Abstract
Background
This work reports, for the first time, a validated methodology for qualitative and quantitative melatonin analyses directly from a complex matrix, namely rat milk, using matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI), in a useful strategy for fast assessment of this molecular species.
Methods
This novel approach uses the imaging feature of a MALDI instrument to create bidimensional images of a selected area. Results for sample concentration are expressed as a function of pixel-by-pixel intensity in grayscale, which are quantified according to pixel intensity through ImageJ open source software. For the extent of this work, two conditions were monitored: light- and dark-obtained milk.
Results
Quantification was performed using the selected reaction monitoring (SRM) transitions m/z 233→174 for melatonin. Limit of detection and limit of quantification have presented lower values (picomolar) when compared to other melatonin quantification strategies, indicating that this method presents high sensitivity. Furthermore, this method has a total analysis time of less than 1 min.
Conclusions
Melatonin concentrations are significantly higher in samples obtained from dark conditions, consistent with previous results, and thus proving that MALDI-MSI is a suitable, simple, and highly sensitive methodology for direct melatonin analysis from complex biological samples.
Similar content being viewed by others
Background
Melatonin is a secretory product biosynthesized in the pineal gland, using serotonin as its precursor molecule, and secreted in a photoperiod-dependent manner (Axelrod and Weissbach 1960; Reiter 1992; Ackermann et al. 2007). In most mammals, blood concentrations of melatonin are increased during dark periods and present decreased levels during the daylight hours (Goldman 2001). Functionally, there is great interest surrounding this molecule, since it is known to be a powerful antioxidant and the main endogenous antihypertensive agent of the cardiovascular system (K-Laflamme et al. 1998; Galano et al. 2011; Lanoix et al. 2012). Furthermore, two high-affinity G protein-coupled receptors, MT1 and MT2, mediate some melatonin’s effects, such as placental cell survival (Dubocovich and Markowska 2005).
Several studies reported its importance in regulating circadian rhythm and reproductive events (Tamura et al. 2008; Reiter 1998; Olcese 1995; Malpaux et al. 1999; Gerlach and Aurich 2000). It is already known, for example, that there is effective transfer of photoperiod information from a mother to her fetus, since melatonin can cross the placental barrier (Yellon et al. 1985; Okatani et al. 1998). Recent studies have shown radiolabeled melatonin distributed in the brain of hamsters during late fetal life, located in the same sites that present high number of melatonin receptors in adult hamsters. This information is supported by another study, where pinealectomized mother hamsters were not able to transmit photoperiod information to their fetuses (Carlson et al. 1991; Weaver and Reppert 1986). Following this logic, melatonin transfer from the fetus to the mother has also proven to be feasible. A study performed melatonin level measurements in pregnant women blood serum by radioimmunoassay (RIA); individuals with twin pregnancies have presented maternal nighttime melatonin serum levels higher than single pregnancies. These findings suggest that the feto-placental unit may contribute to maternal circulating melatonin (Nakamura et al. 2001).
In the late years, the detection and measurement of this molecule in biological samples has been performed using several different analytical strategies. Gas chromatography (GC) and liquid chromatography (LC) coupled to mass spectrometry (MS) have been the most widely and effectively employed (Jensen et al. 2011; Wang et al. 2011; Kennaway et al. 1977; Lewy and Markey 1978; Covaci et al. 1999). Among others, mass spectrometry stands out as a simple and versatile approach, as it can be used to identify, characterize, and quantify endogenous substances from biological samples. Its many resources for structural elucidation notably increase the assertiveness of the analysis, as any molecular identity can be confirmed in just a few steps. Additionally, MS can be coupled to chromatographic techniques; these previous separation processes can be widely modified and/or enhanced in order to assist the analyst to have great control among the many different species present in any complex samples, such as biological fluids (Bonnefont-Rousselot and Collin 2010). Hybrid MS techniques, namely matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) have been explored in melatonin research (Rizzi et al. 2006). For example, Rizzi et al. (2006) have conducted an investigation in order to understand the role of melatonin in melanogenesis. They were able to identify melatonin and its oxidation products by fluorescence, UV-visible spectrophotometry, and MALDI-TOF.
