Abstract
Background
Bilateral symmetry flower (zygomorphy) is the ancestral state for Gesneriaceae species. Yet independent reversions to actinomorphy have been parallelly evolved in several lineages. Conandron ramondioides is a natural radially symmetrical species survived in dense shade mountainous habitats where specialist pollinators are scarce. Whether the mutations in floral symmetry genes such as CYC, RAD and DIV genes, or their expression pattern shifts contribute to the reversion to actinomorphy in C. ramondioides thus facilitating shifts to generalist pollinators remain to be investigated. To address this, we isolated putative orthologues of these genes and relate their expressions to developmental stages of flower actinomorphy.
Results
Tissue specific RT-PCR found no dorsal identity genes CrCYCs and CrRADs expression in petal and stamen whorls, while the ventral identity gene CrDIV was expressed in all petals. Thus, ventralized actinomorphy is evolved in C. ramondioides. However, CrCYCs still persists their expression in sepal whorl. This is congruent with previous findings that CYC expression in sepals is an ancestral state common to both actinomorphic and zygomorphic core Eudicot species.
Conclusions
The loss of dorsal identity genes CrCYCs and CrRADs expression in petal and stamen whorl without mutating these genes specifies that a novel regulation change, possibly on cis-elements of these genes, has evolved to switch zygomorphy to actinomorphy.
Similar content being viewed by others
Background
Evolutionary reversal to actinomorphy (flower radial symmetry) from zygomorphy (flower bilateral symmetry) have occurred multiple times independently across flowering plant diversification (Hileman 2014). Although zygomorphy enhances pollination specialization, these reversals may have evolved in a benefit of increased pollinator generalization when pollinators are scarce or in harsh conditions (Cronk and Möller 1997; Donoghue et al. 1998). The frequent transitions of floral symmetry reversals along many flowering plant lineages imply that a similar or modified developmental program has been independently recruited for floral symmetry transition (Zhang et al. 2013; Hileman 2014).
Gesneriaceae species are predominantly bilateral flower symmetry (zygomorphy) and exhibit a great diversity of floral forms (Endress 1999, 2001; Weber 2004). Their flowers have been adaptively evolved with bee, fly, moth, birds and even bat into a variety of pollination syndromes (Harrison et al. 1999; Perret et al. 2007). However, there are also certain lineages evolved with flower reversion to actinomorphy. There are at least five and four independent reversals to actinomorphy events occurred in old world and new world Gesneriaceae lineages, respectively (Wang et al. 2010; Smith et al. 2004; Clark et al. 2011). This frequency of reversals to actinomorphy in Gesneriaceae is the highest among Lamiales, probably because it is the basal most lineage of Lamiales which is just derived from the ourgroup actinomorphic Oleaceae species (Endress 1999). The repeated reversals to actinomorphy in Gesneriaceae species also implied there are perhaps similar yet modified developmental programs repeatedly recruited among these reversals.
It has been argued that the reversals to actinomorphy may contain selection disadvantage by losing its specific pollinators (Cronk and Möller 1997). However, in the aforementioned examples, the actinomorphic floral forms caused by reversals in Gesneriaceae species are usually compromised by pollinator shifts and pollination strategies switched from nectar to pollen rewards (Weber 2004). The reversal to actinomorphy with corolla tube fully opened to attract every kinds of general pollinators may be selected for when only few pollinators are available (Cronk and Möller 1997). For example, the European relict actinomorphic species in Pyrenees, Ramonda myconi, has been inferred that reversion to actinomorphy by opening of the corolla tube provides adaptive advantage to attract more available general pollinators in harsh alpine habitat (Cronk and Möller 1997; Picó et al. 2002). Additionally, its pollination syndrome has shifted from nectar reward which is usually favored by specific pollinators to pollen reward allowing general pollinators to visit (Weber 2004; Wang et al. 2010).
Conandron ramondioides is another relic and paleoendemic genus in East Asia which is also apparently evolved as an actinomorphic reversal from zygomorphic ancestor (Kokubugata and Peng 2004; Wang et al. 2010; Xiao et al. 2012). Like R. myconi, its corolla tube is lost and five petals are equally large with five stamens fully developed. These indicate they are natural peloria with complete actinomorphy. Nectary glands of C. ramondioides are lost. Also, its stamens are dehisced by apical pores and pollens are powdery suggest they are pollinated by pollen-collecting bees (Wang, pers. obs.). Conandron ramondioides can only survive in limestone cliff, often in deep shade forest, where pollinators are scarce. Taken together, the reversal to actinomorphy in C. ramondioides could prevent it from relying only on specific pollinators because insect activity is relative low under this habitat.
The establishment of flower zygomorphy requires CYCLOIDEA (CYC) gene specifically expressed in dorsal side of the flower to promote growth difference between dorsal and ventral petals and retarding dorsal stamen (Luo et al. 1996, 1999; Corley et al. 2005). Thus the reversal to actinomorphy in C. ramondioides could probably results from loss of CYC function or a CYC expression shifts in flower bud. In Antirrhinum majus, mutation of CYC and its paralog DICH can result in complete actinomorphy with all petals equal in size resembling the ventral one and no retardation on stamens. CYC in C. ramondioides may also be mutated thus becoming actinomorphy. However, CYC and DICH’s effect is through activating a downstream MYB family gene RADIALIS (RAD) at dorsal side, whose encoded protein restricts another MYB-like protein DIVARICATA (DIV) to the ventral region (Corley et al. 2005; Costa et al. 2005; Raimundo et al. 2013). Thus in addition to CYC, fully functional RAD is also needed to develop flower zygomorphy. In cyc dich double mutant, RAD could not be activated to restrict ventral identity gene DIV to ventral region resulting in all petals resembling to ventral petal of wild type. This type of actinomorphic reversal due to mutation of CYC and/or its downstream RAD therefore is often called abaxialized (ventralized) effect (Cronk 2006; Zhang et al. 2013; Hileman 2014; Spencer and Kim 2017).
