Abstract
The discovery of an expanded (GGGGCC)n repeat (termed G4C2) within the first intron of C9orf72 in familial ALS/FTD has led to a number of studies showing that the aberrant expression of G4C2 RNA can produce toxic dipeptides through repeat-associated non-AUG (RAN-) translation. To reveal canonical translation factors that impact this process, an unbiased loss-of-function screen was performed in a G4C2 fly model that maintained the upstream intronic sequence of the human gene and contained a GFP tag in the GR reading frame. 11 of 48 translation factors were identified that impact production of the GR-GFP protein. Further investigations into two of these, eIF4B and eIF4H, revealed that downregulation of these factors reduced toxicity caused by the expression of expanded G4C2 and reduced production of toxic GR dipeptides from G4C2 transcripts. In patient-derived cells and in post-mortem tissue from ALS/FTD patients, eIF4H was found to be downregulated in cases harboring the G4C2 mutation compared to patients lacking the mutation and healthy individuals. Overall, these data define eIF4B and eIF4H as disease modifiers whose activity is important for RAN-translation of the GR peptide from G4C2-transcripts.
Similar content being viewed by others
Introduction
In Amyotrophic Lateral Sclerosis (ALS) and Frontotemporal Degeneration (FTD), the presence of a hexanucleotide expansion of > 30 GGGGCC repeats (termed G4C2) within the C9orf72 gene is the most prominent mutation in familial disease [17, 65]. The mechanisms underlying potential toxicity associated with G4C2 are still being defined with two leading hypotheses centering around gain-of-function mechanisms [5, 93]: sequestration of RNA-binding proteins by the aberrant expression of sense- and antisense- G4C2 RNA [30, 90]; repeat-associated non-AUG (RAN-) translation of repeat-containing transcripts produce dipeptides that are toxic to neurons [4, 25, 47,48,49, 56, 58]. Five dipeptides can be produced from these transcripts, depending on the reading frame: GA and GR (sense strand associated), PA and PR (antisense strand associated), and GP (produced from both sense and antisense strands).
In recent years, it has become clear that dipeptides produced from G4C2 RNA transcripts cause neurodegenerative effects [5, 93]. Of the 5 potential RAN-translation products, GR and PR cause particularly strong degenerative phenotypes in multiple model systems, including Drosophila [22, 53]. Therefore, increasing understanding of the mechanisms underlying expression of these dipeptides would highlight potential therapeutic avenues centered around preventing their expression.
Many mechanistic questions remain regarding RAN-translation in G4C2-associated disease. Recent investigations have drawn a number of parallels between mechanisms underlying general translation [10, 77, 78] and RAN-translation [37, 96], finding that dipeptide production is sensitive to the inhibition/downregulation of canonical translation factors: eIF4E, eIF4G, eIF4A, eIF2α, eIF2A [12, 27, 84, 97]. Of interest, eIF4A is a DEAD-Box helicase [3], and thus may be important for the unwinding of G4C2-RNA for translation. While eIF4A has relative weak helicase activity alone, this can be significantly stimulated by accessory proteins eIF4B and eIF4H [24, 31, 59, 68, 70, 74, 82, 91]. These latter factors contain RRM-domains and, importantly, have been reported to interact directly with the G4C2 RNA [14, 29, 72].
In an unbiased, directed screen for canonical translation factors, we identified 11 potential translation factors that modulate GR-production in G4C2-expressing flies. Further investigations into two of these, eIF4B and eIF4H1 (fly orthologue to eIF4H), further defined them as modifiers of G4C2-toxicity. Their downregulation significantly reduced GR-levels in animals expressing the repeat. Further investigations into eIF4B and eIF4H in C9+ derived cells revealed that eIF4H was significantly downregulated. eIF4H downregulation also occurred in post-mortem tissue from C9+ ALS/FTD compared to C9- ALS/FTD and healthy individuals. This work identifies eIF4B and eIF4H as important disease modifiers that alter RAN-translation of the GR-reading frame.