A recent variation of the latter has expanded MS limits to the field of molecular imaging. The technique, known as MALDI mass spectrometry imaging (MSI), was developed to identify the spatial distribution of compounds in any physical sample, such as tissue sections (Solon 2007), drug tablets (Earnshaw et al. 2010), and cosmetic products (de Oliveira et al. 2013a). Despite its ingenuity, MSI’s operating principle is very straightforward: an infrared or ultraviolet laser beam is focused on a sample and “scans” all over its extension in quick shots, generating a mass spectrum in each spot, just like a pixel, forming a molecular image at the end of the process (Caprioli et al. 1997). Our group has proposed an innovative study applied to cosmetic products, which changed the concept of “qualitative analysis-only” for MALDI-MSI. We were able to perform a relative quantification of the Sudan III dye directly in nail polishes, employing ImageJ, a software commonly used as a tool for western blot quantification (de Oliveira et al. 2013a; Gassmann et al. 2009).
In the present work, we describe a new and validated method for melatonin quantification in maternal rat milk using MALDI-MSI. This methodology comprises both melatonin detection and quantification with minimal sample preparation. Method validation was performed using the melatonin chemical standard, following its application in samples of maternal milk from rats subjected to photoperiod.
Methods
Standard preparation
A 50 mM melatonin (Sigma-Aldrich, MO, USA) stock solution was prepared in ethanol. It was further diluted with H2O MilliQ to obtain a working solution of 500 μM. Then, seriated dilutions were performed in H2O milliQ for recovery experiments and analytical curves.
Animal experimentation
Female Wistar rats at the 10th day of lactation (n = 10) received an intraperitoneal injection of 500 μL oxytocin (Sigma-Aldrich, MO, USA) (500 IU), 2 h before milking. Four to six breasts of each rat were milked, and the contents were pooled to a single sample for each rat. The milk was collected 4 h before (dark) and 6 h after (light—control sample) light turned on, corresponding at 1 a.m. and 1 p.m., comprising a total of 10 samples for each condition (n = 10 for dark and n = 10 for light) All animal procedures were in agreement with the Ethical Principles in Animal Research, adopted by the Brazilian College for Animal Experimentation according to the American Psychological Association Guidelines for Ethical Conduct in the Care and Use of Animals. All the study protocols were approved by the Ethical Committee of the University of Campinas (Protocol 2804-1).
MALDI-MSI analyses
To perform MALDI-MSI analyses, 1 μL of each sample and/or standard were mixed in 20 μL of α-cyano-4-hydroxycinnamic acid (CHCA) matrix (Sigma-Aldrich, PA, USA) (10 mg/mL in 1:1 acetonitrile/methanol solution) and spotted in a stainless steel plate suitable for MALDI-MSI (Thermo Scientific, CA, USA). Chemical images and mass spectra were acquired in a MALDI-LTQ-XL instrument equipped with imaging feature (Thermo Scientific, CA, USA). The instrument uses an ultraviolet laser as the ionization source and a quadrupole-ion-trap mass analyzing system. All data were acquired in the positive ion mode with a sample size of 500 × 500 μm (50 pixels). Each sample was run in triplicates. For image acquisition, a 50-μm raster width was selected. Selected reaction monitoring (SRM) data were acquired with transition m/z 233 → 174 for melatonin, using the collision-induced energy (CID) to 45 eV. Helium was used as the collision gas. Fragmentation pathways were proposed using Mass Frontier expert system software (Thermo Scientific, CA, USA). All imaging data were then processed using ImageQuest software v.1.0.1 (Thermo Scientific, CA, USA).
