Variants of unknown significance
References
American Kennel Club. Breeds by Year Recognized [Available from: https://www.akc.org/press-center/articles/breeds-by-year-recognized/. Accessed May 2019.
Parker HG, Shearin AL, Ostrander EA. Man's best friend becomes biology's best in show: genome analyses in the domestic dog. Annu Rev Genet. 2010;44:309–36.
Ostrander EA, Franklin H. Epstein lecture. Both ends of the leash--the human links to good dogs with bad genes. N Engl J Med. 2012;367(7):636–46.
Switonski M. Dog as a model in studies on human hereditary diseases and their gene therapy. Reprod Biol. 2014;14(1):44–50.
Evans JP, Brinkhous KM, Brayer GD, Reisner HM, High KA. Canine hemophilia B resulting from a point mutation with unusual consequences. Proc Natl Acad Sci U S A. 1989;86(24):10095–9.
Grall A, Guaguere E, Planchais S, Grond S, Bourrat E, Hausser I, et al. PNPLA1 mutations cause autosomal recessive congenital ichthyosis in golden retriever dogs and humans. Nat Genet. 2012;44(2):140–7.
Lin L, Faraco J, Li R, Kadotani H, Rogers W, Lin X, et al. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell. 1999;98(3):365–76.
van De Sluis B, Rothuizen J, Pearson PL, van Oost BA, Wijmenga C. Identification of a new copper metabolism gene by positional cloning in a purebred dog population. Hum Mol Genet. 2002;11(2):165–73.
Jonasdottir TJ, Mellersh CS, Moe L, Heggebo R, Gamlem H, Ostrander EA, et al. Genetic mapping of a naturally occurring hereditary renal cancer syndrome in dogs. Proc Natl Acad Sci U S A. 2000;97(8):4132–7.
Dobson JM. Breed-predispositions to cancer in pedigree dogs. ISRN Vet Sci. 2013;2013:941275.
Chandler MR, Bilgili EP, Merner ND. A review of whole-exome sequencing efforts toward hereditary breast Cancer susceptibility gene discovery. Hum Mutat. 2016;37(9):835–46.
Schmidt LS, Warren MB, Nickerson ML, Weirich G, Matrosova V, Toro JR, et al. Birt-Hogg-Dube syndrome, a genodermatosis associated with spontaneous pneumothorax and kidney neoplasia, maps to chromosome 17p11.2. Am J Hum Genet. 2001;69(4):876–82.
Nickerson ML, Warren MB, Toro JR, Matrosova V, Glenn G, Turner ML, et al. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer Cell. 2002;2(2):157–64.
Lingaas F, Comstock KE, Kirkness EF, Sorensen A, Aarskaug T, Hitte C, et al. A mutation in the canine BHD gene is associated with hereditary multifocal renal cystadenocarcinoma and nodular dermatofibrosis in the German shepherd dog. Hum Mol Genet. 2003;12(23):3043–53.
Goebel K, Merner ND. A monograph proposing the use of canine mammary tumours as a model for the study of hereditary breast cancer susceptibility genes in humans. Vet Med Sci. 2017;3(2):51–62.
Egenvall A, Bonnett BN, Ohagen P, Olson P, Hedhammar A, von Euler H. Incidence of and survival after mammary tumors in a population of over 80,000 insured female dogs in Sweden from 1995 to 2002. Prev Vet Med. 2005;69(1–2):109–27.
Jitpean S, Hagman R, Strom Holst B, Hoglund OV, Pettersson A, Egenvall A. Breed variations in the incidence of pyometra and mammary tumours in Swedish dogs. Reprod Domest Anim. 2012;47(Suppl 6):347–50.
Liu D, Xiong H, Ellis AE, Northrup NC, Rodriguez CO Jr, O'Regan RM, et al. Molecular homology and difference between spontaneous canine mammary cancer and human breast cancer. Cancer Res. 2014;74(18):5045–56.
Rivera P, Melin M, Biagi T, Fall T, Haggstrom J, Lindblad-Toh K, et al. Mammary tumor development in dogs is associated with BRCA1 and BRCA2. Cancer Res. 2009;69(22):8770–4.