Results
GFP-tagged GR dipeptides are produced in LDS-(G4C2)n flies with expanded (> 30) repeats
We previously identified a number of translation factors as modifiers of G4C2-toxicity [26]. To investigate these and other factors in the context of RAN-translation, a new C9orf72 fly model for ALS/FTD was designed (Fig. 1a). This model contained the 114-base pair sequence immediately upstream of the repeat in intron 1 of C9orf72 in ALS/FTD patient genomes (termed a “leader” sequence; LDS). The addition of this sequence puts the repeat in a more patient-relevant context while this region is likely to influence pathological mechanisms, including RAN-translation [36, 73, 87, 96]. G4C2 expansions can produce three sense-strand associated dipeptides: GA, GR, and GP. Importantly, of these GR is associated with extreme toxicity in multiple models, including flies [22, 53]. To facilitate investigations into genes that may impact RAN-translation of GR, a GFP tag (lacking an ATG initiation codon) was added 3′-prime of the repeat in the GR-reading frame.
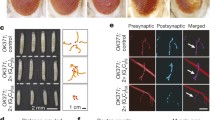
Expanded G4C2 transgenes produce GFP-tagged GR. a. A new transgenic (G4C2)n model was developed to look at RAN-translation of the GR reading frame. A “leader” sequence (LDS) was added 5′ of the repeat: 114 bp of intronic sequence found upstream of the repeat in patient samples. The GR reading frame has an in-frame GFP coding sequence 3′ of the repeat that lacks an ATG initiation. b. To define the number of repeats inserted into genomes of w1118 transgenic flies, PCR reactions were developed that amplified the repeat and its flanking region. The number of repeats were calculated from PCR product lengths measured on a Bioanalyzer and agarose gel. Shown: the maximum number of repeats found in the control (CTRL) or expanded (EXP) G4C2 fly lines; individual data points from 2 independent DNA preps with mean. c. qPCR analysis for RNA levels between control and expanded G4C2 fly lines. Shown: individual data points with mean ± SD. Statistics: unpaired student t-test, p-value **** < 0.0001. d. Western immunoblots confirmed GR is produced and successfully tagged with GFP in EXP-G4C2 flies. No GR/GFP was detected in control G4C2 flies, even with overexposure (Additional file 1: Figure S1). Uncropped westerns (Additional file 1: Figure S4) e. External and internal eye analysis in animals expressing G4C2 or control (DSRED) transgenes using GMR-GAL4. Degeneration was seen only in LDS-(G4C2)EXP animals: external pigment loss and reduced integrity of internal retina tissue. f. External eye imaging for fluorescence caused by transgene expression. Positive (DSRED) control flies show uniform, diffuse signal. Control G4C2 flies show no signal, even with increased exposure (data not shown). Expanded G4C2 flies show GFP puncta. Shown (e-f): representative images while all conditions were tested 2+ times. For full genotypes see Additional file 7: Table S1
LDS-G4C2 transgenes were randomly inserted into the genome of w1118 animals. To define the number of repeats inserted into individual fly lines, primers that flanked the G4C2 repeat were used to PCR amplify the region [26]. The number of repeats was then calculated from the length of the PCR products, resolved by agarose gel electrophoresis and Bioanalyzer. Two primary fly transgenic lines were defined: a control (CTRL) line containing short (G4C2)≤12 repeats and an expanded (EXP) line containing (G4C2)≤44 repeats (Fig. 1b). By quantitative-real time PCR (qPCR), these two lines expressed significantly different G4C2 RNA levels (Fig. 1c), most likely the result of variability in insertion site within the fly genome [43].
Western immunoblots were used to determine whether a (GR)n dipeptide was produced from the LDS-G4C2 transgenes, despite the absence of an AUG-start codon in the GR-reading frame. LDS-G4C2 transgenes were expressed in the fly eye using GMR-GAL4 and protein lysates were prepared from heads. Using an antibody designed to target the GR-dipeptide [46], we found that a GR peptide was produced only from the expanded LDS-(G4C2)EXP fly line (Fig. 1d). Re-probing with an anti-GFP antibody confirmed that the GR-dipeptides produced were tagged with GFP. GFP expressing control flies confirmed that the molecular weight of the GR/GFP band in LDS-(G4C2)EXP animals was higher than GFP alone. As LDS-(G4C2)EXP lines had 2.5-fold higher RNA expression than the LDS-(G4C2)CTRL, longer exposure times were also evaluated and continued to show no GR/GFP signal in LDS-(G4C2)CTRL expressing animals (Additional file 1: Figure S1).