In contrast to ventralization, actinomorphy can be established through expanded expression of CYC and its homologues in petal whorl, an adaxialized (dorsalized) effect. In actinomorphic species such as Cadia of legumes and certain Malpighiaceae species, the expression of CYC extended from dorsal regions to lateral and ventral regions of the corolla (Citerne et al. 2006; Zhang et al. 2013). It thus appears that both the loss of CYC-like gene expression (ventralization) and the expansion (dorsalization) of CYC-like gene expression are two major mechanisms in creating flower actinomorphic reversal in angiosperm.
In Gesneriaceae, reversal to actinomorphy through both ventralization or dorsalization were reported (e.g. Bournea leiophylla, Tengia scopulorum and Saintpaulia ionantha) (Zhou et al. 2008; Pang et al. 2010; Hsu et al. 2018). In B. leiophylla, the BlCYC1 and BlRAD genes were transiently expressed in floral meristem initiation stage and then quickly vanished at latter developmental stages. The loss of CYC expression at later stages correlates with the fact that all petals resembling ventral ones, demonstrating a ventralization form of reversal (Zhou et al. 2008). Unlike B. leiophylla, CYC-like expression in T. scopulorum has ubiquitously expressed in all petals across dorsoventral axis, a dorsalized form of actinomorphic reversal (Pang et al. 2010). Partial CYC sequences have been isolated from both C. ramondioides and R. myconi but no apparent SNP mutations result in premature stop codon in their coding regions (Xiao and Wang 2007; Picó et al. 2002). These imply CYC in C. ramondioides may still function. It would therefore essential to investigate whether expression patterns of CYC together with downstream RAD and DIV shifts, which may correlate and explain the developmental switch of actinomorphic reversal in C. ramondioides.
To examine possible roles of floral symmetry genes (homologues of CYC, RAD and DIV) involving in establishment of actinomorphic in C. ramondioides, we examined their expression patterns along floral developmental stages and separated floral organs. In order to ascertain the developmental process of actinomorphy in C. ramondioides, we also observed the bud development using scanning electron microscope. From these results, we hope to find whether there is a correlation between shifts of expression patterns among these floral symmetry genes and corresponded floral symmetry transition.
Materials and methods
Floral development in C. ramondioides
Floral buds of C. ramondioides were collected from top of inflorescence containing developing floral organ primordia for SEM examination. The materials were fixed in FAA overnight then transfer to 70% EtOH for preservation. Fixed materials were pre-dissected under stereo microscope S8APO (Leica) then dehydrated through an ethanol series (85%, 95%, and 100% twice) with each step for 20 min. After dehydration in 100% ethanol, materials were dehydrated through ascending gradients of acetone, dried with molecular sieve, and finally dried in a critical point dryer (Hitachi E101). Dried samples were mounted on aluminum stubs and then coated with gold–palladium using Hitachi E1011I sputter. Specimens were viewed using FEI SEM at working distance at 10 mm, and operating at 15 kV. Stages of flower development were summarized in Additional file 1: Table S1.
Isolation and characterization of CYC, RAD and DIV homologues
CYC, RAD and DIV homologues were isolated from C. ramondioides total genomic DNA by using degenerate primer pairs. We used either previously published primers which have been claimed able to amplify all possible gene copies, or newly designed primers located at conserve domain of each gene to isolate these floral symmetry gene copies. For example, for isolating CYC homologues from C. ramondioides, a pair of Geseneriaceae specific primers, GcycFS and GcycR, were used for amplifying CYC homologues (Möller et al. 1999). For RAD, primers were design from conserved MYB domain with all available GenBank sequences from Gesneriaceae species and Antirrhinum. Similarly, DIV primers were design from conserved R2 and R3 domain. To isolate CYC homologues from C. ramondioides, The PCR products were then cloned into pGEM-T easy vector system (Promega, USA) and 8 clones were sequenced for checking numbers of CYC homologues of C. ramondioides. Then, to isolate RAD homologues of C. ramondioides, degenerate primer pair QAL-F (5′-RTTRGCRGTKTAYGACA-3′) and FPN-R (5′-TTYCCYAAYTACWGGACCA-3′) locating at conserve MYB domain and conserve 3′ end were designed according to available RAD homologues of other Gesneriaceae species. Last, to isolate DIV homologues of C. ramondioides, degenerate primer pair DIV-F MEI (5′-ATGGAGATTTTRDCMCCAAGTT-3′) and DIV-YGK-R1 (5′-CTCCARTCYCCYTTYCCATA-3′) locating at R2 end and R3 domain were designed based on available Gesneriaceae sequences and DIV form A. majus respectively. Both RAD and DIV homologue PCR products were cloned and sequenced (8 clones for examining RAD homologue; 7 clones for examining DIV homologue) following the same process as previously mentioned above. To extend into the 5′ and 3′ unknown sequence region of amplified RAD and DIV partial sequence above, rapid amplification of cDNA ends (5′- and 3′-RACE, SMART RACE cDNA amplification kit, Clontech) technique is applied for obtaining full length cDNA according to manufacturer suggestion. To investigate the homology of isolated CYC, RAD and DIV of C. ramondioides, we aligned full length sequences of them with their genbank available homologs from subfamily Didymocarpoideae where it belongs to and those from closely related model species Antirrhinum majus (Scrophulariaceae) (Luo et al. 1996; Almeida et al. 1997; Zhou et al. 2008; Yang et al. 2012).
