Abstract
Background
Within multi-male and multi-female mammalian societies, paternity assignment is crucial for evaluating male reproductive success, dominance hierarchy, and inbreeding avoidance. It is, however, difficult to determine paternity because of female promiscuity during reproduction. Noninvasive molecular techniques (e.g., fecal DNA) make it possible to match the genetic father to his offspring. In the current study, a troop of free-ranging Taihangshan macaques (Macaca mulatta tcheliensis) in Mt. Taihangshan area, Jiyuan, China, was selected for studying the paternity. We successfully screened a set of microsatellite loci from fecal DNA and evaluated the efficiency of these loci for paternity testing using clearly recorded data of maternity.
Results
The results showed that: 1) ten loci out of 18 candidate microsatellite loci were amplified successfully in the fecal samples of Taihangshan macaques. The error probability in maternity assignments and paternity testing was very low as indicated by their power of discrimination (0.70 to 0.95), power of exclusion (0.43 to 0.84), and the values of polymorphic information content ranging from 0.52 to 0.82; 2) the combined probability of exclusion in paternity testing for ten qualified loci was as high as 99.999%, and the combined probability of exclusion reached 99.99% when the seven most polymorphic loci were adopted; 3) the offspring were assigned to their biological mother correctly and also matched with their genetic father.
Conclusions
We concluded that the ten polymorphic microsatellite loci, especially a core set of seven most polymorphic loci, provided an effective and reliable tool for noninvasive paternity testing in free-ranging rhesus macaques.
Similar content being viewed by others
Background
Within multi-male and multi-female mammalian societies, maternal rather than paternal care plays a crucial role in offspring development (Rosenblatt 2003). Although rare, paternal care (e.g., male-infant affiliation and the selective support of offspring in agonistic interactions) in many cercopithecine species have been documented (Buchan et al. 2003, Fernandez-Duque et al. 2009, Langos et al. 2013). Multiple-mating by promiscuous females, however, masks paternity certainty (Soulsbury 2010), which consequently could obscure the true paternal care (Buchan et al. 2003, Widdig 2007). Paternal care is expected to result in fitness benefits for sires (Buchan et al. 2003), and known paternity is thus critical for evaluating male reproduction success in primates with a promiscuous mating system (Smith 1993, Berard et al. 1994, Bercovitch 1997). Therefore, paternity identification serves as the cornerstone of understanding male reproductive ecology. Moreover, paternity testing could clarify many important issues in the field of male reproductive ecology, including male reproductive skew (Pereira et al. 2000) and the effect of dominance hierarchy on male reproductive success (Bergman et al. 2008). However, it is debatable whether the relationship between male non-human primates and their genetic offspring and unrelated individuals can be determined through epigenetic and/or transgenerational effects (Widdig 2007, Fernandez-Duque et al. 2009). As female mating promiscuously leads to paternity confusion, it is hard for researchers to determine the consanguinity between the father and his offspring through morphological and/or behavioral clues in the natural environment (Inoue et al. 1993). Fortunately, advancing molecular techniques (e.g., noninvasive genotyping) help to overcome many limitations in the field and broaden the scope of studies of free-ranging non-human primates, including paternity determination (Woodruff 1993, Constable et al. 2001, Taberlet et al. 1999).
With the development of non-invasive sampling techniques, DNA sources such as fecal DNA samples have been used in examining paternity (Constable et al. 2001). Reports of primate studies utilizing feces have become more and more prevalent in recent years (Buchan et al. 2003, Liu et al. 2008, Bergman et al. 2008, Langos et al. 2013, Liu et al. 2013). Among various molecular markers, microsatellites have been demonstrated to be ideal for analyses of genotyping, parentage, and pedigree (Estoup et al. 1998, Constable et al. 2001, Balloux and Lugon-Moulin 2002, Luna-González et al. 2012, Charpentier et al. 2012, Yang et al. 2014). A growing body of studies has examined the validity of microsatellite in parentage assessment (Vigilant et al. 2001, Reis et al. 2008, Luna-González et al. 2012, Wojtas et al. 2013) and pedigree analysis (Sefc et al. 1998, Wang et al. 2009, Li et al. 2010). Given a sufficient number of alleles, sound results for individual identification and paternity testing could be obtained with relatively small number of microsatellite loci (less than seven) (Bernatchez and Duchesne 2000). For instance, using only five microsatellite loci, the success rates of parentage identification in Plateau pike (Ochotona curzoniae) reached over 99.99% (Li et al. 2010). In captive rhesus macaques (Macaca mulatta), the probability of paternity assignment success was 99.53% when six markers were adopted (Xu et al. 2013). Along with the number of microsatellite loci, the validity of the statistics analyses also relies on the polymorphism of loci and sample size (Cooper et al. 1997). Highly polymorphic loci and fresh samples are crucial for reducing the ratio of genotype mismatch because fecal samples generally provide a low DNA concentration, and these DNA could be highly degraded due to numerous inhibitive factors (Constable et al. 2001, Liu et al. 2008). In rhesus macaques, many studies on identifying polymorphism of microsatellite loci have been reported. However, none of the markers were assured to be highly polymorphic in all rhesus macaque populations including subspecies distributed in various regions (Xu et al. 2013). For instance, different patterns of allele frequency distribution for some loci (e.g., D6S291, D6S276, and D6S2741) were documented between Indian- and Chinese-origin rhesus macaques (Penedo et al. 2005).