Borge KS, Melin M, Rivera P, Thoresen SI, Webster MT, von Euler H, et al. The ESR1 gene is associated with risk for canine mammary tumours. BMC Vet Res. 2013;9:69.
Melin M, Rivera P, Arendt M, Elvers I, Muren E, Gustafson U, et al. Genome-wide analysis identifies germ-line risk factors associated with canine mammary Tumours. PLoS Genet. 2016;12(5):e1006029.
Sayyab S, Viluma A, Bergvall K, Brunberg E, Jagannathan V, Leeb T, et al. Whole-Genome Sequencing of a Canine Family Trio Reveals a FAM83G Variant Associated with Hereditary Footpad Hyperkeratosis. G3 (Bethesda). 2016;6(3):521–7.
Gilliam D, O'Brien DP, Coates JR, Johnson GS, Johnson GC, Mhlanga-Mutangadura T, et al. A homozygous KCNJ10 mutation in Jack Russell terriers and related breeds with spinocerebellar ataxia with myokymia, seizures, or both. J Vet Intern Med. 2014;28(3):871–7.
Guo J, Johnson GS, Brown HA, Provencher ML, da Costa RC, Mhlanga-Mutangadura T, et al. A CLN8 nonsense mutation in the whole genome sequence of a mixed breed dog with neuronal ceroid lipofuscinosis and Australian shepherd ancestry. Mol Genet Metab. 2014;112(4):302–9.
Kolicheski AL, Johnson GS, Mhlanga-Mutangadura T, Taylor JF, Schnabel RD, Kinoshita T, et al. A homozygous PIGN missense mutation in soft-coated wheaten terriers with a canine paroxysmal dyskinesia. Neurogenetics. 2017;18(1):39–47.
Fyfe JC, Hemker SL, Frampton A, Raj K, Nagy PL, Gibbon KJ, et al. Inherited selective cobalamin malabsorption in Komondor dogs associated with a CUBN splice site variant. BMC Vet Res. 2018;14(1):418.
Meurs KM, Friedenberg SG, Kolb J, Saripalli C, Tonino P, Woodruff K, et al. A missense variant in the titin gene in Doberman pinscher dogs with familial dilated cardiomyopathy and sudden cardiac death. Hum Genet. 2019;138(5):515–24.
Plassais J, Rimbault M, Williams FJ, Davis BW, Schoenebeck JJ, Ostrander EA. Analysis of large versus small dogs reveals three genes on the canine X chromosome associated with body weight, muscling and back fat thickness. PLoS Genet. 2017;13(3):e1006661.
Liu YH, Wang L, Xu T, Guo X, Li Y, Yin TT, et al. Whole-genome sequencing of African dogs provides insights into adaptations against tropical parasites. Mol Biol Evol. 2018;35(2):287–98.
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11 0 1–33.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20.
Hoeppner MP, Lundquist A, Pirun M, Meadows JR, Zamani N, Johnson J, et al. An improved canine genome and a comprehensive catalogue of coding genes and non-coding transcripts. PLoS One. 2014;9(3):e91172.
Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25(14):1754–60.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164.
Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–8.
Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, et al. Ensembl 2018. Nucleic Acids Res. 2018;46(D1):D754–D61.
Lemke JR, Lal D, Reinthaler EM, Steiner I, Nothnagel M, Alber M, et al. Mutations in GRIN2A cause idiopathic focal epilepsy with rolandic spikes. Nat Genet. 2013;45(9):1067–72.
EVS. NHLBI GO Exome Sequencing Project (ESP). Seattle: WA; 2019.
Guo MH, Plummer L, Chan YM, Hirschhorn JN, Lippincott MF. Burden testing of rare variants identified through exome sequencing via publicly available control data. Am J Hum Genet. 2018;103(4):522–34.
Smith BN, Ticozzi N, Fallini C, Gkazi AS, Topp S, Kenna KP, et al. Exome-wide rare variant analysis identifies TUBA4A mutations associated with familial ALS. Neuron. 2014;84(2):324–31.