To define potential toxicity associated with LDS-(G4C2)EXP , transgenes were expressed in the fly optic system using GMR-GAL4 (Fig. 1e). LDS-(G4C2)CTRL animals showed external and internal eye morphologies similar to controls, supporting that the short repeat was not toxic [22, 26, 41, 53]. In contrast, expression of LDS-(G4C2)EXP caused mild pigment loss externally and dramatic loss of retinal tissue internally, indicative of neurodegeneration.
To further assess GR production, fluorescence imaging of the fly eyes revealed that the LDS-(G4C2)EXP expressing animals produced GFP-positive puncta (Fig. 1f). In contrast, a control fluorescence protein (DSRED) did not show puncta formation but rather had a uniform diffuse signal, indicating that the unique punctate fluorescence pattern seen with LDS-(G4C2)EXP was the result of the GR. LDS-(G4C2)CTRL animals were also imaged and showed no GFP signal, even with 5-10x longer exposure time (data not shown).
Overall, these data indicate that expression of LDS-(G4C2)EXP in flies can induce toxicity and that GFP-tagged GR are produced by an expanded LDS-G4C2 transcript.
A loss of function screen for candidate RAN-translation factors
Despite recent advances into mechanisms underlying G4C2-associated RAN-translation, a full understanding of which canonical translation factors are involved remains unclear [12, 27, 37, 84, 96]. To define translation factors that may mediate GR-associated RAN-translation, we designed a loss-of-function (LOF) fly screen utilizing external eye imaging for toxicity, and GR-GFP fluorescence of the eyes for protein, in LDS-(G4C2)EXP expressing animals (Fig. 2a). 48 RNAi [57, 60] or LOF mutant [6, 7, 80, 81] fly lines were obtained that target specific translation factors, covering 86% of the 56 known translation factors in the fly [50]. 40 of 48 (83.3%) lines were RNAi and 8 of 48 (16.6%) were mutant lines.
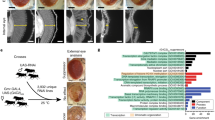
A screen of translation factors reveals those important for expression of GR from (G4C2)EXP. a. To identify canonical translation factors that may be involved in RAN translation of G4C2 in the GR reading frame, a loss-of-function (LOF) based screen was designed utilizing previously developed RNAi [57, 60] or LOF mutant fly lines [6, 7, 80, 81] targeting 48 of 56 (86%) known translation factors [50]. Individual translation factors were downregulated in animals expressing LDS-(G4C2)EXP and any that altered the external eye phenotype and/or GR-GFP levels were defined (Step 1). These 28 LOF lines were further tested in (GR)36 expressing animals, defining 6 that acted similarly on GR-associated toxicity (Step 2). Additional quality control experiments excluded LOF lines that altered expression from a control (LacZ) transgene by western immunoblot and/or altered a WT eye morphology when expressed alone, as described [13, 26, 41] (Step 3). b. Summary of screen results. Overall, 11 translation factors were identified as candidate RAN translation factors as their depletion reduced GR-GFP levels. Overall, excluded LOF lines either: altered toxicity of G4C2 flies but did not alter GR-GFP levels, similarly altered toxicity in a non-G4C2, GR fly model arguing that these acted downstream of GR production, had no effect on G4C2 toxicity or GR-GFP levels, or were “unspecific” modifiers identified by quality control experiments. Shown: representative images while all RNAi were tested 2+ times for effects under each condition. Details on LOF lines used and complete results with each line can be found in Additional file 2: Table S2. For full genotypes and RNAi lines see Additional file 7: Table S1
28 of the 48 tested LOF lines altered toxicity and/or GR-GFP levels caused by LDS-(G4C2)EXP expression in the fly eye, assessed by comparing images with controls (Fig. 2a, step 1). These 28 lines were further examined in a fly model that expresses (GR)36 from a non-G4C2 transcript [53], to determine if they acted downstream of toxic GR-production in the LDS-(G4C2)EXP animals (Fig. 2a, step 2). 6 of the LOF lines targeting translation factors were found to similarly alter GR-induced toxicity in this model and were not further studied. The remaining 22 LOF lines were further tested for unspecific effects using quality control experiments (Fig. 2a, step 3) [13, 26, 41, 55]. Specifically, lines were examined to eliminate those that cause an effect when expressed on their own in the eye and tested to confirm no effect on the protein levels of a control (LacZ) transgene.