Taihangshan macaques, M. mulatta tcheliensis, are currently confined to the southern part of Mt. Taihangshan and considered as the northernmost natural population of rhesus macaques in the world (Zhang et al. 1989, Lu et al. 2007). Although studies on reproductive biology (Song and Qu 1996, Hou et al. 1998, Tian et al. 2013), behavioral ethogram and social structure (Tian et al. 2011a, 2011b) of Taihangshan macaques have been carried out, little is known about the variation in reproductive contribution among adult males in a given group. In the present study, by using noninvasive genotyping with fecal DNA, we aimed to: 1) screen for highly polymorphic and reliable microsatellite markers, 2) evaluate the effectiveness of these microsatellite markers through matching the maternity assignment to observed maternity, and then 3) determine the paternity of offspring.
Methods
Target troop and sampling
A free-ranging troop of Taihangshan macaque (Troop Wangwu 1 (WW-1)), occurring in Taihangshan Macaque National Nature Reserve (TMNNR) (35°05′ to 35°42′ N, 112°02′ to 112°45′ E) in Mt. Taihangshan area, Jiyuan, China, was assigned as our target troop. During the study period, there were six adult males (HB, BB, ZM, PZ, XBL, and SQ) and three matrilineal units (HL unit, CM unit, and BD unit; Figure 1), and HB held the highest dominance rank in WW-1 (Tian et al. 2011b). We determined mother-offspring relationship by identifying infant-to-mother attachment (e.g., infant breastfed by adult female). The majority of parental cares for infants within troop WW-1 were performed by mothers. During mating season (mainly from October to December), (adult) males (single or ‘bachelor troop’) from outside of WW-1 but surrounding the troop were commonly observed, and the adult males of them occasionally were observed to mate with adult females within troop WW-1.
During March 2010 and June 2011, fresh feces were collected from individually identified macaques shortly after defecation (in general, less than 5 min), which including adult males, adult females, and offspring born in 2009 (CX, JS, GX, XXC, LM, CL, and TT) and 2010 (ZY). The outer layer of the feces were harvested and then shifted into 50-mL conical tubes, one in each, filled with 95% ethanol (sample: ethanol = 1:3). The samples were kept at −20°C for further testing. In total, 136 fecal samples were collected from six adult males (mean ± SE, 6.0 ± 1.0 samples per individual), eight adult females (mean ± SE, 8.0 ± 1.5 samples per individual), and eight offspring (mean ± SE, 4.5 ± 0.5 samples per individual). The maternity within WW-1, serving as the ruler of evaluating the effectiveness of screened microsatellite markers (see results), were identified (e.g., infant breastfed by adult female) and recorded based on long-term field observation.
DNA extractions
QIAamp® DNA Stool Kit (Qiagen, GmbH, Germany) was employed in extracting DNA from fecal samples following the manual with some modifications. To remove impurities dissolved in water and ethanol, and homogenize cells in samples, fecal samples were firstly centrifuged at 10,000 g, 4°C for 10 min, and the supernatant was abandoned. The residual samples were centrifuged with 97% ethanol and sterile water (sample: liquid = 1:3) three times at 10,000 g (4°C for 15 min each time), respectively, and the supernatants were abandoned each time. Two negative extractions (no sample) controls were processed along with each set of eight samples (in total 17 sets). All extracted fecal DNA per sample was quantified via spectrophotometry (DNA concentration ranging from 0 to 12.30 ng/μL), while samples with less than 5.00 ng/μL were not adopted because they rarely worked (Constable et al. 2001).
Microsatellite loci screening and sequence analysis
In total, 18 microsatellite markers (D12S372, D9S934, D16S403, D1S548, D3S1768, D5S820, D6S311, D5S1457, D6S2741, D14S306, D3S3045, D21S1246, D6S2419, D7S513, D4S1645, D15S644, D12S67, and D18S536) were chosen as the candidate loci based on previous genetic studies of macaques, which amplified these loci from blood or tissue samples (Kanthaswamy and Smith 1998, Kanthaswamy et al. 2006, Li et al. 2009) (Table 1).
PCR amplifications were carried out in 20 μL reaction mixtures, consisted of 1 μL template DNA, 50 mM KCl, 20 mM Tris-HCl (pH 8.3), 4.0 mM MgCl2, 0.4 mM of dNTP, 0.3 μL of each forward and reverse primer (10 μM), and 0.5 U of Hotstart Taq DNA polymerase (Qiagen GMBH, Germany). Amplification conditions were as follows. Initial denaturing of 95°C for 5 min, followed by 45 cycles of 1 min at 95°C, 55°C to 60°C for 30 s, and 72°C for 1 min followed by a final 7 min extension at 72°C (see Table 2 for the optimum annealing temperature). Two negative PCR controls were employed in each set of PCRs. We repeated the amplification for two to three times.