Phylogenetic analysis of isolated CYC, RAD and DIV homologues in C. ramondioides
To check the homology of these isolated CYC, RAD and DIV genes from C. ramondioides, available NCBI homologues of Gesneriaceae species, Antirrhinum majus and Arabidopsis thaliana were downloaded and used for reconstructing their phylogenies respectively. Sequences used to reconstruct phylogeny were listed in Additional file 2: Table S2. Nucleotide sequences were first translated into amino acid and aligned using default settings in CLUSTALX (Thompson et al. 1997) with major domains specified, then manually aligned afterward. We apply both neighbor joining (NJ) and maximum likelihood (ML) algorithm for testing the robustness of reconstructed CYC, RAD and DIV phylogeny. The NJ tree of each gene dataset was reconstructed using MEGA 6 (Tamura et al. 2013). For ML tree, the web interface PhyML 3.0 was applied (Guindon et al. 2010). Best-fit nucleotide substitution model of each dataset was evaluated by smart model selection (SMS) which is implementing in PhyML 3.0 (Lefort et al. 2017). For CYC-like gene dataset, the best-fit model is HKY + G model. For RAD, TN93 + G model is suggested and GTR + I + G model for DIV.
Locus-specific RT-PCR
Flower buds were categorized into three stages (Fig. 1): early stage (Stage 10, 2–4 mm in diameter, sepal longer than petal), middle stage (Stage 13, 5–7 mm in diameter, petal longer than sepal), and late stage (Stage 15, anthesis) (see Additional file 1: Table S1). They were collected, in the field through fixing in RNAlater (Ambion, Life technologies, USA), or freshly collected from individuals grown in the greenhouse. Next, to detect expression locations of CYC, RAD and DIV homologues on petals, a single petal was dissected from flower buds at early developmental stage. Because RNA yield may be low in single petal, pooled sepals, petals, stamen and gynoecium were dissected from flower buds at early stage to confirm the expression locations of CYC, RAD and DIV homologues in C. ramondioides. Total RNA of floral buds and dissected floral organs were extracted following TRIzol® Reagent (Invitrogen, USA) protocol. Single-strand cDNA (20 ng/μl) were reverse transcribed from total RNA of these samples by SuperscriptIV Reverse Transcriptase (Invitrogen). Gene specific primer pairs were used to examine each candidate gene’s expression level: CrCYC1C (forward: 5′-AGACATGCTTTCTGGCCACT-3′, Reverse: 5′-CTTCTTCGCCTTCTGAATGC-3′), CrCYC1D (Forward: 5′-CAGGTGCAGATTCGATGAGA-3′, Reverse: 5′-GTTCCATTGCAGTCTCCCAT-3′), CrCYC2 (Forward: 5′-TCTTGCTTCATCAGCACCAC-3′, Reverse: 5′-GTGATGCCCCTACTTGCACT-3′), CrRAD1 (Forward: 5′-ATGGCCTCAAGTTCCTTGACCGCT, Reverse: 5′-GTGGTCCAGTAGTTGGGAAATGGT), CrRAD2 (Forward: 5′-GCAATGAGCTCCATGTCTAGTGGT, Reverse: 5′-CCCACTTCGATATTCTTCACATCCTCC-3′) and CrDIV (Forward: 5′-CGATCGATGGCAAAGAGTGG-3′, Reverse: 5′-TACTCGAACACCAACCCAGG-3′). The RT-PCR mixture contains 12 μl of Ampliqon master mix Red III (Denmark), 6.25 mM of each primer, 9.5 μl ddH2O and 1 μl first-strand cDNA. The PCR condition used for amplifying CrCYC1C, CrCYC1D and CrCYC2 genes were 94 °C for 3 min followed by 35 cycles of 94 °C for 30 s, 53 °C for 40 s, 72 °C for 20 s and a 2 min final extension at 72 °C. To amplify higher Tm CrRAD1 and CrRAD2 genes, the program is 94 °C for 3 min followed by 35 cycles of 94 °C for 30 s, 57 °C for 40 s, 72 °C for 20 s and a 2 min final extension at 72 °C. For CrDIV gene, the program is 94 °C for 3 min followed by 35 cycles of 94 °C for 30 s, 53 °C for 40 s, 72 °C for 20 s and a 2 min final extension at 72 °C. Cr18S was used as positive control. Two biological replicates were carried out to validate the reproducibility of the results (see Additional file 3: Figure S1).
The flower of C. ramondioides from early to late stage. Early, middle and late stage corresponding to Stage 10, 13, 15 in Additional file 1: Table S1. Actinomorphy was observed in late stage C. ramondioides flower. Scale bar represented 1 cm
Results
Floral development
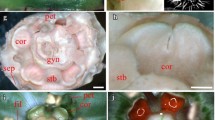
Development of flower in C. ramondioides can be divided into 15 milestone stages (Additional file 1: Table S1). From Stage 3 SEM picture (Fig. 2a), all five sepals were already initiated but dorsal and lateral sepals were slightly smaller than ventral ones (a residual zygomorphy). During Stage 7 (sepal removed), petals and stamens appeared to be equaled in size when initiated (Fig. 2c). This is more evident during Stage 7A when gynoecium started to emerged, in which all petals and stamens were grown in equal size (Fig. 2d). Petals and stamens continued to grow in equal rate thus all 5 petals and stamens are the same size toward anthesis (Fig. 2e). The floral diagram of fully developed C. ramondioides thus can be drawn as Fig. 2f, showing complete actinomorphy of C. ramondioides flower at anthesis.