In summary, 20 of the lines did not alter toxicity or GR-GFP levels in LDS-(G4C2)EXP expressing animals (Fig. 2b). 17 lines were excluded from further study because they either caused increased GR-GFP levels (1 line), altered LDS-(G4C2)EXP toxicity but not GR-GFP signal (4 lines), could alter GR-toxicity independent of G4C2-RNA (6 lines), or failed quality control experiments (6 lines; termed “unspecific modifiers”). Thus, from the screen of 48 factors, 11 candidate RAN-translation factors were identified (Table 1, Additional file 2: Table S2).
Depletion of eIF4B or eIF4H1 mitigates toxicity in LDS-(G4C2)EXP animals
Of the 11 factors that reduced GR-GFP levels, eIF4B and eIF4H1 (fly orthologue to human eIF4H) were intriguing. These two factors have independent and redundant roles in activating eIF4A [24, 31, 59, 68, 70, 74, 82, 91] which was recently identified as a RAN-translation factor in a G4C2-model [27, 84]. Further, eIF4B and eIF4H had previously been reported to bind G4C2 RNA through RNA recognition motifs (RRMs) [14, 29, 72].
To further investigate eIF4B and eIF4H1 as modifiers of LDS-(G4C2)EXP in flies, a second, independent set of RNAi lines targeting these genes was obtained (termed RNAi-2). All RNAi lines were confirmed to downregulate the expected targets, eIF4B or eIF4H1 (Fig. 3a, Additional file 1: Figure S3A). Further, eIF4B RNAi did not cause reduced expression of eIF4H1, and vice versa, indicating that expression of these two genes is independent and that the RNAi lines are specific. Interestingly, ubiquitous downregulation of eIF4B or eIF4H1 by RNAi produced viable adults with no obvious phenotype (Fig. 3b), supporting that these genes are not essential in the fly (also [33]).
Analysis of eIF4B and eIF4H1 RNAi in control flies. a. RNA levels produced from eIF4B or eIF4H1 were assessed by qPCR in flies ubiquitously expressing RNAi (by Daughterless-GAL4). Statistics: one-way ANOVAs with Tukey’s multiple comparison correction, p-values **** < 0.0001, *** < 0.001, ** < 0.01, * < 0.05, no significance > 0.05. Shown: individual data points from 2 independent experiments with mean ± SEM. b. Viability studies in Drosophila reveal that RNAi-depletion of eIF4B or eIF4H1 does not significantly alter the number of adult flies expected to eclose. Shown: ratio of progeny from two individual crosses with RNAi compared to sibling animals with the balancer chromosome that reach adulthood (1-2d adult animals). Comparing the # progeny that eclose from a single vial compensates for differences in mating variability, fertilized eggs laid, among other variables. However, we note that the presence of the balancer chromosome could potentially cause mild sub-viability to adulthood. Crosses: RNAi/CyO x Da-GAL4 (III); counted progeny: RNAi/+; Da-GAL4/+ and CyO/+; Da-GAL4/+. RNAi lines: control (JF01355), eIF4B RNAi (HMS04503), eIF4H1 RNAi (HMS04504). For full genotypes see Additional file 7: Table S1
The effects of co-expressing eIF4B, eIF4H1, or control (Luc) RNAi with LDS-(G4C2)EXP using GMR-GAL4 were analyzed in the eye. Externally, eIF4B or eIF4H1 RNAi caused reduced toxicity compared to the control RNAi, seen by recovered red pigment and ommatidial organization (Fig. 4a). Internally, retinal tissue loss caused by LDS-(G4C2)EXP was also mitigated by depletion of eIF4B or eIF4H1. Blinded quantification of the total surface area for retina tissue or of tissue depth (at the point of the optic chiasm) revealed that suppression was consistent and significant (Fig. 4b). Suppression was recapitulated with a second set of RNAi lines, supporting that the effects seen are the result of downregulating these target genes (Additional file 1: Figure S4B-C).