Olesen MS, Andreasen L, Jabbari J, Refsgaard L, Haunso S, Olesen SP, et al. Very early-onset lone atrial fibrillation patients have a high prevalence of rare variants in genes previously associated with atrial fibrillation. Heart Rhythm. 2014;11(2):246–51.
Ryman N, Jorde PE. Statistical power when testing for genetic differentiation. Mol Ecol. 2001;10(10):2361–73.
Fisher's Exact Test for Single Variant Analysis [Available from: http://varianttools.sourceforge.net/Association/ExactTest. Accessed May 2020.
Kim HY. Statistical notes for clinical researchers: sample size calculation 2. Comparison of two independent proportions. Restor Dent Endod. 2016;41(2):154–6.
Borge KS, Borresen-Dale AL, Lingaas F. Identification of genetic variation in 11 candidate genes of canine mammary tumour. Vet Comp Oncol. 2011;9(4):241–50.
Yoshikawa Y, Morimatsu M, Ochiai K, Nagano M, Tomioka Y, Sasaki N, et al. Novel variations and loss of heterozygosity of BRCA2 identified in a dog with mammary tumors. Am J Vet Res. 2008;69(10):1323–8.
Yoshikawa Y, Morimatsu M, Ochiai K, Nagano M, Yamane Y, Tomizawa N, et al. Insertion/deletion polymorphism in the BRCA2 nuclear localization signal. Biomed Res. 2005;26(3):109–16.
Pritzlaff M, Summerour P, McFarland R, Li S, Reineke P, Dolinsky JS, et al. Male breast cancer in a multi-gene panel testing cohort: insights and unexpected results. Breast Cancer Res Treat. 2017;161(3):575–86.
Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12(11):745–55.
Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438(7069):803–19.
Frischknecht M, Niehof-Oellers H, Jagannathan V, Owczarek-Lipska M, Drogemuller C, Dietschi E, et al. A COL11A2 mutation in Labrador retrievers with mild disproportionate dwarfism. PLoS One. 2013;8(3):e60149.
Jagannathan V, Bannoehr J, Plattet P, Hauswirth R, Drogemuller C, Drogemuller M, et al. A mutation in the SUV39H2 gene in Labrador retrievers with hereditary nasal parakeratosis (HNPK) provides insights into the epigenetics of keratinocyte differentiation. PLoS Genet. 2013;9(10):e1003848.
Owczarek-Lipska M, Jagannathan V, Drogemuller C, Lutz S, Glanemann B, Leeb T, et al. A frameshift mutation in the cubilin gene (CUBN) in border collies with Imerslund-Grasbeck syndrome (selective cobalamin malabsorption). PLoS One. 2013;8(4):e61144.
Ahonen SJ, Arumilli M, Lohi H. A CNGB1 frameshift mutation in Papillon and Phalene dogs with progressive retinal atrophy. PLoS One. 2013;8(8):e72122.
Broeckx BJ, Hitte C, Coopman F, Verhoeven GE, De Keulenaer S, De Meester E, et al. Improved canine exome designs, featuring ncRNAs and increased coverage of protein coding genes. Sci Rep. 2015;5:12810.
Belkadi A, Bolze A, Itan Y, Cobat A, Vincent QB, Antipenko A, et al. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc Natl Acad Sci U S A. 2015;112(17):5473–8.
van Steenbeek FG, Hytonen MK, Leegwater PA, Lohi H. The canine era: the rise of a biomedical model. Anim Genet. 2016;47(5):519–27.
Forman OP, Hitti RJ, Boursnell M, Miyadera K, Sargan D, Mellersh C. Canine genome assembly correction facilitates identification of a MAP 9 deletion as a potential age of onset modifier for RPGRIP1-associated canine retinal degeneration. Mamm Genome. 2016;27(5–6):237–45.
Viluma A, Sayyab S, Mikko S, Andersson G, Bergstrom TF. Evaluation of whole-genome sequencing of four Chinese crested dogs for variant detection using the ion proton system. Canine Genet Epidemiol. 2015;2:16.