The 5'-end of the forward primer was fluorescently labeled, and successfully amplified products were added to 3.0-μL methanamid and run in an ABI PRISM 3730 Genetic Analyser (Applied Biosystems Inc., Foster City, CA, USA). The analysis of allele sizes against the internal size standard and genotyped individuals was performed using GeneScan™-500LIZ™ Size Standard and GeneMapper™ V4.0 software.
Data analysis
Paternity analysis was mainly performed with CERVUS program version 3.0, which was also used to estimate allele frequencies, observed and expected heterozygosities (Marshall et al. 1998). This program calculates the paternity inference likelihood ratio and generates a statistic variable, delta (Δ), defined as the difference in positive log likelihood ratios (LODs) between the top two candidate fathers. If only one candidate father with a positive LOD score exists, his delta score equals his LOD score. CERVUS uses a simulation based on the observed allele frequencies, which takes typing error rates and incomplete sampling into account, to determine the statistical significance of the delta value generated for each paternity. CERVUS simulation parameters include number of offspring = 10,000, number of candidate fathers = 3, prop. sample = 1, proportion of loci typed depends on allele frequency analysis, rate of typing error = 0.01, relaxed confidence level = 80%, strict confidence level = 95%.
Moreover, following the instructions of programs, numeric errors and null alleles at 95% confidence interval were checked by the allelic size data using Micro-Checker (van Oosterhout et al. 2004). GENEPOP version 3.4 was adopted to test the Hardy-Weinberg equilibrium (Raymond and Rousset 1995). Power of exclusion (PE) and combined power of exclusion (CPE) of the ten loci were calculated from allele frequencies using PowerStats V1.2 (Promega Corporation, Madison, WI, USA). Detailed information is listed within Table 3.
Ethics statement
The present study was conducted with noninvasive method which was aimed to reduce the anthropogenic impact. Rare direct contact between the observers and the study subjects occurred, though the observers were able to view the macaques from a distance as short as 0.5 m. All protocols for the field work portion of this study complied with protocols approved by the Life Sciences Ethics Committee of Zhengzhou University and adhered to the legal requirements for animal research in China and the American Society of Primatologists principles for the ethical treatment of primates.
Results
Variation of microsatellites
In this study, we extracted DNA samples from fresh feces of 22 individuals belonging to a free-ranging rhesus macaque population. Within 136 fecal samples, 78 of them (DNA concentration >5.00 ng/μL) were adopted (mean ± SE, 3.0 ± 0.5 samples per individual). We tried to amplify 18 candidate microsatellite loci compiled from previous publications on captive macaques but derived from various samples (e.g., blood and tissues). Ten out of these 18 candidate microsatellite loci were successfully amplified, while the remaining 8 loci were discarded because of their poor amplification. Within this set of ten loci, the average success rate of amplification with fecal DNA of Taihangshan macaques was 64.20% (763 attempts) and the success rate ranged from 35.63% for locus D16S403 (31/87 attempts) to 85.71% (84/98 attempts) for locus D5S820 (Table 2).
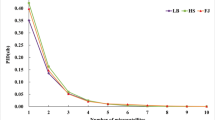
For ten microsatellite loci, the number of alleles per locus ranged from 4 to 9 (mean 6.8 alleles per locus), and 9 loci had ≥5 alleles. Of the target monkeys, 99.55% was genotyped. The observed heterozygosity (H o) ranged from 0.14 to 0.86 (mean 0.66), while the expected heterozygosity (H e) ranged from 0.63 to 0.86 (mean 0.76). The distribution of alleles at D16S403 was found to deviate from the Hardy-Weinberg expectations (P < 0.001), while all other loci conformed to expectations (Table 4). The power of exclusion (PE) for individual loci was from 0.43 to 0.84 (mean 0.66), and the polymorphism information content (PIC) ranged from 0.52 to 0.82 (mean 0.70). The PIC was an ideal indicator for evaluating the polymorphism of fragment. The loci could be determined with high polymorphism when PIC > 0.50. For the seven most polymorphic loci (D12S372, D9S934, D3S1768, D5S820, D6S2741, D5S1457, and D14S306), the observed heterozygosity (H o) ranged from 0.62 to 0.86 (mean 0.77), while the expected heterozygosity (H e) ranged from 0.64 to 0.86 (mean 0.79). The PE for individual locus was from 0.43 to 0.84 (mean 0.64), and the PIC ranged from 0.55 to 0.82 (mean 0.73) (Table 4). The combined probability of exclusion (CPE) in paternity testing for ten reliable loci in this study was as high as 99.999%. Moreover, the CPE could reach 99.99% when we used the core set of seven most polymorphic loci (Table 4), and the CPE increased slowly when added the remaining loci one by one (Figure 2).