Depletion of eIF4B and eIF4H1 selectively suppresses LDS-(G4C2)EXP associated toxicity. a. Using GMR-GAL4, RNAi-mediated depletion of eIF4B or eIF4H1 in LDS-(G4C2)EXP expressing flies results in reduced toxicity in both the external and internal eye: seen externally by recovered pigment and ommatidial structure, seen internally by recovered retinal tissue integrity. b. Blinded quantification of internal retina tissue was done by measuring the total surface area of tissue present and by measuring the depth of the tissue at the position where the optic chiasm occurs. n = 9–10 animals per genotype. c. eIF4B or eIF4H1 RNAi was expressed in (GR)36 flies (GMR-GAL4) and effects on GR-associated toxicity were observed in the external and internal eye. d. Blinded quantification of internal retina tissue. n = 5–9 animals per genotype. e. eIF4B or eIF4H1 RNAi was expressed in control flies (GMR-GAL4) and effects on the normal eye were observed externally and internally. f. Blinded quantification of internal retina tissue. n = 4 animals per genotype. For graphs, shown are individual data points representing 1 animal with mean ± SD. Statistics: one-way ANOVAs with Tukey’s multiple comparison correction, p-values **** < 0.0001, *** < 0.001, ** < 0.01, * < 0.05, no significance > 0.05. RNAi lines: control (JF01355), eIF4B (HMS04503), eIF4H1 (HMS04504). For full genotypes see Additional file 7: Table S1
To further assess if eIF4B and eIF4H1 could be acting downstream of toxic GR-production, RNAi lines targeting eIF4B, eIF4H1, or control (Luc) were co-expressed with (GR)36 in the fly eye using GMR-GAL4. The (GR)36 transgene produces a GR dipeptide from a non-G4C2 repeat transcript [53]. In contrast to the effect in LDS-(G4C2)EXP animals, eIF4B and eIF4H1 RNAi increased GR-toxicity in both the external and internal eye (Fig. 4c-d). This argues that these genes do not act on the same pathway in GR animals as in LDS-(G4C2)EXP animals. Depletion of either eIF4B or eIF4H1 on their own did not alter normal eye morphology (Fig. 4e-f).
Depletion of eIF4B or eIF4H1 reduces GR-production in LDS-(G4C2)EXP animals
As eIF4B and eIF4H1 are canonical translation factors we hypothesized that they modified LDS-(G4C2)EXP toxicity by mediating translation from the G4C2 transcript. To further test if depletion of these factors reduced GR production, eIF4B, eIF4H1, or control (Luc) RNAi were co-expressed with LDS-(G4C2)EXP in the fly eye and fluorescence imaging was performed (Fig. 5a). Blinded quantification of GR-GFP signal in LDS-(G4C2)EXP flies revealed that eIF4B depletion caused a 48.3 ± 13% decrease in total GR-GFP fluorescence (Fig. 5b, grey). Further, eIF4H1 depletion caused a 65.5 ± 3.7% decrease in GR-GFP fluorescence levels. As puncta formation was associated with fluorescently tagged GR (see Fig. 1.), additional analyses were performed to define changes in the number of bright GR-GFP puncta and the average size of these puncta (Fig. 5b, black). eIF4B RNAi reduced the number of puncta from 233 per eye to 44 per eye, an 81% reduction. Additionally, the average size of the puncta per eye was reduced by 63% (7.6μm2 to 2.8μm2). eIFH1 RNAi caused a 91% reduction in the number of GR-GFP puncta per eye (233 to 20) and the size of the puncta was reduced from 7.6μm2 to 2.4 μm2, a 68% reduction. Effects on GR-GFP levels and puncta were also seen using the second set of RNAi lines targeting eIF4B and eIF4H1 (Additional file 1: Figure S3D-E).