The SEM photos of morphological development process of Conandron ramondioides flowers. Zygomorphy was observed at Stage 3 of sepal whorl. Definition of developmental stages of C. ramondioides based on (Harrison et al. 1999). a Stage 3, the sepals form as bulges at the points of the pentagon. b Stage 4, the sepals grow, while the floral meristem remains undifferentiated. c Stage 6, the corolla and androecium grow, and the gynoecium initiates. d Stage 7A, petal growth. e Stage 7B, androecium and gynoecium development. f Floral diagram of C. ramondioides. Se Sepals, Pe petals, S stamen, G gynoecium. Scale bar represents 50 μm
Characterization and phylogeny of CrCYC, CrRAD and CrDIV genes
Among CYC clones, three out of 8 belonged to CrCYC1C, two belongs to CrCYC1D and three belongs to CrCYC2. For CrRADs, four clones belonged to CrRAD1, while the other four were CrRAD2. For CrDIV, all seven clones belonged to CrDIV. We believed our approach can effectively isolate all possible copies of each gene because we designed the primers in most conserved domain of each gene (see “Material and methods” section). Full-length cDNA of CrCYC genes, CrRAD genes and CrDIV were isolated from developing floral tissues and dissected tissues. The CrCYC, CrRAD and CrDIV sequences we isolated from C. ramondioides have been deposited in NCBI database (Accession number MH366524 to MH366529, detailed information see Additional file 2: Table S2). There are three CYC homologs, CrCYC1C, CrCYC1D, CrCYC2 identified in C. ramondioides (Fig. 3a). Their full length amino acid sequences are 339, 338, and 335, respectively. According to phylogeny (Fig. 4a), we designated them as CrCYC1C, CrCYC1D and CrCYC2. Sequence analysis shows that CrCYC1C, CrCYC1D and CrCYC2 are 26.1%, 17% and 23% identical to Antirrhinum CYC, respectively. When comparing the TCP domain, R domain, and ECE domains, CrCYC1C, CrCYC1D, and CrCYC2 shared 92.6%, 90.2% and 92.6% amino acid sequence identity with Antirrhinum CYC, suggesting these genes are functionally related. When compared with CYC-like genes from available closely related Didymocarpoid Gesneriaceae species, CrCYC1C and CrCYC1D are 86% and 78.4% identical to BlCYC1 of Bournea leiophylla, respectively, and CrCYC2 is 88.1% identical to BlCYC2.
Alignments of protein sequences of CrCYC, CrRAD and CrDIV genes with homologs from Antirrhinum majus (CYC, RAD and DIV) and Bournea leiophylla (BlCYC1, BlCYC2, BlRAD, BlDIV1 and BlDIV2). a Alignmentes of CYC homologs. TCP, ECE and R domain are outlined. Identical amino acids are in black and similar amino acids are in gray. b Alignments of RAD homologs. MYB domain are outlined. c Alignments of DIV homologs. Two MYB domains (R2 and R3) are outlined. A highly conserved SHAQKY motif in R3 MYB domain is identified and labelled in white box. Arrows indicate sequence region used in phylogeny analysis. Sequence region used in phylogenetic analysis coving almost all important domains in all three gene dataset except for PlDIV in DIV dataset
Neighbour-joining trees of CYC-like, RAD-like and DIV-like genes. Trees from a to c are reconstructed based on amino acid sequences. a CYC is from A. majus, others are CYC-like genes from Gesneriaceae species (see Additional file 2: Table S2). Trees show CrCYCs cluster into three groups with high support. b The tree shows CrRADs cluster into two distinct clade with high support. c Bootstrap values from NJ and ML are listed above and below branches respectively. Bootstrap values > 70 are shown. Detailed sequences information is listed in Additional file 2: Table S2
The sequence difference between these CYC-like genes are mainly located in the intervening regions of CYC domains mentioned above. Phylogenetic analysis shows that CrCYC1C, CrCYC1D (first isolated in this study) and CrCYC2 formed three monophyletic clades (GCYC1C, GCYC1D and GCYC2) with other Gesneriaceae CYC homologs at amino acid level, confirming to previous phylogenetic trees (Fig. 4a).
Next, two RAD homologues isolated from C. ramondioides were CrRAD1 and CrDAD2. They shared 76% and 68% amino acid identity with RAD from A. majus, respectively. Both CrRAD1 and CrDAD2 have one conserved 55aa-MYB-domain as RAD does (Fig. 3b). Phylogenetic analyses based on neighbor-joining method showed that CrRAD1 and CrRAD2 formed two distinct clades with high support (bootstrap/ML: 96/99 for RAD1 clade; 100/100 for RAD2 clade) (Fig. 4b). With A. majus RAD as outgroup, CrRAD1 formed one monophyletic clade with Didymocarpoid Generiaceae species such as RAD1 of Saintpaulia ionantha with high support, while CrRAD2 formed another monophyletic clade with Saintpaulia RAD2 and RAD-like gene of B. leiophylla according to nucleotide and amino acid NJ tree (Fig. 4b).
We only isolated one DIV homolog (CrDIV) from C. ramondioides which encode a protein of 296 amino acids. CrDIV is 54% identical to DIV from A. majus at amino acid level, and 90% and 89% identical to BlDIV1 and BlDIV2 from B. leiophylla. Conserved R2 and R3 domain and MYB-specific motif “SHAQKY” were found in CrDIV (Fig. 3c). Phylogenetic trees reconstructed from nucleotide and amino acid showed that CrDIV forms a monophyletic clade with BlDIV1 (Fig. 4c).
Tissue-specific expressions of CrCYC, CrRAD and CrDIV
Transcripts of CrCYC1C and CrCYC1D were first detected in the early developmental stage (E) of the floral bud and then the expression of CrCYC1C and CrCYC1D were gradually reduced to almost invisible from middle (M) to late stage (L) of entire floral buds (Fig. 5). To further confirm organ expression pattern of CrCYC1C and CrCYC1D, pooled tissues including petals, gynoecium, stamens and sepals dissected from early development stage were used. Surprisingly, transcripts of CrCYC1C and CrCYC1D were both only detected in sepals (Fig. 5). This unique expression pattern of CrCYC1C and CrCYC1D in C. ramondioides seems to correlate with the residual zygomorphy found in sepal whorl (see Fig. 2a). As to CrCYC2, its expression was undetectable throughout all flower developmental stages and dissected organs. In summary, our RT-PCR results showed that no CrCYCs transcripts were detectable at petal and stamen whorl.
