Abstract
Mutations in the fused-in-sarcoma (FUS) gene have been linked to familial amyotrophic lateral sclerosis (fALS). FUS aggregates in the cytosol and associates with stress granules (SGs) in pathological cases, whereas FUS is normally found in the nucleus. However, little is known about how FUS mutations cause neurodegeneration in ALS, which is distinguished by FUS-positive inclusion and stress granules. In this study, we investigated the mechanism of abnormal cytoplasmic aggregate formation caused by ALS-linked FUS mutations. FUS R521C interacted more with ubiquitin-associated protein 2-like (UBAP2L) and protein arginine methyltransferase 1 (PRMT1) than FUS WT, and PRMT1 and UBAP2L are sequestered into FUS R521C-positive stress granules under oxidative stress. PRMT1 asymmetrically demethylates UBAP2L, which is required for both SG assembly and disassembly. Furthermore, in FUS R521C, arginine methylation of UBAP2L is reduced, and the loss of PRMT1 increases FUS-positive SGs in oxidative stress. These results imply that an aberrant interaction between FUS-R521C and PRMT1 causes insufficient arginine methylation of UBAP2L, resulting in abnormal FUS-R521C-positive SGs remaining in the cytoplasm. This study could lead to the identification of a new pathogenic mechanism and therapeutic targets for the FUS mutation, which has been associated with abnormally increased protein interactions in ALS.
Graphical abstract

Similar content being viewed by others
Introduction
Amyotrophic lateral sclerosis (ALS) is a disease of the motor neurons, which are the nerve cells that connect the brain to the spinal cord and the spinal cord to the peripheral nerves (Kiernan et al. 2011; Chiò et al. 2009). Up to 90% of ALS patients are sporadic (sALS), with genetics accounting for only 10% (fALS) (Tiryaki and Horak 2014). Surprisingly, the majority of ALS-linked mutation genes are related to RNA binding protein (RNP)-encoding genes like TARDBP and FUS, and form aggregates known as resident stress granule (SG) components or SG-associated proteins (Kim et al. 2021; Wolozin and Ivanov 2019).
Stress granules (SGs) are cytoplasmic ribonucleoprotein (RNP) granules formed in response to stresses that cause a temporal pause in protein translation to prevent the accumulation of misfolded proteins during cell stress conditions such as heat shock, hypoxia, oxidative stress, ultraviolet irradiation, and viral infection (Anderson and Kedersha 2009; Kedersha et al. 2013; Kedersha and Anderson 2002). Many post-translational modifications (PTMs), such as glycosylation (Ohn et al. 2008), phosphorylation (Stoecklin et al. 2004; Courchet et al. 2008), dephosphorylation (Tourrière et al. 2003), poly (ADP)-ribosylation (Leung et al. 2011), and deacetylation (Kwon et al. 2007), can influence stress granule composition and dynamics. Recent research indicates that methylation is an important PTM that regulates SG dynamics (Leeuw et al. 2007; Wall and Lewis 2017).
Ubiquitin-associated protein 2-like (UBAP2L), which has an ubiquitin-associated (UBA) domain near its N-terminus, is an important regulator of SG formation (Wilde et al. 2011; Youn et al. 2018; Markmiller et al. 2018; Cirillo et al. 2020). Recently, it was discovered that UBAP2L participates in SG formation via arginine methylation in the Arg-Gly-Gly (RGG) motif (Youn et al. 2018; Huang et al. 2020). Arginine methylation is a post-translational protein modification that is important in eukaryotic cells and is catalyzed by a family of protein arginine methyltransferases (PRMTs), nine of which have been identified in mammals to date (Bedford and Richard 2005; Blanc and Richard 2017; Hwang et al. 2021). PRMT1 is the most common PRMT subtype among the nine and is widely expressed in mammalian cells, accounting for the majority of asymmetrical dimethylarginines in human cells (Tang et al. 2000). The ability of PRMT1 to regulate the cellular distribution of FUS, the formation of FUS-positive stress granules, and the cellular toxicity of ALS-linked FUS mutants has been studied (Scaramuzzino et al. 2013; Yamaguchi and Kitajo 2012; Tradewell et al. 2011).
FUS is a DNA/RNA binding protein, and genetic mutations in this protein have been linked to ALS (Kwiatkowski et al. 2009; Vance et al. 2009). Previous research discovered that the majority of FUS mutations are found in the nuclear localization signal (NLS) in the FUS C-terminus (Kwiatkowski et al. 2009; Vance et al. 2009; Corrado et al. 2010; Lattante et al. 2013). These FUS mutations cause FUS mislocalization from the nucleus to the cytoplasm and the formation of FUS-positive stress granules, which sequester various proteins and RNA (Takanashi and Yamaguchi 2014; Shelkovnikova et al. 2014; Dormann et al. 2010). In response to oxidative stress, the ALS-linked FUS mutant associated with PRMT1 more avidly than FUS WT and was found in stress granules with PRMT1 (Jun et al. 2017).