eIF4B and eIF4H1 RNAi selectively reduce GR-GFP levels produced from LDS-(G4C2)EXP. a. Fluorescence imaging in LDS-(G4C2)EXP expressing flies shows that depletion of eIF4B or eIF4H by RNAi results in reduced GR-GFP levels (GMR-GAL4). b. Blinded quantification of GR-GFP signal in LDS-(G4C2)EXP animals relative to the signal in control RNAi animals. Analysis of GR-GFP puncta number and size are shown in black. Total GR-GFP signal is shown in grey. n = 6–7 animals per genotype. Further, qPCR was used to quantify RNA levels in LDS-(G4C2)EXP flies co-expressing control, eIF4B, or eIF4H1 RNAi, shown in light grey. c. A control fluorescent protein, DsRed, was similarly expressed in the fly eye with RNAi to control, eIF4B, or eIF4H. d. Blinded quantification of DsRed fluorescence shows no effect by RNAi. n = 6 animals per genotype. e. A representative western immunoblot image for β-Galactosidase in LacZ flies co-expressing control, eIF4B, or eIF4H1 RNAi. Uncropped westerns (Additional file 1: Figure S4). Blinded quantification of β-Galactosidase western immunoblots normalized to the loading control, Tubulin. For graphs, shown are individual data points from 2 independent assays with mean ± SEM. Statistics: one-way ANOVAs with Tukey’s multiple comparison correction, p-values **** < 0.0001, *** < 0.001, ** < 0.01, * < 0.05, no significance > 0.05. RNAi lines: control (JF01355), eIF4B (HMS04503), eIF4H1 (HMS04504). For full genotypes see Additional file 7: Table S1
While we had already determined that eIF4B and eIF4H1 RNAi did not alter toxicity downstream of GR production (see Fig. 4c-d), we considered whether their depletion could modify LDS-(G4C2)EXP upstream of translation, on the transcriptional level. To assess this, we used qPCR to measure transcript levels of the LDS-(G4C2)EXP transgene in animals co-expressing control RNAi (Luc), eIF4B or eIF4H1 RNAi (Fig. 5b, light grey). Depletion of eIF4B or eIF4H1 did not alter LDS-(G4C2)EXP RNA levels, further supporting that they act on the translational level.
To test the specificity of eIF4B and eIF4H1 to translation of a G4C2 transcript, we first confirmed specificity of the effect of eIF4B and eIF4H1 RNAi, by assessing whether their depletion had an effect on eye fluorescence of a control DsRed transgene in the fly optic system (GMR-GAL4) (Fig. 5c). Blinded quantification supported that total fluorescence was unchanged, arguing that the effect in LDS-(G4C2)EXP animals was specific to the GR-tagged fluorescent protein (Fig. 5d). We further used western immunoblots to analyze protein levels produced from a control (LacZ) transgene in animals co-expressing eIF4B, eIF4H1, or control (Luc) RNAi (Fig. 5e). Transgenes were expressed using GMR-GAL4 and protein was extracted from whole heads. Consistent with fluorescence data using DsRed, no significant difference in the amount of β-galactosidase protein translated from the LacZ transcript was seen. The second set of RNAi lines targeting eIF4B and eIF4H1 also did not alter protein expression from a control (LacZ) gene (Additional file 1: Figure S3F). These data are consistent with previous reports that these factors are not essential for general translation [2, 11, 15, 33].
Overall, these data support that eIF4B or eIF4H1 modify LDS-(G4C2)EXP toxicity by mediating toxic GR production. Importantly, translation from the G4C2 transcript is particularly sensitive to their depletion as expression from control transcripts were unaltered under similar conditions.
EIF4H is downregulated in ALS/FTD cases harboring a G4C2 expansion in C9orf72
Data in the fly supported that eIF4B and eIF4H1 were modifiers of LDS-(G4C2)EXP that could alter the amount of GR produced from the repeat-containing transcript. To further investigate these translation factors in disease, we considered that the expression of the human orthologues to these factors, eIF4B and eIF4H, could be dysregulated if they played a critical role in G4C2-associated expression.
eIF4B and eIF4H protein levels were assessed by western immunoblot in primary fibroblast cell lines (Fig. 6.A; lines described in Additional file 3: Table S3). The mean expression from four independent C9 + patient derived lines was compared to the mean expression from five independent lines derived from healthy individuals. Interestingly, eIF4B total levels were unchanged while eIF4H levels were reduced by 47.5%. As eIF4B is inhibited by phosphorylation at Ser422 [64, 69], we further examined levels of phospho-eIF4B to determine whether this factor was dysregulated by protein modification. No obvious changes were observed in phospho-eIF4B levels visually or relative to total eIF4B in C9+ versus healthy cells.