Gene-specific reverse transcriptase polymerase chain reaction (RT-PCR) analysis of CrCYC, CrRAD and CrDIV genes from C. ramondioides buds and dissected flower tissues. E, M, L represent three flower development stage defined in this study. E stands for early flower development stage; M stands for middle flower development stage; L stands for anthesis stage. P1 to P5 represent dissected petal from flower bud at early (E) flower developmental stage. Sep, Pe, Sta and gyn denote pooled sepals, pooled petals, pooled stamens and gynoecium dissected from early flower developmental stage. CrCYC1C, CrCYC1D and CrCYC2 indicate expression of CrCYC1C, CrCYC1D and CrCYC2. CrRAD1 and CrRAD2 indicate expression of CrRAD1 and CrRAD2. CrDIV indicates expression of CrDIV. 18S is included as a positive control. CrCYC1C, CrCYC1D, CrRAD1, CrRAD2 and CrDIV are detected through flower development stages, only CrCYC2 is restricted through flower development stages. CrDIV is detected in all petals, whereas CrCYC1C, CrCYC1D, CrCYC2, CrRAD1 and CrRAD2 are restricted in petals. CrCYC1C and CrCYC2 were detected in pooled sepal tissue
As to CrRADs, CrRAD1 expression can be detected throughout all flower developmental stages but CrRAD2 could only be detected in the early floral development stage (Fig. 5). Their organ expression pattern revealed that both CrRAD1 and CrRAD2 expressed in stamens, while CrRAD2 also expressed in gynoecium (Fig. 5). The CrDIV was continuously expressed throughout all flower developmental stages. Specifically, CrDIV expressed in petals, stamens and gynoecium but not in sepals (Fig. 5).
Discussion
Reversal to actinomorphy correlate to diverse CYC expression shifts in Gesneriaceae and other eudicot lineages
Our SEM pictures showed that all five petals of C. ramondioides seems to grow in consistent rate of enlargement ever since petal primordia initiation stages (Fig. 2). Similarly, this was also observed in the stamen whorl. Development of actinomorphy in C. ramondioides therefore resembles to that in cyc/dich mutant of A. majus in which all petal primordia maintain equal growth rate along flower developmental stages and all stamens are fully developed (Luo et al. 1996).
From RT-PCR results, CrCYCs and CrRADs have no sign of expression in petal whorl (Fig. 5). This suggests C. ramondioides is a ventralized actinomorphy. In Gesneriaceae, ventralized actinomorphy has been reported in the peloric cultivar of Sinningia speciosa where CYC is not expressed in petal whorl because a deletion in CYC coding region (Hsu et al. 2015, 2017; Wang et al. 2015). Loss of CYC expression in late petal developmental stage but not in early stage can also contribute to actinomorphic reversal in B. leiophylla (Zhou et al. 2008). On the other hand, dorsalized actinomorphy due to ubiquitous CYC expression in entire petal whorl has been reported in T. scopulorum and occasionally in Petrocosmea hybrids (Pang et al. 2010; Yang et al. 2015). Other than Gesneriaceae, examples of dorsalized actinomorphy have been reported in Cardia purpurea (Leguminosae), in Aquilegia alpina (Ranunculaceae), and in certain Malpighiaceae lineages (Citerne et al. 2006; Zhang et al. 2010, 2012, 2013; Jabbour et al. 2014). While ventralized actinomorphy where the loss of CYC expression has been documented in Tradescantia (Commelinaceae), in Nigella damascene (Ranunculaceae), in Plantago lanceolata (Plantaginaceae), in Malpighiaceae lineages and in Arabidopsis (Cubas et al. 2001; Reardon et al. 2009; Preston et al. 2011; Preston and Hileman 2012; Zhang et al. 2013; Jabbour et al. 2014).
Although we did not detect any CrCYC1C and CrCYC1D expression in petal and stamen whorls, we did find them distinctively expressed in sepals. Sepal specific expression of CYC has been inferred the ancestral state among Ranunculales including both actinomorphic Papaveraceae species and zygomorphic Fumariaceae species, the basal most core Eudicot lineage (Damerval et al. 2007). Thus, the shifts of CrCYC1C/1D back to ancestral expression in sepals and cease of expression in petals/stamens seem to associate with the actinomorphic reversal in C. ramondioides. Indeed, the recruitment of CYC expression (ECE clade) in sepal was evolved earlier than expression in dorsal-specific manner (Preston and Hileman 2009). It would be interesting to examine whether all those reversals to actinomorphy species (Wang et al. 2010; Clark et al. 2011) in Geseneriaceae also have their CYC expression return to ancestral state of sepal expression.
Duplications of CYC may associate to expression shift and flower shape variation
Our phylogenetic analysis revealed that the three CrCYCs we isolated (CrCYC1C, CrCYC1D and CrCYC2) were resulted from at least two duplication events among Gesneriaceae species, congruent to previous findings (Möller et al. 1999; Citerne et al. 2000; Wang et al. 2004; Du and Wang 2008; Song et al. 2009; Pang et al. 2010; Yang et al. 2015). Although these CrCYCs have lost their expression in petal and stamen whorls, which is correlate to the reversal to actinomorphy in C. ramondioides, their coding sequences contain no frame shift or nonsense mutations. This implies these CrCYCs may still function yet the regulation controls on cis-elements in their promoter regions could have been mutated to become mis-expression (i.e. sepal only expression). It would be interesting to test this hypothesis by ectopically express these CrCYCs separately in Arabidopsis and compare their phenotypic effects.