However, the molecular mechanism underlying the FUS-R521C mutation's strong association with PRMT1 is unknown. As a result, it is critical to investigate how specific ALS-associated mutations interact with physiological protein–protein interactions, resulting in FUS mislocalization in the cytoplasm and subsequent pathological outcomes. Although numerous studies have identified PRMT1 as an essential regulator of proteins implicated in neurodegenerative diseases in general (Scaramuzzino et al. 2013; Yamaguchi and Kitajo 2012; Tradewell et al. 2011; Jun et al. 2017), little is known about how PRMT1 contributes to the formation of FUS aggregates and neurodegeneration.
In this study, we discovered that UBAP2L strongly associates with FUS-R521C but not with FUS WT and that UBAP2L is sequestered into the SGs in response to oxidative stress.
We also found that PRMT1 asymmetrically dimethylates UBAP2L and that FUS R521C expression reduces the arginine methylation of UBAP2L. Furthermore, PRMT1 deficiency increases FUS R521C-positive SGs in response to oxidative stress. These results suggest a new cellular pathogenic mechanism in which FUS-positive SGs are produced due to insufficient UBAP2L methylation caused by PRMT1 sequestered by FUS R521C.
Materials and methods
Cell culture
Human embryonic kidney (HEK293T) cells were cultured in Dulbecco’s Modified Eagle’s Medium with high glucose (DMEM, Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco; Thermo Fisher Scientific, Waltham, MA, USA) and penicillin/streptomycin (Invitrogen; Thermo Fisher Scientific, Waltham, MA, USA). Cells were maintained in an incubator at 37 °C with 5% CO2 and 95% humidity.
Plasmid DNA and siRNA transfection
Constructs encoding human FUS WT, R521C, R521G, R521H and P525L fused to a N-terminal FLAG tag were provided by Jin-A Lee. The cells were transfected with plasmid DNA constructs using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA). The transfection was performed for 24 h at 37 °C. The human UBAP2L gene (Genbank ID: 9898) was commercially cloned by CELANTEC (Republic of Korea) and inserted into the pCMV-HA vector (TAKARA).
To inhibit PRMT1, HEK293T cells were treated with siRNA (10 nM) and the cells were transfected with siRNA using Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific, Waltham, MA, USA) for 48 h at 37 °C. The following siRNAs were used in this study: Human PRMT1 siRNA #3276-1 and -2 (Bioneer, Daejeon, South Korea).
For co-transfection of siRNA for PRMT1 and plasmid DNA for FUS mutants, plasmid DNA was treated on siRNA-transfected cells 24 h after siRNA treatment. Then, the cells were co-transfected with siRNA and plasmid DNA for an additional 24 h. After co-transfection, the cells were washed with ice-cold PBS for following experiments.
Immunoprecipitation and Western blotting.
To obtain the total cell lysates from HEK293T cells expressing FLAG-FUS (WT, R521C, or P525L), cells were lysed in IP buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% NP-40, and protease and phosphatase inhibitors). For immunoprecipitation, the cell lysates were incubated with anti-FLAG (Sigma Aldrich, St. Louis, MO) or anti-HA (Cell Signaling Technology, Danvers, MA) antibodies overnight at 4 °C. The following day, antibody-protein complexed cell lysates were incubated with magnetic beads (Dynabead, Thermo Fisher Scientific, Waltham, MA, USA) for 30 min at RT and then washed three times with 1X PBS (5 min each) at RT.
For western blot analysis, the proteins were resolved by SDS-PAGE, and the immunoblot was performed using an ECL detection system (Thermo Fisher Scientific, Waltham, MA, USA). We used anti-FLAG (1:70,000; Sigma Aldrich, St. Louis, MO), anti-HA (1:10,000, Cell Signaling Technology, Danvers, MA), anti-ADMA (1:1,000, Cell Signaling Technology, Danvers, MA), anti-PRMT1 (1:1,000; Cell Signaling Technology, Danvers, MA), anti-β-actin (1:10,000, Sigma Aldrich, St. Louis, MO), and anti-UBAP2L (1:1,000, Bethyl, Montgomery, TX) antibodies as primary antibodies, and anti-HRP rabbit and mouse ones as secondary antibodies.
Immunocytochemistry and Confocal microscopy
For immunofluorescence labeling, transfected HEK293T cells were fixed with 4% paraformaldehyde (PFA) for 10 min and washed three times with 1X PBS at RT (5 min each). Then the fixed cells were permeabilized with 0.1% Triton-X 100 for 10 min. After several washes with PBS, the cells were blocked with 3% bovine serum albumin (BSA) for 1 h at RT. After blocking, cells were incubated with primary antibodies (1:100) overnight at 4 °C. We used anti-FLAG (Sigma Aldrich, St. Louis, MO), anti-eIF4G (Novus Biologicals, Minneapolis, MN, USA), and anti-UBAP2L (Bethyl, Montgomery, TX) antibodies as primary antibodies. Next day the cells were then washed with 1X PBS and subsequently incubated with Alexa 405, 488, and 568 fluorescein-conjugated anti-mouse IgG or anti-rabbit IgG secondary antibodies (1:200, Thermo Fisher Scientific, Waltham, MA, USA) for 1 h at RT. Following several washes with PBS, the cells were mounted onto glass slides with a mounting medium. Immunofluorescence images were captured and processed using a confocal laser scanning microscope (Carl Zeiss, LSM800, Germany).