Paternity assignment
To evaluate the effectiveness and reliability of paternity assignment, we firstly matched the maternity assigned according to the screened ten microsatellite loci with recorded maternity and found that all the offspring was assigned correctly to the observed mother. The reliability of the assignment was estimated by the LOD ranged from 2.38 to 5.01 (mean ± SE, 3.56 ± 0.32), while there were two mismatched loci, D6S311 (between ZY and HL) and D16S403 (between LM and HT).
Using the evaluated ten microsatellite loci, we assigned the paternity within the studied offspring. The LOD values for candidate paternity varied from −13.1 to 9.66 (mean ± SE, 1.94 ± 2.08), which suggested that mismatch occurred between offspring and ‘genetic father’ because of negative values (Table 5). We found that the estimation of only one paternity assignment (between JS and ZM) was negative (−13.1), therefore all the other offspring were assigned to their genetic fathers (Table 5). In addition, we detected a paternity relationship between two adult males that the alpha adult male (HB) was the genetic father of another adult male (BB) with 3.73 of LOD value (Table 5), but the later did not sire in the studied offspring or other target adult males (Table 5).
Discussion
From our study, ten microsatellite loci, screened with fecal DNA, exhibited higher polymorphism and higher success rate of amplification and stability. By evaluating the amplified microsatellite loci based on recorded mother-offspring pairs, we could assign all the offspring but JS to their genetic fathers. The screened markers provide sound molecular support for the study of reproductive success and parental care in free-ranging rhesus macaques. Due to the high level of polymorphism, microsatellite markers have been used for paternity testing and population genetics studies of non-human primates successfully (Chambers et al. 2004, Bradley et al. 2007, Xu et al. 2013). In addition, the relatively new methods of noninvasive genotyping for studying free-ranging primates offer possibilities to characterize the patterns of reproduction and migration in organisms with a poor knowledge of their reproductive biology (Taberlet et al. 1999, Bergman et al. 2008, Langos et al. 2013).
Screening of microsatellite loci with fecal DNA
In published works, tri- or tetranucleotide repeat markers have been proved to reduce occurrence rate of stutter bands and provide convenience in consistent allele identification by using automated techniques (Edwards et al. 1991, Bradley et al. 2000, Liu et al. 2008). In the current study, six of the seven most polymorphic loci were tetranucleotide repeat loci (D12S372, D9S934, D3S1768, D5S820, D6S2741, and D14S306). For the other three loci, two of them (D16S403, D6S311) were dinucleotide repeats (Table 2). Considering the effectiveness of DNA extraction from blood and other tissues, microsatellite genotyping has been increasingly used in paternity testing and population genetic analysis (Domingo-Roura et al. 1997, Li et al. 2010, Luna-González et al. 2012, Wojtas et al. 2013). Fecal samples generally serve as poor DNA resource (e.g., low DNA concentration and highly degraded/fragmented DNA) with repressive factors for amplification, which results in a low success rate and a high allelic dropout rate (Gerloff et al. 1995, Kohn and Wayne 1997, Lathuillière et al. 2001, Liu et al. 2008). In this study, the incidence of allelic dropout was low and had no effect on paternity testing results. Previous studies have indicated that the incidence of allelic dropout would decrease with increased temple concentration (Taberlet et al. 1996). Morin et al. (2001) indicated that, for fecal samples of wild chimpanzees (Pan troglodytes verus), when amounts of template DNA being >121 pg, only two replications were needed to score with high confidence homozygous loci amplified and just 5.20% (34/656) incidence of allelic dropout occurred. In our study, the template of which the concentration was higher than 5.00 ng/μL (extracted DNA samples >250 pg/μL in 20 μL PCR reaction mixtures) was accepted for amplification. Moreover, a multiple-tube approach to genotyping was recommended for reducing the influences of allele dropout on results (Taberlet et al. 1996). However, in practical situation, few investigators working with low concentration DNA from noninvasive samples follow this approach to the recommended extent (Gerloff et al. 1999, Kohn et al. 1999). Because the multiple-tube approach would exhaust finite sample material while also add significantly the time and expense of a project. Instead, non-independent assessments of reliability are often performed, such as comparisons of mother-offspring genotypes or comparison of the results from multiple extracts (Morin et al. 2001). We found that all genotypes used in these analyses were the results of using multiple DNA extracts for each individual. For eight mother-offspring pairs studied, the sharing of alleles did not reveal any deviations from the expected Mendelian inheritance pattern.
We successfully obtained ten microsatellite loci from fecal samples with rates of 35.63% to 85.71%, and the most polymorphic seven of ten loci reached >60% success rates (Table 2). We hereafter suggested that loci with moderate or higher amplification rates can be used in future noninvasive genetic research on free-ranging rhesus macaque populations. Nonetheless, two dinucleotide repeated loci, D16S403 and D6S311, were not recommended for selection because of their low success rates (Table 2).