In snapdragon, CYC evolved as major effect copy (higher and broader dorsal-specific expression) and its duplicates DICH as helper function (Luo et al. 1996, 1999). Similarly, CYC tend to duplicate in most Angiosperm lineages with either both copies retain similar expression pattern (major/helper) or expressions become diversified thus under different selection pressures (Ree et al. 2004; Chapman et al. 2008; Bello et al. 2017). In C. ramondioides, CrCYC1C and CrCYC1D both have sepal only expression but expression level of CrCYC1C is higher than CrCYC1D. This is similar to the case of CYC and DICH in snapdragon. Duplication could allow one copy to maintain the essential function but the other to evolve into novel or modified function. CYC duplications may therefore link to the evolution of diverse floral shape in angiosperms, although yet to be determined. There are reports, however, indicating shifts of expression between CYC paralogs correlate to floral symmetry transitions and/or flower shape variations (Bartlett and Specht 2011; Zhang et al. 2012; Jabbour et al. 2014).
Specific expression patterns of CrRADs and CrDIV may suggest loss of antagonistic expression pattern between CrRADs and CrDIV following loss of CYC expression in C. ramondioides
In cyc/dich double mutant A. majus, RAD does not express in dorsal region, thus, allowing ventral region restricted DIV to spread to whole flower (Corley et al. 2005). In B. leiophylla, once dorsal region restricted BlCYC1 and BlRAD are downregulated, BlDIV spread to corolla and stamen whorl (Zhou et al. 2008). To sum up, RAD-like genes and DIV-like genes seem have antagonistic function in A. majus and B. leiophylla. However, expression patterns of CrRADs and CrDIV are different from those gene expression pattern in A. majus and B. leiophylla. In C. ramondioides, both CrRADs and CrDIV expressed in stamens (including CrRAD1, CrRAD2 and CrDIV) and gynoecium (including CrRAD2 and CrDIV) at the same development stage (Fig. 5). Comparing expression patterns of RAD-like and DIV-like genes among these three species, antagonistic function seem have lost in C. ramondioides. Based on expression patterns from these three species, we postulate that loss of expression of upstream gene (e.g. CYC and its homologue) may provide opportunity for its downstream gene (e.g. RAD and its homologue, DIV and its homologue) releasing from genetic constraint. To conclude, antagonistic expression pattern between RAD-like and DIV-like genes was maintained in B. leiophylla, which resembling to CYC-mediated regulatory pathway in A. majus. However, this antagonistic expression pattern was lost in C. ramondioides. Since the RAD-like and DIV-like genes were rarely studied in Gesneriaceae, it would be interesting to examine whether the maintaining or loss of antagonistic function between RAD-like and DIV-like genes is a common pattern or not in Gesneriaceae.
Reversal to actinomorphy may help to attract general pollinator visiting C. ramondioides
In C. ramondioides, reversal to actinomorphy coupling with very short corolla tube at anthesis may facilitate generalist (e.g. bees, small beetles) visitation because they can obtain pollen from any direction (Fig. 1). Similarly, it has been postulated that reversal to actinomorphy in R. myconii could allow visits from a wider range of pollinators in alpine extreme habitats (Cronk and Möller 1997). In alpine or harsh conditions, plants may suffer low pollinator visiting. If certain zygomorphic species still rely on their specific pollinator visiting in harsh condition, their reproductive success may be low. But if species which can reverse to actinomorphy by opening the corolla, such as C. ramondioides and R. myconii, they could have pollinator shifts to a variety of pollinators to increase visiting rate. Although C. ramondioides is not distributed in alpine environment like R. myconii, its deep-forest dense shape habitat may discourage insect pollinator visiting (Peat and Goulson 2005). Reversal to actinomorphy to attract more general pollinators in pollinator scarce habitats may actually compensate for maintaining the reproductive success in C. ramondioides. To support this idea, detailed pollination experiment and pollinator observation in the field are necessary in the future.
Abbreviations
- Sep:
-
pooled sepals
- Pe:
-
pooled petals
- Sta:
-
pooled stamens
- gyn:
-
gynoecium
References
Almeida J, Rocheta M, Galego L (1997) Genetic control of flower shape in Antirrhinum majus. Development 124:1387
Bartlett ME, Specht CD (2011) Changes in expression pattern of the teosinte branched1-like genes in the Zingiberales provide a mechanism for evolutionary shifts in symmetry across the order. Am J Bot 98:227–243
Bello MA, Cubas P, Álvarez I, Sanjuanbenito G, Fuertes-Aguilar J (2017) Evolution and expression patterns of CYC/TB1 genes in anacyclus: phylogenetic insights for floral symmetry genes in Asteraceae. Front Plant Sci 8:589
Chapman MA, Leebens-Mack JH, Burke JM (2008) Positive selection and expression divergence following gene duplication in the sunflower cycloidea gene family. Mol Biol Evol 25:1260–1273
Citerne HL, Möller M, Cronk QCB (2000) Diversity of cycloidea-like genes in gesneriaceae in relation to floral symmetry. Ann Bot 86:167–176