Transmission electron microscopy
For transmission electron microscopy, transfected HEK293T cells were fixed for 2 h at 4 °C in PBS containing 2.5% glutaraldehyde. The cells were detached from the culture dish with a cell scraper and pelleted by centrifugation at 2000 × g for 2 min at 4 °C. The cell pellets were resuspended in warm agar (1% in PBS) and re-centrifuged. After three washes in PBS, the agar-embedded cell pellets were embedded in an Epon 812 mixture and polymerized for 24 h at 60 °C, after dehydration in a graded series of ethanol and propylene oxide solutions. For TEM imaging experiments, ultrathin (80-nm) sections of polymerized cell pellet that had been deposited on 200 mesh copper grids were counterstained with uranyl acetate (7 min) and lead citrate (2 min) and viewed under a JEM-1400 Plus electron microscope (JEOL Ltd., JAPAN).
Results
UBAP2L associates more avidly with ALS-linked FUS R521C than with FUS-WT
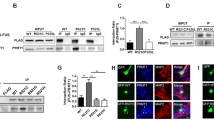
According to recent research, UBAP2L, a gene related to SG dynamics, is required for SG formation by binding to SG components (Cirillo et al. 2020; Huang et al. 2020). As a result, we looked at the relationship between ALS-causing FUS mutations and UBAP2L. To begin, HEK293T cells were transfected with FLAG-FUS R521C, FLAG-FUS WT, or FLAG-empty vectors to examine if UBAP2L expression was altered when ALS-linked FUS mutants were produced. A Western blot of cell lysates was done 24 h after transfection with anti-FLAC and anti-UBAP2L antibodies (Fig. 1A). The level of UBAP2L protein expression was not significantly different whether FUS WT or FUS R521C was expressed (Fig. 1B). Following that, to determine the relationship between FUS and UBAP2L, HA-tagged UBAP2L was co-expressed in FUS WT or FUS R521C. Then, using anti-FLAG and anti-HA antibodies, we performed a co-IP and western blot. HA-tagged UBAP2L was used to pull down FLAG-FUS WT or FLAG-FUS R521C. The R521C point mutation in FUS specifically increased its interaction with UBAP2L in comparison to the WT (Fig. 1C, D), indicating the possibility that the FUS-UBAP2L relationship could be altered by the specific ALS-linked mutation, R521C.
UBAP2L was more associated with ALS-linked FUS R521C than with FUS WT. A Western blot analysis demonstrating UBAP2L expression in HEK293T cell lysates expressing FLAG-empty vector, FLAG-FUS WT, and FLAG-FUS R521C. B A bar graph depicting the relative UBAP2L expression ratios of the WT and R521C mutants, normalized to actin expression. Data from independently replicated experiments are presented as the mean SEM (n = 4). ns: not significant, one-way ANOVA, Tukey's multiple-comparison test. C Co-Immunoprecipitation (Co-IP) analysis of UBAP2L-FUS interaction in HEK293T cells co-expressing HA-UBAP2L and FLAG-FUS WT or FLAG-FUS R521C. Following Co-IP, anti-HA (for UBAP2L) or anti-FLAG (for FUS WT or FUS R521C) antibodies were used for Western blotting. D A bar graph depicting the relative interaction ratio of UBAP2L with FUS WT versus FUS R521C mutant. Data from independently replicated experiments are presented as mean SEM (n = 3). **P < 0.01, Students t-test. E Confocal microscopy images showing the cellular localization of FLAG-FUS and HA-UBAP2L in HEK293T cells expressing FLAG-FUS WT or FLAG-FUS R521C with sodium arsenite (SA, 0.5 mM, 1 h) or without (DW) oxidative stress. The arrows indicate the co-localization of FUS with UBAP2L in the peri-nuclear cytoplasm of SA-treated HEK293T cells. Scale bar: 10 μm. F Co-localization of FLAG-FUS and UBAP2L with stress granule marker eIF4G in SA-treated HEK293T cells. The arrows indicate the co-localization of FUS with UBAP2L at elF4G positive cytoplasmic aggregates. Scale bar: 10 μm. SA: sodium arsenite. G Representative transmission electron micrographs of stress granules (SGs) imaged from HEK293T cells expressing Flag-FUS-R521C with (SA) or without (DW) oxidative stress. Scale bars: 1 um (a-d) and 500 nm (e and f, high magnification images of the marked area in SA-treated cells). Nu: nucleus, M: mitochondria, SG: stress granule. H Representative transmission electron micrographs of mitochondria (M) imaged from HEK293T cells expressing Flag-FUS-R521C with (SA) or without (DW) oxidative stress. Scale bars: 1 um (a and b, d-f) and 2 um (c). Nu: nucleus, M: mitochondria, SG: stress granule, ER: endoplasmic reticulum, asterisks: mitochondria showing structural abnormalities in DW or SA treated cells
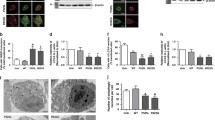
Since the R521C point mutation in FUS specially increased its interaction with UBAP2L, we next investigated the cellular localization of FUS WT or FUS R521C with UBAP2L in HEK293T cells expressing FLAG-FUS WT or FLAG-FUS R521C with HA-UBAP2L. In the absence of oxidative stress (DW), in both FUS WT and FUS R521C, FUS was mostly found in the nucleus (Fig. 1E a, i), whereas UBAP2L was mostly found in the cytoplasm (Fig. 1E b, j) and did not co-localize without forming cytosolic aggregates. In contrast, when HEK293T cells were treated with the oxidative stress inducer sodium arsenite (SA), aggregates positive for FUS (Fig. 1E e, m) and UBAP2L (Fig. 1E f, n) were formed in the cytoplasm. In addition, FUS and UBAP2L were co-localized in the aggregates (Fig. 1E h, p). Furthermore, when FUS R521C was compared to FUS WT, the number of cytosolic aggregates positive for both FUS and UBAP2L increased significantly (Fig. 1E m, n, p). These FUS and UBAP2L co-localizing cytosolic aggregates were also shown to be co-localized with the stress granule marker, elF4G (Fig. 1F e–h). This finding clearly shows that the R521C point mutation in FUS promotes cytosolic sequestration of UBAP2L into stress granules in HEK293T cells.