Quantification by MALDI-MSI
Imaging data were analyzed using ImageQuest software (Thermo Scientific, CA, USA). The quantification was performed using ImageJ (National Institutes of Health, USA—Open Source) on grayscale images. The area was standardized in number of pixels for all the replicates, and the ImageJ software assigned a value for the selection based on the intensity of each pixel.
SST
The system suitability test (SST) (Guideline, ICH Harmonized Tripartite 2005) was performed under the optimized MALDI conditions for the direct detection of melatonin. The standard solution was analyzed 10 times in two different days, to verify the instrumental precision (intra- and interday), determined as relative standard deviation (%RSD) of values obtained by ImageJ. Linearity, expressed as the determination coefficient (R2), was checked from 0.5 to 2.5 pM and calculated as values of each concentration against the respective known concentration.
Method validation
The validation was performed according to the International Conference on Harmonization Guidance for the Validation of Analytical Methods (Guideline, ICH Harmonized Tripartite 2005). It was based on the following criteria: limit of detection (LOD), limit of quantification (LOQ), precision (intra- and interday variability), recovery, and linearity. LOD and LOQ were determined in blank samples of MALDI matrix (CHCA) and/or three mixed samples of light-exposed maternal rat milk (CHCA + milk), using the melatonin standard analytical curve as the response method, with the formulas [LOD = (3.3σ)/S] and [LOQ = (10σ)/S], where σ is the standard deviation of the blank and S the slope of the analytical curve. Standard deviations of the blanks were calculated by analyzing the background image of CHCA matrix and the three pooled control samples in CHCA matrix using ImageJ. External analytical curves were built using standard solutions with different concentration levels. Recoveries (%R) were determined by comparing the absolute response of the melatonin standard spiked in control samples, using 30 determinations (three concentrations/ten replicates each). Method precision (intraday and interday precision) was determined as the relative standard deviation (%RSD) with three replicates and five concentrations, on two different days by the same analyst.
Method applicability
The method was applied to maternal milk samples from rats subjected to photoperiod. Samples were prepared and analyzed exactly as described above. Melatonin concentrations were calculated with external analytical curves (minimum of 5 points). The latter was built by spiking control milk with melatonin standard solution diluted in CHCA matrix.
Statistical analyses
All statistical analyses were performed in GraphPad Prism v.5.0 for Windows (GraphPad Software, CA, USA). For standard curve, the linear regression test was applied, and unpaired T test for establishing the significance for light-dark differentiation.
Results and discussion
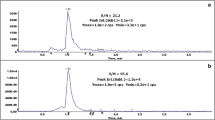
Melatonin mass spectrum was determined by performing MSn reactions using a melatonin standard, as described in “Methods” section. The standard was used as the comparison parameter for molecule identification by both MS2 and MS3 (Fig. 1). Fragment structures are available in Supplemental data (Additional file 1: Figure S1). SST showed satisfactory precision for pixel value determination, both intra- and interday (Table 1). Detector response linearity was checked using linear regression. The coefficient of determination (R2) was >0.99 from 0.5 to 2.5 pM. Thus, MALDI-MSI has proven to be a suitable approach for melatonin quantification directly from rat milk samples, with no previous sample preparation. With high precision parameters (<3%RSD, Table 1) for all cases, this analytical strategy is specific and precise enough to carry out melatonin quantitative analyses with total time less than 1 min. Specificity in this case can be assigned to the possibility of fragmentation reactions (MSn), which are supported by using an external standard for comparative purposes. This powerful combination of standards and tandem mass spectrometry for assertive compound identification in complex matrices has already been extensively discussed in recent literature (de Oliveira et al. 2013b).