Citerne HL, Pennington RT, Cronk QCB (2006) An apparent reversal in floral symmetry in the legume Cadia is a homeotic transformation. Proc Natl Acad Sci 103:12017–12020
Clark JL, Roalson EH, Pritchard RA, Coleman CL, Teoh V-H, Matos J (2011) independent origin of radial floral symmetry in the Gloxinieae (Gesnerioideae: gesneriaceae) is Supported by the rediscovery of Phinaea pulchella in Cuba. Syst Bot 36:757–767
Corley SB, Carpenter R, Copsey L, Coen E (2005) Floral asymmetry involves an interplay between TCP and MYB transcription factors in Antirrhinum. Proc Natl Acad Sci USA 102:5068–5073
Costa MMR, Fox S, Hanna AI, Baxter C, Coen E (2005) Evolution of regulatory interactions controlling floral asymmetry. Development 132:5093
Cronk QCB (2006) Legume flowers bear fruit. Proc Natl Acad Sci USA 103:4801–4802
Cronk Q, Möller M (1997) Genetics of floral symmetry revealed. Trends Ecol Evol 12:85–86
Cubas P, Coen E, Zapater JMMN (2001) Ancient asymmetries in the evolution of flowers. Curr Biol 11:1050–1052
Damerval C, Guilloux ML, Jager M, Charon C (2007) Diversity and evolution of CYCLOIDEA-Like TCP genes in relation to flower development in Papaveraceae. Plant Physiol 143:759
Donoghue MJ, Ree RH, Baum DA (1998) Phylogeny and the evolution of flower symmetry in the Asteridae. Trends Plant Sci 3:311–317
Du Z-Y, Wang Y-Z (2008) Significance of RT-PCR expression patterns of CYC-like genes in Oreocharis benthamii (Gesneriaceae). J Syst Evol. https://doi.org/10.1007/s00427-008-0227-y
Endress PK (1999) Symmetry in flowers: diversity and evolution. Int J Plant Sci 160:S3–S23
Endress PK (2001) Evolution of floral symmetry. Curr Opin Plant Biol 4:86–91
Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Harrison CJ, MÖller M, Cronk QCB (1999) Evolution and development of floral diversity in Streptocarpus and Saintpaulia. Ann Bot 84:49–60
Hileman LC (2014) Trends in flower symmetry evolution revealed through phylogenetic and developmental genetic advances. Philos Trans R Soc B Biol Sci. https://doi.org/10.1098/rstb.2013.0348
Hsu H-C, Chen C-Y, Lee T-K, Weng L-K, Yeh D-M, Lin T-T, Wang C-N, Kuo Y-F (2015) Quantitative analysis of floral symmetry and tube dilation in an F2 cross of Sinningia speciosa. Sci Hortic 188:71–77
Hsu H-C, Wang C-N, Liang C-H, Wang C-C, Kuo Y-F (2017) Association between petal form variation and CYC2-like genotype in a hybrid line of Sinningia speciosa. Front Plant Sci 8:558
Hsu HJ, He CW, Kuo WH, Hsin KT, Lu JY, Pan ZJ, Wang CN (2018) Genetic analysis of floral symmetry transition in African violet suggests the involvement of trans-acting factor for CYCLOIDEA expression shifts. Front Plant Sci. 9:1008. https://doi.org/10.3389/fpls.2018.01008
Jabbour F, Cossard G, Le Guilloux M, Sannier J, Nadot S, Damerval C (2014) Specific duplication and dorsoventrally asymmetric expression patterns of cycloidea-like genes in Zygomorphic species of Ranunculaceae. PLoS ONE 9:e95727
Kokubugata G, Peng CI (2004) Floral morphology and recognition of varieties in Conandron ramondioides (Gesneriaceae) from Japan and Taiwan. Edinburgh J Bot 61:21–30
Lefort V, Longueville J-E, Gascuel O (2017) SMS: smart model selection in PhyML. Mol Biol Evol 34:2422–2424
Luo D, Carpenter R, Vincent C, Copsey L, Coen E (1996) Origin of floral asymmetry in Antirrhinum. Nature 383:794
Luo D, Carpenter R, Copsey L, Vincent C, Clark J, Coen E (1999) Control of organ asymmetry in flowers of Antirrhinum. Cell 99(4):367–376
Möller M, Clokie M, Cubas P, Cronk QCB (1999) Integrating molecular and developmental genetics: a Gesneriaceae case study. In: Hollingsworth PM, Bateman RJ, Gornall RJ (eds) Molecular systematics and plant evolution. Taylor & Francis, London, pp 375–402
Pang H-B, Sun Q-W, He S-Z, Wang Y-Z (2010) Expression pattern of CYC-like genes relating to a dorsalized actinomorphic flower in Tengia (Gesneriaceae). J Syst Evol 48:309–317
Peat J, Goulson D (2005) Effects of experience and weather on foraging rate and pollen versus nectar collection in the bumblebee, Bombus terrestris. Behav Ecol Sociobiol 58:152–156
Perret M, Chautems A, Spichiger R, Barraclough TG, Savolainen V (2007) The geographical pattern of speciation and floral diversification in the neotropics: THE Tribe Sinningieae (Gesneriaceae) as a case study. Evolution 61:1641–1660
Picó FX, Möller M, Ouborg NJ, Cronk QCB (2002) Single nucleotide polymorphisms in the coding region of the developmental gene Gcyc in natural populations of the relict Ramonda myconi (Gesneriaceae). Plant Biol 4:625–629
Preston JC, Hileman LC (2009) Developmental genetics of floral symmetry evolution. Trends Plant Sci 14(3):147–154
Preston JC, Hileman LC (2012) Parallel evolution of TCP and B-class genes in Commelinaceae flower bilateral symmetry. EvoDevo 3:6
Preston JC, Martinez CC, Hileman LC (2011) Gradual disintegration of the floral symmetry gene network is implicated in the evolution of a wind-pollination syndrome. Proc Natl Acad Sci 108:2343–2348
Raimundo J, Sobral R, Bailey P, Azevedo H, Galego L, Almeida J, Coen E, Costa Maria Manuela R (2013) A subcellular tug of war involving three MYB-like proteins underlies a molecular antagonism in Antirrhinum flower asymmetry. Plant J 75:527–538
Reardon W, Fitzpatrick DA, Fares MA, Nugent JM (2009) Evolution of flower shape in Plantago lanceolata. Plant Mol Biol 71:241–250