In addition, we examined the ultrastructure of cytoplasmic stress granules in the same conditions in which the stress granule formation was determined using immunofluorescence confocal microscopy (Fig. 1E, F). In brief, HEK293T cells were treated with SA to induce oxidative stress and DW for control. The cells were then fixed in glutaraldehyde and osmium tetroxide, dehydrated in ethanol and propylene oxide, embedded in Epon, and ultrathin sections of control and oxidative stress-induced cells were analyzed in high resolution by electron microscopy. Stress granules were not seen in the cytoplasm of DW-treated control HEK293T cells (Fig. 1G a, b). Stress granules, on the other hand, were observed in the perinuclear cytoplasmic area of the cells after SA treatment (Fig. 1G c, d, labeled SG in the boxed areas), as validated by immunofluorescence imaging of the stress granules with stress granule marker elF4G. (Fig. 1F g). The cytoplasmic stress granules had an aggregate-like appearance in the higher magnification TEM images, with tiny granular formations firmly packed (Fig. 1G e, f, labeled SG). As reported in a recent paper on the TEM microstructure of SGs formed by SA treatment in HEK293T and HeLa cells (Souquere et al. 2009), the SGs we found were not surrounded by membrane structures and were not adjacent to other organelles such as mitochondria or endoplasmic reticulum (Fig. 1G c, d, labeled SG). As the relevance of mitochondrial structural damage and dysfunction in the progression of neurodegenerative diseases has been reported in FUS-expressed cellular model and other animal models (Deng et al. 2018; Umeda et al. 2011; Choi et al. 2014), we further focused on the ultrastructural alteration of mitochondria in FUS R521C expressing HEK293T Cells by oxidative stress-inducing SA treatment. Even if some mitochondria's cristae are partially damaged by FUS expression as previously reported (Deng et al. 2018), the majority of mitochondria in DW-treated control cells showed a typical structure, with a dense matrix compacted by regularly spaced cristae and encircled by clear inner and outer membranes (Fig. 1H a and b, labeled M, structural abnormalities are marked with asterisks). In contrast, numerous mitochondria in the SG-formed cell by SA treatment showed various structural abnormalities, including the loss of cristae and membrane (Fig. 1H c-h, labeled M, structural abnormalities are marked with asterisks), which is consistent with mitochondrial structural abnormalities reported in other neurodegenerative diseases (Umeda et al. 2011; Choi et al. 2014).
Arginine methylation regulates the formation of FUS R521C-positive SGs
It has been reported that FUS-R521C interacts more strongly with PRMT1 than FUS-WT, FUS-P525L, R521G, or R521H, however, PRMT1 did not influence the level of arginine methylation in FUS (Jun et al. 2017; Tradewell et al. 2012). Therefore, we explored whether FUS R521C expression altered arginine methylation in the target of PRMT1. Previous research has revealed that UBAP2L can regulate SG formation caused by arginine methylation by PRMT1 (Huang et al. 2020). The interaction between PRMT1 and UBAP2L was studied to investigate whether post-translational modifications (PTMs) influence the formation of FUS mutant-positive SGs.
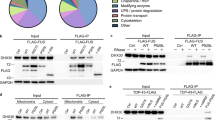
First, we transfected HA-UBAP2L into HEK 293 T cells and allowed it to express for 24 h. The cell extract was then immunoprecipitated with an anti-HA antibody, followed by western blotting with an anti-HA and anti-PRMT1 antibodies. As a result, it was discovered that HA-UBAP2L and PRMT1 interact (Fig. 2A). Then, we employed PRMT1 siRNA to suppress the methylation function of PRMT1. To begin, siRNA suppression of PRMT1 expression in HEK293T cells was performed for 48 h using PRMT1 siRNAs (siPRMT1s), followed by western blot analysis of PRMT1 siRNAs-treated cell lysates with an anti-PRMT1 antibody. Regardless of whether siPRMT1 #1 or siPRMT1 #2 was expressed, PRMT1 protein expression was drastically reduced when compared to the negative control siRNA (siNC). As a result, the efficacy of siPRMT1s #1 and #2 was confirmed (Fig. 2B, C).