Our validated methodology has demonstrated adequate values in each of the assessed parameters: recovery, LOQ, LOD, intra- and interday precision, and linearity. As shown in Table 2, there was high recovery of melatonin in all spiked concentration levels. The percentage of recovery increases as a function of concentration, which can be expected, since higher concentrations facilitate analyte recovery, and results with high linearity indicate high method accuracy (Guideline, ICH Harmonized Tripartite 2005; González et al. 1999). A good linear relationship was found between the pixel area of concentrations of melatonin ranging 0.5–2.5 pM. Linear regression analysis indicated that the correlation coefficient (r2) was greater than 0.99, and the regression equation was Y = 13,422X + 63,366 with Y as the pixel area and X as the concentration of melatonin.
Table 3 presents the LOD and LOQ obtained in two different conditions: (1) CHCA matrix and (2) maternal milk + CHCA matrix. The latter presents both limits higher than the ones obtained only with CHCA matrix. Both LOD and LOQ for spiked samples in CHCA matrix alone have presented lower levels when compared to the ones obtained in Milk + CHCA (Table 3). This is probably due to the fact that milk is a much more complex system than a simple solution of MALDI matrix and hence molecular ionization in easier in simpler environments. However, both were detected at the picomolar level, so regardless of the system complexity, LOD and LOQ are very satisfactory for both cases. The intraday and interday precisions for melatonin in milk are summarized in Table 4 and reveal good precision of the proposed method, with %RSD values under 9% intraday and less than 4% interday, expressed as functions of concentrations. Low variations (<15% RSD) in these measurements significantly increase the method reliability (Guideline, ICH Harmonized Tripartite 2005).
Quantification by imaging is a very interesting feature that has been described by Grey et al. (Grey et al. 2011) and recently implemented in our group in as a relative quantification approach (de Oliveira et al. 2013a). The present work has aimed to provide absolute values for compound quantification by using external standards and analytical curves. Although this is a usual approach, a thorough method validation was necessary to be performed. The obtained data not only have presented satisfactory results in all the analyzed parameters but have also indicated that MALDI-MSI is a sensitive methodology, with lower limit of detection when compared to classical techniques, such as LC-MS/MS and RIA (Rowe and Kennaway 2002; Gomez et al. 2013; Voultsios et al. 1997).
The above-described method was applied to milk samples obtained from rats in lactation period subjected to 12 h of photoperiod. All samples were directly analyzed. Spectra of standard, day and night samples are available in Fig. 2. All spectra were similar, indicating that the same compound is being analyzed. Results of melatonin quantification in light- and dark-period milk are shown in Table 5 and Fig. 3. Although there was a small detection of melatonin in control samples (light period milk), the concentration in dark period milk was higher.
Conclusions
The present study has demonstrated the detection of melatonin in maternal milk of rats, both in light and dark periods. Our results are in accordance with other already reported, in which melatonin levels in dark period are higher than light period (Rowe and Kennaway 2002; Eriksson et al. 1998). We have also proposed an interesting and innovative validated methodology using MSI, proving that imaging data can be a useful tool for quantitative purposes.
Abbreviations
- CHCA:
-
α-Cyano-4-hydroxycinnamic acid
- CID:
-
Collision-induced energy
- GC:
-
Gas chromatography
- LC:
-
Liquid chromatography
- LOD:
-
Limit of detection
- LOQ:
-
Limit of quantification
- MALDI-MSI:
-
Matrix-assisted laser desorption/ionization mass spectrometry imaging
- MS:
-
Mass spectrometry
- SRM:
-
Selected reaction monitoring
- TOF:
-
Time-of-flight
References
Ackermann K, Dehghani F, Bux R, Kauert G, Stehle JH. Day-night expression patterns of clock genes in the human pineal gland. J Pineal Res. 2007;43(2):185–94.
Axelrod J, Weissbach H (1960) Enzymatic O-methylation of N-acetylserotonin to melatonin. Science 131 (3409):1312-1312
Bonnefont-Rousselot D, Collin F. Melatonin: action as antioxidant and potential applications in human disease and aging. Toxicology. 2010;278(1):55–67.
Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69(23):4751–60.