Previous studies have shown that reliability of paternity testing basing on the number of loci and their probability of exclusion is highly correlated with the number of alleles (Estoup et al. 1998, Bernatchez and Duchesne 2000). In this study, the average number of alleles per locus (6.80, 10 loci out of 18 loci) approximated to the study of white-handed gibbons (Hylobates lar) (7.00 alleles per locus, 8 loci out of 47 loci with fecal samples) (Chambers et al. 2004) but was relatively higher than 4.54 alleles per locus (13 loci out of 34 loci with fecal samples) in Yunnan snub-nosed monkeys (Rhinopithecus bieti) (Liu et al. 2008). Expected heterozygosity values of 10 microsatellite loci (0.76) were slightly higher than that of 11 loci used in Yugoslav pied cattle (0.75) (Stevanovic et al. 2010) and 15 loci used in Marajoara horses (0.75) and Puruca ponies (0.70) (Reis et al. 2008). We investigated seven most polymorphic loci and found that our estimate (H o = 0.77, H e = 0.79) showed a slightly higher level of diversity than that of the result (H o = 0.72, H e = 0.72) as reported by Newman et al. (2002). It indicated that the free-ranging Taihangshan macaques had the higher genetic variation coefficient. Furthermore, in our study, in several cases the observed heterozygosity exceeded the expected heterozygosity, and one locus significantly deviated from the Hardy-Weinberg equilibrium (HWE). This deviation and the variations in heterozygosity levels may be explained by the facts that macaques from a structured population were sampled and artificially treated as a randomly mating population. In our study, the PIC of 10 screened loci ranged from 0.52 to 0.82, with an average of 0.70 for 10 highly polymorphic loci and an average of 0.73 for 7 most polymorphic loci, which approximate the estimate of PIC (0.74) across 15 loci from blood samples (Kanthaswamy et al. 2006). Because the frequency of heterozygotes at a particular locus is strongly correlated with the estimate of its PIC value, microsatellites with higher PIC value are especially useful for genetically characterizing a given individuals (Ude et al. 2003).
Those ten screened microsatellite loci exhibited polymorphism and stability, and the combined probability of exclusion achieved 99.99% when the seven most polymorphic loci were employed for paternity testing (Figure 1). To evaluate the effectiveness of a paternity testing system, PE value or CPE value are the major factors in determining the genetic sire of an offspring (Sherman et al. 2004). The degree of relatedness of the candidate sire is also reflected by PE value. Therefore, PE/CPE value reflects the ability of an assay to exclude a candidate sire which is influenced by the number of loci genotyped and heterozygosity of each locus (Xu et al. 2013). The combined probability of exclusion for 10 microsatellite loci exceeded 99.99% (Table 4), which was slightly higher than that (99.62%) of 11 microsatellite loci used in paternity testing of Yugoslav pied cattle (Stevanovic et al. 2010) and relatively higher than that of 8 loci (96.60%) in white-handed gibbon (Hylobates lar) (Chambers et al. 2004). Comparison of our study showed that our selected microsatellite markers had greater power of exclusion, and we could reach a high level of exclusion with only seven loci.
Effectiveness of screened microsatellite loci
We amplified ten microsatellite loci which could be employed to identify the parentage and also found that seven loci of them displayed the most polymorphic. We evaluated the effectiveness of these screened microsatellite loci via matching the maternity assigned following the current study to the maternity identified by our long-term records in the field. All the studied offspring were successfully assigned to their biological mothers, which was consistent with our field observations, except for two mismatched loci between ZY and HL and between LM and HT. The mismatching was probably due to a null allele or a mutation in loci D6S311 and D16S403.
Given the positive effectiveness of our amplified loci, we could assign all the offspring and an adult male to their genetic fathers via the current methodology; however, we failed to find the genetic father for one offspring (JS). To ensure the reliability of testing, we analyzed the data of JS five times, but four mismatched loci in the paternity-offspring pair were found (LOD values < 0). This mismatch plausibly resulted from the disappearance of fecal samples of the genetic father of JS, which could be possible because of male dispersal (Southwick et al. 1996, Thierry 2004). This was true that we did observe adult males, single or from ‘bachelor troop’ but surrounding the studied troop, mating with adult females within WW-1 during copulating season (from October to December). Unfortunately, we failed to collect their fecal samples because these individuals were difficult to observe and follow.
Conclusions
We screened ten microsatellite markers, especially a core set of the most polymorphic seven of ten loci with fecal samples from free-ranging rhesus macaques. We verified the screened microsatellite loci by matching the maternity assignment via the current methodology to recorded maternity and assigned the paternity of our target subjects. We concluded that screened markers offer potential to characterize reproductive success and mating efforts of adult males in free-ranging rhesus macaques.