FUS-positive SGs were regulated by arginine methylation. A HEK293T cells were transfected with the HA-empty vector or HA-UBAP2L. Co-immunoprecipitation (Co-IP) was performed with an anti-HA antibody, followed by a western blot with an anti-HA and anti-PRMT1 antibody. B and C PRMT1 siRNAs were used to knock down PRMT1 expression in HEK293T cells, and an anti-PRMT1 antibody was used to perform western blot analysis of PRMT1 siRNA-treated cell lysates (B). The relative knockdown efficiency of PRMT1 siRNAs compared to that of negative control siRNA. A bar graph was used to show the relative knockdown efficiency of PRMT1 siRNAs in comparison to a negative control siRNA (C). **P < 0.01; ***P < 0.001. The values represent the mean SEM (n = 3), one-way ANOVA, and Tukey's multiple-comparison test. (D) Anti-HA antibodies were used to immunoprecipitate HEK293T cell lysates expressing HA-UBAP2L in conjunction with either negative control siRNA or PRMT1 siRNA. Following that, western blot analysis was performed using anti-ADMA, anti-HA, or anti-PRMT1 antibodies, as appropriate. The UBAP2L arginine methylation band is denoted by an asterisk. (E) Co-IP with an anti-HA antibody was performed on HEK293T cell lysates expressing FLAG-FUS WT or FLAG-FUS R521C. Following that, western blots were performed using anti-HA, anti-FLAG, anti-PRMT1, or anti-ADMA antibodies. (F) Confocal microscopy analysis was performed to demonstrate the cellular localization of FLAG-FUS, HA-UBAP2L, and eIF4G in HEK293T cells expressing either a negative control siRNA (siNC) or PRMT1 siRNA (siPRMT1) with oxidative stress-inducing SA (0.5 mM, 1 h). FLAG-FUS and UBAP2L co-localize at eIF4G positive SGs, as indicated by the arrows in each image. Scale bar: 10 μm. G A bar graph comparing the number of FUS R521C positive SGs in siPRMT1 transfected HEK293T cells expressing FLAG-FUS R521C to siNC transfected cells. Results of siNC or siPRMT1 transfected cells are shown as mean SEM (n = 15 each). ***P < 0.001, Student’s t-test
Following that, siNC or siPRMT1 were co-expressed in HEK293T cells expressing HA-tagged UBAP2L to assess the degree of arginine methylation of UBAP2L by siPRMT1. Then, using an anti-HA antibody, immunoprecipitation was done on HEK293T cell lysates expressing HA-UBAP2L in conjunction with siNC or siPRMT1. Subsequently, western blot analysis was performed using anti-ADMA (Asymmetric Dimethyl Arginine), anti-HA, or anti-PRMT1 antibodies, as appropriate. First, it was confirmed that siPRMT1 treatment lowered PRMT1 expression. Furthermore, when detected with an ADMA antibody, the band (marked with an Asterisk) between 130 and 170 kDa in siPRMT1 was lower than in siNC, indicating that this region is the expression position of arginine methylated HA-UBAP2L protein. This data revealed that siPRMT1 reduced the arginine methylation of HA-UBAP2L. (Fig. 2D). Following that, when HA-UBAP2L and FLAG-FUS WT or -FUS R521C co-expressed HEK293T cell lysates were immunoprecipitated with an anti-HA antibody, the amount of methylation of HA-UBAP2L was reduced in cells expressing FUS R521C.(Fig. 2E).
Furthermore, confocal microscopy imaging and quantitative analysis was performed to examine how the arginine methylation of UBAP2L influences the development of SGs when FUS mutants are produced (Fig. 2F and G). In detail, HEK293T cells co-expressing FLAG-FUS WT or R521C, HA-UBAP2L, and PRMT1 siRNA with oxidative stress-inducing SA (0.5 mM, 1 h) were immunostained with anti-FLAG, anti-HA, and anti-elF4G antibodies, respectively, and analyzed by confocal microscopy. There was no significant difference in the production of FUS WT-positive SGs between the siPRMT1 (Fig. 2F e–h) and siNC groups (Fig. 2F a-d) in HEK293T cells expressing FUS WT. On the contrary, in the instance of FUS R521C-expressing HEK293T cells, the production of FUS R521C-positive SGs was greater in the siPRMT1 group (Fig. 2F m-p, white arrows) than in the siNC group (Fig. 2F i-l, white arrows). Quantitative counting analysis revealed that the number of sg in the siPRMT1 group (8.83 ± 0.64) was more than double that in the siNC group (4.08 ± 0.43) (Fig. 2G). Therefore, as seen the SGs in the triple-positive merging pictures of FLAG-FUS R521C, UBAP2L, and elF4G (Fig. 2F l, p, white arrows) together with the results of quantitative SG number analysis (Fig. 2G), the arginine methylation of UBAP2L appears to be a crucial factor regulating the development of FUS R521C-positive SGs.