Ree RH, Citerne HL, Lavin M, Cronk QCB (2004) Heterogeneous selection on LEGCYC paralogs in relation to flower morphology and the phylogeny of Lupinus (Leguminosae). Mol Biol Evol 21:321–331
Smith JF, Hileman LC, Powell MP, Baum DA (2004) Evolution of GCYC, a Gesneriaceae homolog of CYCLOIDEA, within Gesnerioideae (Gesneriaceae). Mol Phylogenet Evol 31:765–779
Song C-F, Lin Q-B, Liang R-H, Wang Y-Z (2009) Expressions of ECE-CYC2 clade genes relating to abortion of both dorsal and ventral stamens in Opithandra (Gesneriaceae). BMC Evol Biol 9:244
Spencer V, Kim M (2017) Re“CYC”ling molecular regulators in the evolution and development of flower symmetry. Semin Cell Dev Biol. https://doi.org/10.1016/j.semcdb.2017.08.052
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Wang C-N, Möller M, Cronk QCB (2004) Phylogenetic position of Titanotrichum oldhamii (Gesneriaceae) inferred from four different gene regions. Syst Bot 29:407–418
Wang Y-Z, Liang R-H, Wang B-H, Li J-M, Qiu Z-J, Li Z-Y, Weber A (2010) Origin and phylogenetic relationships of the Old World Gesneriaceae with actinomorphic flowers inferred from ITS and trnL-trnF sequences. Taxon 59:1044–1052
Wang C-N, Hsu H-C, Wang C-C, Lee T-K, Kuo Y-F (2015) Quantifying floral shape variation in 3D using microcomputed tomography: a case study of a hybrid line between actinomorphic and zygomorphic flowers. Front Plant Sci 6:724
Weber A (2004) Gesneriaceae. In: Kubitzki K, Kadereit JW (eds) The families and genera of vascular plants. vol. 7. Flowering plants, Dicotyledons: Lamiales (except Acanthaceae including Avicenniaceae). Springer, Berlin, pp 63–158
Xiao LH, Wang YZ (2007) Single nucleotide polymorphisms of Gcyc1 (Cycloidea) in Conandron ramondioides (Gesneriaceae) from Southeast China. Plant Syst Evol 269:145–157
Xiao L-H, Li Z, Wang R, Wang Y-Z (2012) Population differentiation and phylogeographic pattern of a relict species, Conandron ramondioides (Gesneriaceae), revealed from sequence polymorphism and haplotypes of the CYCLOIDEA gene. J Syst Evol 50:45–57
Yang X, Pang H-B, Liu B-L, Qiu Z-J, Gao Q, Wei L, Dong Y, Wang Y-Z (2012) Evolution of double positive autoregulatory feedback loops in CYCLOIDEA2 clade genes is associated with the origin of floral zygomorphy. Plant Cell 24:1834
Yang X, Zhao X-G, Li C-Q, Liu J, Qiu Z-J, Dong Y, Wang Y-Z (2015) Distinct regulatory changes underlying differential expression of TEOSINTE BRANCHED1-CYCLOIDEA-PROLIFERATING CELL FACTOR genes associated with petal variations in zygomorphic flowers of Petrocosmea spp. of the family Gesneriaceae. Plant Physiol 169:2138–2151
Zhang W, Kramer EM, Davis CC (2010) Floral symmetry genes and the origin and maintenance of zygomorphy in a plant-pollinator mutualism. Proc Natl Acad Sci USA 107:6388–6393
Zhang W, Kramer EM, Davis CC (2012) Similar genetic mechanisms underlie the parallel evolution of floral phenotypes. PLoS ONE 7:e36033
Zhang W, Steinmann VW, Nikolov L, Kramer EM, Davis CC (2013) Divergent genetic mechanisms underlie reversals to radial floral symmetry from diverse zygomorphic flowered ancestors. Front Plant Sci 4:302
Zhou X-R, Wang Y-Z, Smith JF, Chen R (2008) Altered expression patterns of TCP and MYB genes relating to the floral developmental transition from initial zygomorphy to actinomorphy in Bournea (Gesneriaceae). New Phytol 178:532–543
Authors’ contribution
CNW conceived the context of the manuscript. CNW and KTH designed the experiment. KTH performed the experiment. Both authors read and approved the final manuscript.
Acknowledgements
We are grateful to the staffs including Dr. Shiang-Jiuun Chen, Ms. Yi-Chun Chuang, Ms. Ya-Chan Yang and Ms. Pei-Yin Wu of TC5 Bio-image Tools, Technology Commons, College of Life science, National Taiwan University for help with SEM operation and observation. We also thank Mr. Chun-Hsien Wu for help with isolating target genes in this study and Mr. Hong-Wen Ma for help with collecting floral buds of C. ramondioides in the field.
Competing interests
The authors declare that they have no competing interests. And there have neither financial competing interests nor other competing interests.
Availability of data and materials
Not applicable.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
We thank fundings from Ministry of Science and Technology of Taiwan MOST-106-2313-B-002-035-MY3 and National Science Council of Taiwan 95-2311-B-002-014-MY3 were granted to Chun-Neng Wang.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1: Table S1.
Development procedure of C. ramondioides flower.
Additional file 2: Table S2.
Sequences used to reconstruct CYC, RAD and DIV tree.
Additional file 3: Figure S1.
Biological repeats of gene-specific reverse transcriptase polymerase chain reaction (RT-PCR) analysis of CrCYC, CrRAD and CrDIV genes from C. ramondioides buds and dissected flower tissues. (a), (b) represent two biological repeats respectively. All abbreviations correspond to descriptions in Fig. 4.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Hsin, KT., Wang, CN. Expression shifts of floral symmetry genes correlate to flower actinomorphy in East Asia endemic Conandron ramondioides (Gesneriaceae). Bot Stud 59, 24 (2018). https://doi.org/10.1186/s40529-018-0242-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40529-018-0242-x