Carlson LL, Weaver DR, Reppert SM. Melatonin receptors and signal transduction during development in Siberian hamsters (Phodopus sungorus). Dev Brain Res. 1991;59(1):83–8.
Covaci A, Doneanu C, Aboul‐Enein HY, Schepens P. Determination of melatonin in pharmaceutical formulations and human plasma by gas chromatography-electron impact mass spectrometry. Biomed Chromatogr. 1999;13(6):431–6.
de Oliveira DN, de Bona SS, Ferreira MS, Catharino RR. Cosmetic analysis using matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI). Materials. 2013a;6(3):1000–10.
de Oliveira DM, Pinto CB, Sampaio GR, Yonekura L, Catharino RR, Bastos DHM. Development and validation of methods for the extraction of phenolic acids from plasma, urine, and liver and analysis by UPLC-MS. J Agric Food Chem. 2013b;61(25):6113–21. doi:10.1021/jf401534d.
Dubocovich ML, Markowska M. Functional MT1 and MT2 melatonin receptors in mammals. Endocrine. 2005;27(2):101–10.
Earnshaw CJ, Carolan VA, Richards DS, Clench MR. Direct analysis of pharmaceutical tablet formulations using matrix-assisted laser desorption/ionisation mass spectrometry imaging. Rapid Commun Mass Spectrom. 2010;24(11):1665–72.
Eriksson L, Valtonen M, Laitinen JT, Paananen M, Kaikkonen M. Diurnal rhythm of melatonin in bovine milk: pharmacokinetics of exogenous melatonin in lactating cows and goats. Acta Vet Scand. 1998;39(3):301–10.
Galano A, Tan DX, Reiter RJ. Melatonin as a natural ally against oxidative stress: a physicochemical examination. J Pineal Res. 2011;51(1):1–16.
Gassmann M, Grenacher B, Rohde B, Vogel J. Quantifying Western blots: pitfalls of densitometry. Electrophoresis. 2009;30(11):1845–55.
Gerlach T, Aurich JE. Regulation of seasonal reproductive activity in the stallion, ram and hamster. Anim Reprod Sci. 2000;58(3):197–213.
Goldman BD. Mammalian photoperiodic system: formal properties and neuroendocrine mechanisms of photoperiodic time measurement. J Biol Rhythm. 2001;16(4):283–301.
Gomez FJ, Hernandez IG, Martinez LD, Silva MF, Cerutti S. Analytical tools for elucidating the biological role of melatonin in plants by LC-MS/MS. Electrophoresis. 2013;34(12):1749–56.
González AG, Herrador MA, Asuero AG. Intra-laboratory testing of method accuracy from recovery assays. Talanta. 1999;48(3):729–36. doi:10.1016/S0039-9140(98)00271-9.
Grey AC, Crouch RK, Koutalos Y, Schey KL, Ablonczy Z. Spatial localization of A2E in the retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2011;52(7):3926–33. doi:10.1167/iovs.10-7020.
Guideline, ICH. Harmonized Tripartite. Validation of analytical procedures: text and methodology. 2005. Q2(R1)1.
Jensen MA, Hansen ÅM, Abrahamsson P, Nørgaard AW. Development and evaluation of a liquid chromatography tandem mass spectrometry method for simultaneous determination of salivary melatonin, cortisol and testosterone. J Chromatogr B. 2011;879(25):2527–32.
Kennaway D, Frith R, Phillipou G, Matthews C, Seamark R. A specific radioimmunoassay for melatonin in biological tissue and fluids and its validation by gas chromatography-mass spectrometry. Endocrinology. 1977;101(1):119–27.
K-Laflamme A, Wu L, Foucart S, de Champlain J. Impaired basal sympathetic tone and alpha1-adrenergic responsiveness in association with the hypotensive effect of melatonin in spontaneously hypertensive rats. Am J Hypertens. 1998;11(2):219–29.