References
Balloux F, Lugon-Moulin N (2002) The estimation of population differentiation with microsatellite markers. Mol Ecol 11:155–165
Berard JD, Nürnberg P, Epplen JT, Schmidtke J (1994) Alternative reproductive tactics and reproductive success in male rhesus macaques. Behaviour 129:177–201
Bercovitch FB (1997) Reproductive strategies of rhesus macaques. Primates 38:247–263
Bergman TJ, Phillips-Conroy JE, Jolly CJ (2008) Behavioral variation and reproductive success of male baboons (Papio anubis × Papio hamadryas) in a hybrid social group. Am J Primatol 70:136–147
Bernatchez L, Duchesne P (2000) Individual-based genotype analysis in studies of parentage and population assignment: how many loci, how many alleles? Can J Fish Aquat Sci 57:1–12
Bradley BJ, Boesch C, Vigilant L (2000) Identification and redesign of human microsatellite markers for genotyping wild chimpanzee (Pan troglodytes verus) and gorilla (Gorilla gorilla gorilla) DNA from faeces. Conserv Genet 1:289–292
Bradley BJ, Doran-Sheehy DM, Vigilant L (2007) Potential for female kin associations in wild western gorillas despite female dispersal. Proc R Soc B Biol Sci 274:2179–2185
Brenner C, Morris JW (1990) Paternity index calculations in single locus hypervariable DNA probes: validation and other studies. In: Proceedings for the International Symposium on Human Identification. Promega Corporation, Madison, USA, pp 21–59
Buchan JC, Alberts SC, Silk JB, Altmann J (2003) True paternal care in a multi-male primate society. Nature 425:179–181
Chambers KE, Reichard UH, Möller A, Nowak K, Vigilant L (2004) Cross-species amplification of human microsatellite markers using noninvasive samples from white-handed gibbons (Hylobates lar). Am J Primatol 64:19–27
Charpentier MJE, Fontaine MC, Cherel E, Renoult JP, Jenkins T, Benoit L, Barthès N, Alberts SC, Tung J (2012) Genetic structure in a dynamic baboon hybrid zone corroborates behavioural observations in a hybrid population. Mol Ecol 21:715–731
Constable JL, Ashley MV, Goodall J, Pusey AE (2001) Noninvasive paternity assignment in Gombe chimpanzees. Mol Ecol 10:1279–1300
Cooper SJB, Bull CM, Gardner MG (1997) Characterization of microsatellite loci from the socially monogamous lizard Tiliqua rugosa using a PCR-based isolation technique. Mol Ecol 6:793–795
Domingo-Roura X, López-Giráldez T, Shinohara M, Takenaka O (1997) Hypervariable microsatellite loci in the Japanese macaque (Macaca fuscata) conserved in related species. Am J Primatol 43:357–360
Edwards A, Civitello A, Hammond HA, Caskey CT (1991) DNA typing and genetic mapping with trimeric and tetrameric tandem repeats. Am J Hum Genet 49:746–756
Estoup A, Gharbi K, SanCristobal M, Chevalet C, Haffray P, Guyomard R (1998) Parentage assignment using microsatellites in turbot (Scophthalmus maximus) and rainbow trout (Oncorhynchus mykiss) hatchery populations. Can J Fish Aquat Sci 55:715–723
Fernandez-Duque E, Valeggia CR, Mendoza SP (2009) The biology of paternal care in human and nonhuman primates. Annu Rev Anthropol 38:115–130
Gerloff U, Schlötterer C, Rassmann K, Rambold I, Hohmahh G, Fruth B, Tautz D (1995) Amplification of hypervariale simple sequence repeats (microsatellite) from excremental DNA of wild living bonobos (Pan paniscus). Mol Ecol 4:515–518
Gerloff U, Hartung B, Fruth B, Hohmann G, Tautz D (1999) Intracommunity relationships, dispersal pattern and paternity success in a wild living community of Bonobos (Pan paniscus) determined from DNA analysis of faecal samples. Proc R Soc Lond Ser B Biol Sci 266:1189–1195
Hou JH, Qu WY, Chen L, Zhang HX (1998) Study of the reproduction eco-behavior of Macaca mulatta in Taihang Mountains. Chin J Ecol 17:22–25
Hsu YC, Huang CC, Severinghaus LL (2010) Microsatellites for the hoopoes (Upupa epops) and their application to the parentage analysis. Conserv Genet Resour 2:187–189
Inoue M, Mitsunaga F, Nozaki M, Ohsawa H, Takenaka A, Sugiyama Y, Shimizu K, Takenaka O (1993) Male dominance rank and reproductive success in an enclosed group of Japanese macaques: with special reference to post-conception mating. Primates 34:503–511
Kanthaswamy S, Smith DG (1998) Use of microsatellite polymorphisms for paternity exclusion in rhesus macaques (Macaca mulatta). Primates 39:135–145
Kanthaswamy S, von Dollen A, Kurushima JD, Alminas O, Rogers J, Ferguson B, Lerche NW, Allen PC, Smith DG (2006) Microsatellite markers for standardized genetic management of captive colonies of rhesus macaques (Macaca mulatta). Am J Primatol 68:73–95