Enginler SO, Ates A, Diren Sigirci B, Sontas BH, Sonmez K, Karacam E, et al. Measurement of C-reactive protein and prostaglandin F2alpha metabolite concentrations in differentiation of canine pyometra and cystic endometrial hyperplasia/mucometra. Reprod Domest Anim. 2014;49(4):641–7.
Murray ML, Cerrato F, Bennett RL, Jarvik GP. Follow-up of carriers of BRCA1 and BRCA2 variants of unknown significance: variant reclassification and surgical decisions. Genet Med. 2011;13(12):998–1005.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9.
Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013; Chapter 7:Unit7 20. https://doi.org/10.1002/0471142905.hg0720s76.
Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32(4):358–68.
Wu K, Hinson SR, Ohashi A, Farrugia D, Wendt P, Tavtigian SV, et al. Functional evaluation and cancer risk assessment of BRCA2 unclassified variants. Cancer Res. 2005;65(2):417–26.
So MK, Jeong TD, Lim W, Moon BI, Paik NS, Kim SC, et al. Reinterpretation of BRCA1 and BRCA2 variants of uncertain significance in patients with hereditary breast/ovarian cancer using the ACMG/AMP 2015 guidelines. Breast Cancer. 2019;26(4):510–9. https://doi.org/10.1007/s12282-019-00951-w. Epub 2019 Feb 6.
Canadas A, Santos M, Nogueira A, Assis J, Gomes M, Lemos C, et al. Canine mammary tumor risk is associated with polymorphisms in RAD51 and STK11 genes. J Vet Diagn Investig. 2018;30(5):733–8.
Mavaddat N, Michailidou K, Dennis J, Lush M, Fachal L, Lee A, et al. Polygenic risk scores for prediction of breast Cancer and breast Cancer subtypes. Am J Hum Genet. 2019;104(1):21–34.
Godet I, Gilkes DM. BRCA1 and BRCA2 mutations and treatment strategies for breast cancer. Integr Cancer Sci Ther. 2017;4(1). https://doi.org/10.15761/ICST.1000228. Epub 2017 Feb 27.
Acknowledgements
Firstly, we would like to thank the Orthopedic Foundation for Animals for establishing the CHIC DNA Repository, which facilitated these research efforts. We would also like to acknowledge the Office of Information Technology at Auburn University for support and compute time on the Hopper High-Performance Computing Cluster.
Funding
This research was supported by an Auburn University Research Initiative In Cancer (AURIC) Seed Grant for the canine WGS efforts. This research was also supported by the AURIC Graduate Fellowship Program (to A.L.W.H.), the Auburn University Cellular and Molecular Biosciences (CMB) Peaks of Excellence Research Fellowship (to A.L.W.H.; NSF-EPS-1158862, grant G00006750), Auburn University Merial Veterinary Scholars Research Program (to K.G. and C.L-F.), the Department of Drug Discovery and Development in the Auburn University Harrison School of Pharmacy, and the Department of Pathobiology in the Auburn University College of Veterinary Medicine.
Author information
Authors and Affiliations
Contributions
ALWH and NDM wrote the manuscript. ALWH, KG, and CLF performed pedigree analysis. KG extracted DNA from buccal swabs. ALWH performed bioinformatic processing. ALWH and IM validated variants through PCR and Sanger sequencing. ALWH and NDM performed variant and statistical analyses. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not Applicable.
Consent for publication
Not Applicable.
Competing interests
The authors disclose no potential conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
Table summary of offspring information from CHIC repository for the 14 WGS samples.
Additional file 2.
Table of exome Coverage Summary for the 14 canines sequenced. Exome regions according to Ensembl build version 75 for CanFam3.1.
Additional file 3.
Detailed summary of canine coding variants found within orthologs of human breast cancer susceptibility genes.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Huskey, A.L.W., Goebel, K., Lloveras-Fuentes, C. et al. Whole genome sequencing for the investigation of canine mammary tumor inheritance - an initial assessment of high-risk breast cancer genes reveal BRCA2 and STK11 variants potentially associated with risk in purebred dogs. Canine Genet Epidemiol 7, 8 (2020). https://doi.org/10.1186/s40575-020-00084-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40575-020-00084-w