Discussion
Despite multiple studies aimed at elucidating the pathogenic mechanism of ALS-FUS, the question of how FUS mutants exist as cytoplasmic aggregates and why they are found in SGs remains unanswered. Many recent studies have concentrated on the PTM of FUS-pathology, and we believe it is linked to the ALS-FUS pathology mechanism. PTM has been identified as an important protein regulator in neurodegenerative diseases such as Alzheimer's disease, Amyotrophic lateral sclerosis, Parkinson's disease, and Huntington's disease (Schaffert and Carter 2020; Suárez-Calvet et al. 2016; Sternburg et al. 2022). Studies on FUS PTM have revealed hypomethylation of FUS in FTD-FUS patients unrelated to FUS mutations and unmethylation of FUS in ALS-FUS patients (Suárez-Calvet et al. 2016). The majority of these studies have focused on FUS methylation, but it is still unknown why FUS aggregates in the cytoplasm rather than the nucleus.
In a previous study, the me-FUS level was not significantly different from that of WT or ALS-FUS mutants, even though FUS R521C had a strong interaction with PRMT1 (Scaramuzzino et al. 2013; Jun et al. 2017). Therefore, we hypothesized that FUS mutations might affect the methylation of proteins other than their own to explain the mechanism of ALS-FUS pathology. We investigated whether UBAP2L, a protein associated with SGs among PRMT1 targets, is associated with FUS R521C. UBAP2L, which functions upstream of G3BP, has been shown to nucleate SGs in the presence of endoplasmic reticulum stress, oxidative stress, and heat stress (Cirillo et al. 2020; Huang et al. 2020; Tolay and Buchberger 2022). First, we investigated whether FUS R521C expression in cells influenced UBAP2L expression. As a result, when compared to FUS WT, the UBAP2L protein level in FUS R521C was unchanged (Fig. 1A, B). Next, we wondered if FUS R521C had a direct -relationship with UBAP2L. Interestingly, UBAP2L was found to be more strongly associated and pulled down with FUS R521C than with FUS WT (Fig. 1C, D), as well as co-localized with FUS-positive SGs (Fig. 1F), implying that UBAP2L may be associated with FUS R521C.
Under stress conditions, UBAP2L dissociates from PRMT1 while arginine methylation is reduced and SGs are assembled at the same time. After stress recovery, PRMT1 re-methylated UBAP2L and SGs disassembled, indicating that PRMT1 regulates UBAP2L in response to cellular stress (Markmiller et al. 2018; Cirillo et al. 2020). So that, we investigated the level of UBAP2L methylation in the presence of FUS R521C. Because FUS R521C has a strong interaction with PRMT1, UBAP2L methylation may be hindered. In this study, we discovered that me-UBAP2L was reduced in FUS R521C compared to FUS WT (Fig. 2E), implying that FUS R521C disrupts UBAP2L methylation. In this result, one could wonder why the amount of UBAP2L methylation was reduced exclusively in the FUS R521C expressing group, despite the fact that the amount of PRMT1 bound to HA-UBAP2L in HEK293T cells expressing FUS R521C is comparable to that in HEK293T cells expressing FUS WT. Because this IP experiment did not focus solely on direct binding, PRMT1 interacting with FLAG-FUS R521C bound to HA-UBAP2L could have been detected together. It underlines the importance of further research to determine if FUS-R521C can interfere with direct binding between UBAP2L and PRMT1.
Additionally, when PRMT1 function was inhibited with siPRMT1, FUS R521C/UBAP2L-positive SGs increased more than siNC (Fig. 2F), indicating that PRMT1 controls FUS R521C-positive SGs. More research on the direct interaction between UBAP2L and FUSR521C is required, however, because we did not suppress and confirm the function of PRMT1, which acts specifically with UBAP2L in this study. We should look into the relationship between UBAP2L and other FUS mutant proteins besides FUS R521C, but because FUS R521C has a strong association with PRMT1, the findings of this study can give sufficient clues for FUS R521C pathology research.
Taken together, our findings reveal a novel FUS R521C binding target, UBAP2L, as well as an unknown relationship between FUS R521C and PRMT1 that may influence the arginine methylation of UBAP2L and induce SG formation. As a result of our findings, we believe that regulating UBAP2L methylation may provide a therapeutic approach to ALS-FUS pathology.