Kayser M, Nürnberg P, Bercovitch F, Nagy M, Roewer L (1995) Increased microsatellite variability in Macaca mulatta compared to humans due to a large scale deletion/ insertion event during primate evolution. Electrophoresis 16:1607–1611
Kayser M, Ritter H, Bercovitch FB, Mrug M, Roewer L, Nürnberg P (1996) Identification of highly polymorphic microsatellites in the rhesus macaque Macaca mulatta by cross-species amplification. Mol Ecol 5:157–159
Kohn MH, Wayne RK (1997) Facts from feces revisited. Trends Ecol Evol 12:223–227
Kohn MH, York EC, Kamradt DA, Haught G, Sauvajot RM, Wayne RK (1999) Estimating population size by genotyping faeces. Proc R Soc Lond Ser B Biol Sci 266:657–663
Langos D, Kulik L, Mundry R, Widdig A (2013) The impact of paternity on male-infant association in a primate with low paternity certainty. Mol Ecol 22:3638–3651
Lathuillière M, Ménard N, Gautier-Hion A, Crouau-Roy B (2001) Testing the reliability of noninvasive genetic sampling by comparing analyses of blood and fecal samples in Barbary macaques (Macaca sylvanus). Am J Primatol 55:151–158
Li RS, Zhao S, Gao R, Hu ZM (2009) Analysis of microsatellites DNA polymorphism in Macaca mulatta and Macaca irus. Acta Lab Anim Sci Sin 17:57–60
Li KX, Geng JN, Qu JP, Zhang YM, Hu SN (2010) Effectiveness of 10 polymorphic microsatellite markers for parentage and pedigree analysis in plateau pika (Ochotona curzoniae). BMC Genet 11:101
Liu ZJ, Ren BP, Hao YL, Zhang HR, Wei FW, Li M (2008) Identification of 13 human microsatellite markers via cross-species amplification of fecal samples from Rhinopithecus bieti. Int J Primatol 29:265–272
Liu ZJ, Huang CM, Zhou QH, Li YB, Wang YF, Li M, Takenaka O, Takenaka A (2013) Genetic analysis of group composition and relatedness in white-headed langurs. Integr Zool 8:410–416
Lu JQ, Hou JH, Wang HF, Qu WY (2007) Current status of Macaca mulatta in Taihangshan mountains area, Jiyuan, Henan, China. Int J Primatol 28:1085–1091
Luna-González A, Hernández-Arteaga S, Sánchez-Garza M, López-Revilla R, Medina-Esparza L, Cruz-Vázquez C (2012) Microsatellite loci and paternity analysis in Nubia and Boer goats. Arch Med Vet 44:123–127
Marshall TC, Slate J, Kruuk LEB, Pemberton JM (1998) Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7:639–655
Morin PA, Chambers KE, Boesch C, Vigilant L (2001) Quantitative polymerase chain reaction analysis of DNA from noninvasive samples for accurate microsatellite genotyping of wild chimpanzees (Pan troglodytes verus). Mol Ecol 10:1835–1844
Newman TK, Fairbanks LA, Pollack D, Rogers J (2002) Effectiveness of human microsatellite loci for assessing paternity in a captive colony of vervets (Chlorocebus aethiops sabaeus). Am J Primatol 56:237–243
Penedo MCT, Bontrop RE, Heijmans CMC, Otting N, Noort R, Rouweler AJM, de Groot N, Ward T (2005) Microsatellite typing of the rhesus macaque MHC region. Immunogenetics 57:198–209
Pereira ME, Clutton-Brock TH, Kappeler PM (2000) Understanding male primates. In: Kappeler PM (ed) Primate males: causes and consequences of variation in group composition. University Press, Cambridge, pp 271–277
Perwitasari-Farajallah D (2007) Human short tandem repeat (STR) markers for paternity testing in pig-tailed macaques. HAYATI J Biosci 14:39–43
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Reis SP, Gonçalves EC, Silva A, Schneider MPC (2008) Genetic variability and efficiency of DNA microsatellite markers for paternity testing in horse breeds from the Brazilian Marajó archipelago. Genet Mol Biol 31:68–72
Rogers J, Bergstrom M, IV RG, Kaplan J, Arya A, Novakowski L, Johnson Z, Vinson A, Shelledy W (2005) A panel of 20 highly variable microsatellite polymorphisms in rhesus macaques selected for pedigree or population genetic analysis. Am J Primatol 67:377–383
Rosenblatt JS (2003) Outline of the evolution of behavioral and nonbehavioral patterns of parental care among the vertebrates: critical characteristics of mammalian and avian parental behavior. Scand J Psychol 44:265–271
Sefc KM, Steinkellner H, Glössl J, Kampfer S, Regner F (1998) Reconstruction of a grapevine pedigree by microsatellite analysis. Theor Appl Genet 97:227–231
Sherman G, Kachman S, Hungerford L, Rupp GP, Fox CP, Brown MD, Feuz BM, Holm TR (2004) Impact of candidate sire number and sire relatedness on DNA polymorphism-based measures of exclusion probability and probability of unambiguous parentage. Anim Genet 35:220–226
Smith DG (1993) A 15-year study of the association between dominance rank and reproductive success of male rhesus macaques. Primates 34:471–480