Lanoix D, Lacasse AA, Reiter RJ, Vaillancourt C. Melatonin: the smart killer: the human trophoblast as a model. Mol Cell Endocrinol. 2012;348(1):1–11.
Lewy AJ, Markey S. Analysis of melatonin in human plasma by gas chromatography negative chemical ionization mass spectrometry. Science. 1978;201(4357):741–3.
Malpaux B, Thiéry J-C, Chemineau P. Melatonin and the seasonal control of reproduction. Reprod Nutr Dev. 1999;39(3):355.
Nakamura Y, Tamura H, Kashida S, Takayama H, Yamagata Y, Karube A, Sugino N, Kato H. Changes of serum melatonin level and its relationship to feto‐placental unit during pregnancy. J Pineal Res. 2001;30(1):29–33.
Okatani Y, Okamoto K, Hayashi K, Wakatsuki A, Tamura S, Sagara Y. Maternal‐fetal transfer of melatonin in pregnant women near term. J Pineal Res. 1998;25(3):129–34.
Olcese J. The mammalian pineal gland and reproduction. Controversies and strategies for future research. Adv Exp Med Biol. 1995;377:1.
Reiter RJ. The ageing pineal gland and its physiological consequences. Bioessays. 1992;14(3):169–75.
Reiter RJ. Melatonin and human reproduction. Ann Med. 1998;30(1):103–8.
Rizzi A, Comai S, Bertazzo A, Costa CV, Allegri G, Traldi P. An investigation on the possible role of melatonin in melanogenesis. J Mass Spectrom. 2006;41(4):517–26.
Rowe SA, Kennaway DJ. Melatonin in rat milk and the likelihood of its role in postnatal maternal entrainment of rhythms. Am J Phys Regul Integr Comp Phys. 2002;282(3):R797–804.
Solon EG. Autoradiography: high-resolution molecular imaging in pharmaceutical discovery and development. Expert Opin Drug Discov. 2007;2(4):503–14.
Tamura H, Takayama H, Nakamura Y, Reiter RJ, Sugino N. Fetal/placental regulation of maternal melatonin in rats. J Pineal Res. 2008;44(3):335–40.
Voultsios A, Kennaway DJ, Dawson D. Salivary melatonin as a circadian phase marker: validation and comparison to plasma melatonin. J Biol Rhythm. 1997;12(5):457–66. doi:10.1177/074873049701200507.
Wang A-Q, Wei B-P, Zhang Y, Wang Y-J, Xu L, Lan K. An ultra-high sensitive bioanalytical method for plasma melatonin by liquid chromatography-tandem mass spectrometry using water as calibration matrix. J Chromatogr B. 2011;879(23):2259–64.
Weaver DR, Reppert SM. Matkrnal melatonin communicates daylength to the fetus in Djungarian hamsters. Endocrinology. 1986;119(6):2861–3.
Yellon S, Tamarkin L, Goldman B. Maturation of the pineal melatonin rhythm in long-and short-day reared Djungarian hamsters. Experientia. 1985;41(5):651–2.
Acknowledgements
The authors would like to thank FAPESP (process numbers 11/50400-0, 15/06809-1, and 14/00084-2), INCT, CAPES, and CNPq for the financial support that allowed this research to be conducted.
Authors’ contributions
MSF and DNO performed all experiments and validations in mass spectrometry and wrote the paper. CCM and APLB performed the sample collection and provided animal care throughout the experiments. GFA and RRC conceived the experiments, supervised the research teams, and approved the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1: Figure S1.
Fragment structures of (A) MS2 of precursor ion m/z 233 and (B) MS3 of its secondary ion m/z 174 by MALDI-MSI. (PDF 16 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Ferreira, M.S., de Oliveira, D.N., Mesquita, C.C. et al. MALDI-MSI: a fast and reliable method for direct melatonin quantification in biological fluids. J Anal Sci Technol 7, 28 (2016). https://doi.org/10.1186/s40543-016-0106-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40543-016-0106-5