Conclusion
Using confocal and electron microscopy and molecular cell biology, we examined ALS-linked FUS mutation-induced aberrant cytoplasmic stress granules (SGs) aggregation. Under oxidative stress, FUS R521C forms stress granules with UBAP2L, which is required for SG assembly, and PRMT1, which demethylates UBAP2L. FUS R521C decreases UBAP2L arginine methylation, and PRMT1 knockdown increases FUS-positive SGs. These findings suggest that an aberrant interaction between FUS-R521C and PRMT1 causes insufficient arginine methylation of UBAP2L, resulting in abnormal FUS-R521C-positive SGs formation in the cytoplasm. This work may uncover an additional pathogenic mechanism and therapeutic alternatives for the FUS mutation, which has been linked to abnormally enhanced protein interactions in ALS patients.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Abbreviations
- ALS:
-
Amyotrophic lateral sclerosis
- FUS:
-
Fused-in-Sarcoma
- UBAP2L:
-
Ubiquitin-associated protein 2-like
- PRMT1:
-
Protein arginine methyltransferase 1
- SG:
-
Stress granule
References
Anderson P, Kedersha N. Stress granules. Curr Biol. 2009;19(10):R397–8.
Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18(3):263–72.
Blanc RS, Richard S. Arginine methylation: the coming of age. Mol Cell. 2017;65(1):8–24.
Chiò A, Mora G, Calvo A, Mazzini L, Bottacchi E, Mutani R. Epidemiology of ALS in Italy: a 10-year prospective population-based study. Neurology. 2009;72(8):725–31.
Choi KJ, Kim MJ, Je AR, Jun SM, Lee CH, Lee EJ, Jo MJ, Huh YH, Kweon HS. Three-dimensional analysis of abnormal ultrastructural alteratio in mitochondria of hippocampus of APP/PSEN1 transgenic mouse. J Biosci. 2014;39(1):97–105.
Cirillo L, Cieren A, Barbieri S, Khong A, Schwager F, Parker R, et al. UBAP2L forms distinct cores that act in nucleating stress granules upstream of G3BP1. Curr Biol. 2020;30(4):698-707.e6.
Corrado L, Del Bo R, Castellotti B, Ratti A, Cereda C, Penco S, et al. Mutations of FUS gene in sporadic amyotrophic lateral sclerosis. J Med Genet. 2010;47(3):190–4.
Courchet J, Buchet-Poyau K, Potemski A, Brès A, Jariel-Encontre I, Billaud M. Interaction with 14-3-3 adaptors regulates the sorting of hMex-3B RNA-binding protein to distinct classes of RNA granules. J Biol Chem. 2008;283(46):32131–42.
De Leeuw F, Zhang T, Wauquier C, Huez G, Kruys V, Gueydan C. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp Cell Res. 2007;313(20):4130–44.
Deng G, Wang P, Chen X, Cheng H, Liu J, Fushimi K, Zhu Li WuJ. FUS interacts with ATP synthase beta subunit and induces mitochondrial unfolded protein response in cellular and animal models. Proc Natl Acad Sci USA. 2018;115(41):E9678–86.
Dormann D, Rodde R, Edbauer D, Bentmann E, Fischer I, Hruscha A, et al. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010;29(16):2841–57.
Huang C, Chen Y, Dai H, Zhang H, Xie M, Zhang H, et al. UBAP2L arginine methylation by PRMT1 modulates stress granule assembly. Cell Death Differ. 2020;27(1):227–41.
Hwang JW, Cho Y, Bae G-U, Kim S-N, Kim YK. Protein arginine methyltransferases: promising targets for cancer therapy. Exp Mol Med. 2021;53(5):788–808.
Jun M-H, Ryu H-H, Jun Y-W, Liu T, Li Y, Lim C-S, et al. Sequestration of PRMT1 and Nd1-L mRNA into ALS-linked FUS mutant R521C-positive aggregates contributes to neurite degeneration upon oxidative stress. Sci Rep. 2017;7(1):40474.
Kedersha N, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30(6):963–9.
Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci. 2013;38(10):494–506.
Kiernan MC, Vucic S, Cheah BC, Turner MR, Eisen A, Hardiman O, et al. Amyotrophic lateral sclerosis. Lancet (London, England). 2011;377(9769):942–55.
Kim W, Kim DY, Lee KH. RNA-binding proteins and the complex pathophysiology of ALS. Int J Mol Sci. 2021;22(5):589.
Kwiatkowski TJ, Bosco DA, LeClerc AL, Tamrazian E, Vanderburg CR, Russ C, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science (new York, NY). 2009;323(5918):1205–8.
Kwon S, Zhang Y, Matthias P. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 2007;21(24):3381–94.
Lattante S, Rouleau GA, Kabashi E. TARDBP and FUS mutations associated with amyotrophic lateral sclerosis: summary and update. Hum Mutat. 2013;34(6):812–26.
Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. Poly (ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell. 2011;42(4):489–99.
Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, et al. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell. 2018;172(3):590-604.e13.
Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol. 2008;10(10):1224–31.
Scaramuzzino C, Monaghan J, Milioto C, Lanson NA Jr, Maltare A, Aggarwal T, et al. Protein arginine methyltransferase 1 and 8 interact with FUS to modify its sub-cellular distribution and toxicity in vitro and in vivo. PLoS ONE. 2013;8(4):e61576.
Schaffert LN, Carter WG. Do post-translational modifications influence protein aggregation in neurodegenerative diseases: a systematic review. Brain sciences. 2020;10(4):5289.