Smith DG, Kanthaswamy S, Viray J, Cody L (2000) Additional highly polymorphic microsatellite (STR) loci for estimating kinship in rhesus macaques (Macaca mulatta). Am J Primatol 50:1–7
Song CS, Qu WY (1996) Scientific survey of Taihangshan Macaque Nature Reserve. China Forestry Publishing House, Beijing, pp 56–114
Soulsbury CD (2010) Genetic patterns of paternity and testes size in mammals. PLoS ONE 5:e9581
Southwick CH, Zhang YZ, Jiang HS, Liu ZH, Qu WY (1996) Population ecology of rhesus macaques in tropical and temperate habitats in China. In: Fa JE, Lindburg DG (ed) Evolution and ecology of macaque societies. Cambridge University Press, Cambridge, pp 95–105
Stevanovic J, Stanimirovic Z, Dimitrijevic V, Maletic M (2010) Evaluation of 11 microsatellite loci for their use in paternity testing in Yugoslav Pied cattle (YU Simmental cattle). Czech J Anim Sci 55:221–226
Taberlet P, Griffin S, Goossens B, Questiau S, Manceau V, Escaravage N, Waits LP, Bouvet J (1996) Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res 16:3189–3194
Taberlet P, Waits LP, Luikart G (1999) Noninvasive genetic sampling: look before you leap. Trends in Ecology and Evolution 14:323–327
Thierry B (2004) Social epigenesist. In: Thierry B, Singh M, Kaumanns W (ed) Macaque societies: a model for the study of social organization. Cambridge University Press, Cambridge, pp 267–294
Tian JD, Wang ZL, Lu JQ, Guo XB, Liu JD (2011a) PAE coding system-based ethogram of Taihangshan macaques (Macaca mulatta tcheliensis), Jiyuan, Henan Province, China. Acta Theriol Sin 31:125–140
Tian JD, Wang ZL, Lu JQ, Guo XB, Wang BS (2011b) Social structure of wild Macaca mulatta in the Mt. Taihangshan area, Jiyuan, China. Acta Anthropol Sin 30:425–436
Tian JD, Wang ZL, Lu JQ, Wang BS, Chen JR (2013) Reproductive parameters of temperate-dwelling female Macaca mulatta tcheliensis. Am J Primatol 75:605–612
Ude GN, Kenworthy WJ, Costa JM, Cregan PB, Alvernaz J (2003) Genetic diversity of soybean cultivars from China, Japan, North America and North American ancestral lines determined by amplified fragment length polymorphism. Crop Sci 43:1858–1869
van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Vigilant L, Hofreiter M, Siedel H, Boesch C (2001) Paternity and relatedness in wild chimpanzee communities. Proc Natl Acad Sci 98:12890–12895
Wang J, Liu CS, Zhang LP, Wang ZG, Yu FQ, Zhang GX, Song XZ, Han X, Wei YC (2009) Individual identification and paternity testing of bulls using microsatellite. Hereditas 31:285–289
Widdig A (2007) Paternal kin discrimination: the evidence and likely mechanisms. Biol Rev 82:319–334
Wojtas M, Piniewska D, Polańska N, Stawowiak A, Sanak M (2013) Mutations of microsatellite autosomal loci in paternity investigations of the Southern Poland population. Forensic Sci Int Genet 7:389–391
Woodruff DS (1993) Non-invasive genotyping of primates. Primates 34:333–346
Xu YR, Li JH, Zhu Y, Sun BH (2013) Development of a microsatellite set for paternity assignment of captive rhesus macaques (Macaca mulatta) from China. Russ J Genet 49:730–736
Yang BH, Ren BP, Xiang ZF, Yang JY, Yao H, Garber PA, Li M (2014) Major histocompatibility complex and mate choice in the polygynous primate: the Sichuan snub-nosed monkey (Rhinopithecus roxellana). Integr Zool 9:598–612
Zhang YZ, Quan GQ, Lin YL, Southwick CH (1989) Extinction of rhesus monkeys (Macaca mulatta) in Xinglung, North China. Int J Primatol 10:375–381
Acknowledgements
We thank Hou JF and Hou XQ for their sampling assistance and logistic support. We are grateful to the Jiyuan Administration of Taihangshan Macaque National Nature Reserve for their permission to our research. This work was financially supported by the Natural Science Foundation of China (30970378, 31170503) and the Graduate Programme Fund of Zhengzhou University (10L10402).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JQL, ZLW, and BSW designed the study. BSW and JDT collected fecal samples, and BSW carried out the laboratory work. ZLW and ZWC analyzed the data and prepared figures and tables. BSW interpreted the data and drafted the manuscript. JQL and JDT made intellectual contributions to prepare the manuscript and revised it critically. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, BS., Wang, ZL., Tian, JD. et al. Establishment of a microsatellite set for noninvasive paternity testing in free-ranging Macaca mulatta tcheliensis in Mount Taihangshan area, Jiyuan, China. Zool. Stud. 54, 8 (2015). https://doi.org/10.1186/s40555-014-0100-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40555-014-0100-9