Shelkovnikova TA, Robinson HK, Southcombe JA, Ninkina N, Buchman VL. Multistep process of FUS aggregation in the cell cytoplasm involves RNA-dependent and RNA-independent mechanisms. Hum Mol Genet. 2014;23(19):5211–26.
Souquere S, Mollet S, Kress M, Dautry F, Pierron G, Weil D. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J Cell Sci. 2009;122(Pt 20):3619–26.
Sternburg EL, da Gruijs SLA, Dormann D. Post-translational modifications on RNA-binding proteins: accelerators, brakes, or passengers in neurodegeneration? Trends Biochem Sci. 2022;47(1):6–22.
Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, et al. MK2-induced tristetraprolin: 14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23(6):1313–24.
Suárez-Calvet M, Neumann M, Arzberger T, Abou-Ajram C, Funk E, Hartmann H, et al. Monomethylated and unmethylated FUS exhibit increased binding to Transportin and distinguish FTLD-FUS from ALS-FUS. Acta Neuropathol. 2016;131(4):587–604.
Takanashi K, Yamaguchi A. Aggregation of ALS-linked FUS mutant sequesters RNA binding proteins and impairs RNA granules formation. Biochem Biophys Res Commun. 2014;452(3):600–7.
Tang J, Kao PN, Herschman HR. Protein-arginine methyltransferase I, the predominant protein-arginine methyltransferase in cells, interacts with and is regulated by interleukin enhancer-binding factor 3. J Biol Chem. 2000;275(26):19866–76.
Tiryaki E, Horak HA. ALS and other motor neuron diseases. Continuum. 2014;20(5):1185–207.
Tolay N, Buchberger A. Role of the ubiquitin system in stress granule metabolism. Int J Mol Sci. 2022;23(7):3624.
Tourrière H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, et al. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160(6):823.
Tradewell ML, Yu Z, Tibshirani M, Boulanger M-C, Durham HD, Richard S. Arginine methylation by PRMT1 regulates nuclear-cytoplasmic localization and toxicity of FUS/TLS harbouring ALS-linked mutations. Hum Mol Genet. 2011;21(1):136–49.
Tradewell ML, Yu Z, Tibshirani M, Boulanger M-C, Durham HD, Richard S. Arginine methylation by PRMT1 regulates nuclear-cytoplasmic localization and toxicity of FUS/TLS harbouring ALS-linked mutations. Hum Mol Genet. 2012;21(1):136–49.
Umeda T, Tomiyama T, Sakama N, Tanaka S, Lambert MP, Klein WL, Mori H. Interneuronal amyloid beta oligomers cause cell death via endoplasmic reticulum stress, endosomal/lysosomal leakage, and mitochondrial dysfunction in vivo. J Neurosci Res. 2011;89:1031–42.
Vance C, Rogelj B, Hortobágyi T, De Vos KJ, Nishimura AL, Sreedharan J, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science (New York, NY). 2009;323(5918):1208–11.
Wall ML, Lewis SM. Methylarginines within the RGG-motif region of hnRNP A1 affect its IRES trans-acting factor activity and are required for hnRNP A1 stress granule localization and formation. J Mol Biol. 2017;429(2):295–307.
Wilde IB, Brack M, Winget JM, Mayor T. Proteomic characterization of aggregating proteins after the inhibition of the ubiquitin proteasome system. J Proteome Res. 2011;10(3):1062–72.
Wolozin B, Ivanov P. Stress granules and neurodegeneration. Nat Rev Neurosci. 2019;20(11):649–66.
Yamaguchi A, Kitajo K. The effect of PRMT1-mediated arginine methylation on the subcellular localization, stress granules, and detergent-insoluble aggregates of FUS/TLS. PLoS ONE. 2012;7(11):e49267.
Youn J-Y, Dunham WH, Hong SJ, Knight JDR, Bashkurov M, Chen GI, et al. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol Cell. 2018;69(3):517-32.e11.
Youn J-Y, Dunham WH, Hong SJ, Knight JD, Bashkurov M, Chen GI, et al. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol Cell. 2018;69(3):517-32. E11.
Acknowledgements
The authors are grateful to Prof. Jin-A Lee for providing plasmid DNAs (FLAG-Vector, FLAG-FUS WT, FLAG-FUS R521C, and HA-Vector).
Funding
This work was funded by grants from the National Research Foundation of Korea to the Bioimaging Data Curation Center (2022M3H9A2083956) and the Korea Basic Science Institute (KBSI) R&D project (#C330221, #C300200, #C320000).
Author information
Authors and Affiliations
Contributions
M.H.J. and Y.H.H. designed and conducted experiments and analyzed the data. S.H.L. carried out electron microscopy experiments. M.H.J. and Y.H.H. wrote the paper. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jun, MH., Lee, SH. & Huh, Y.H. ALS-linked FUS R521C disrupts arginine methylation of UBAP2L and stress granule dynamics. J Anal Sci Technol 14, 23 (2023). https://doi.org/10.1186/s40543-023-00389-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40543-023-00389-y