EIF4H is downregulated in C9+ ALS/FTD. a. Western immunoblots were used to define changes in eIF4B or eIF4H protein levels from 5 independent control or 4 independent C9+ derived fibroblast cell lines. Data are relative to controls. Quantification of total protein was done after normalizing to loading, using GAPDH. Phospho-eIF4B quantification was further normalized to total eIF4B. Statistics: unpaired student t-tests. Shown: each data point represents 1 cell line with mean ± SEM; the mean data from 2 independent protein preparations is shown per line. b. RNA levels of EIF4B or EIF4H were assessed by qPCR in human cerebellar tissue from healthy individuals or ALS/FTD patients with (C9+) or without (C9-) the G4C2 expansion in C9orf72. n = 22 (healthy), 46 (C9- ALS/FTD), 66 (C9+ ALS/FTD). Statistics: one-way ANOVAs with Dunn’s multiple comparison correction. Shown: individual data points representing 1 individual with mean ± SEM. Cell line details: Additional file 3: Table S3. Patient details: Additional file 4: Table S4. p-values **** < 0.0001, *** < 0.001, ** < 0.01, * < 0.05, no significance > 0.05
Data from patient-derived cells supported that eIF4H is dysregulated in C9+ situations. To further assess this finding in patients, total RNA was extracted from post-mortem, cerebellar tissue from 112 ALS/FTD individuals or 22 healthy individuals and the expression from eIF4B or eIF4H were defined by qPCR (Fig. 6b; individuals described in Additional file 4: Table S4). The ALS/FTD cohort were further broken down based on the presence or absence of the G4C2-repeat expansion in C9orf72 into 46 C9- ALS/FTD and 66 C9+ ALS/FTD cases. Consistent with protein data from fibroblast lines, eIF4B expression was unaltered in disease. Importantly, eIF4H was significantly downregulated by 71.2% in C9+ ALS/FTD compared to healthy controls and 54.4% compared to C9- ALS/FTD cases.
These data indicate a significant decrease in eIF4H expression in response to the presence of expanded G4C2 in ALS/FTD.
Discussion
Mechanisms underlying repeat-associated non-AUG (RAN-) translation remain unclear despite evidence that this form of translation occurs in disease [37, 96]. To help define potential RAN-translation factors, we developed a gain-of-function fly model for C9orf72-associated ALS/FTD that expressed an expanded GGGGCC hexanucleotide repeat (termed G4C2) downstream of the sequence normally found upstream of the repeat in patients (114 bp of intronic DNA found 5′-prime of the repeat in intron 1 of C9orf72 in ALS/FTD); the transgene expressed also contained a GFP tag downstream of the repeat in the GR reading frame (see Fig. 1). Using this model, we screened 48 of 56 canonical translation factors in flies [50] to define those that could impact expression of the GR dipeptide (see Fig. 2 and Additional file 2: Table S2). 11 candidate RAN-translation factors were defined (see Table 1). When depleted, these factors reduced GR-GFP levels, reduced G4C2-induced toxicity, and did not reduce toxicity associated with a non-G4C2 transcript generated toxic GR protein. Further investigations into two of these, eIF4B and eIF4H1 (fly orthologue to eIF4H), revealed that their depletion reduced G4C2-induced toxicity and GR-GFP levels, but did not alter G4C2 RNA levels (see Fig. 5 and Additional file 1: Figure S3). Investigations into eIF4B and eIF4H expression in patient-derived cells and in post-mortem tissue revealed that eIF4H is significantly downregulated in ALS/FTD patients harboring the G4C2 expansion (C9+ ALS/FTD) (see Fig. 6). This effect was not seen in ALS/FTD patients lacking the G4C2 mutation (C9- ALS/FTD), arguing that it is a response to the presence of the repeat. These data highlight eIF4B and eIF4H as novel factors mediating disease-associated pathways.
To our knowledge, this is the first in vivo investigation into canonical translation factors that impact dipeptide production in a C9orf72-associated disease model. The simplest hypothesis is that these factors impact the process of RAN translation from the G4C2 repeat-containing transcript. Interestingly, our data suggest that production of the GR dipeptide requires specific factors as only 11 of 48 canonical translation factors screened altered GR-GFP levels in G4C2-expressing animals. Further, investigations into mammalian systems and in disease-relevant tissue using DPR-specific antibodies will determine if eIF4B or eIF4H depletion disrupts expression of multiple DPR and confirm effects on GR production [98]. Although these factors could function to alter GR-production through alternative means versus RAN-translation (i.e. stability of the protein, altered expression of more direct RAN-translation factors), it is compelling that the factors identified converge at key regulatory steps of translation initiation and that the majority of these factors function together or with previously suggested RAN-translation factors (see Fig. 7). We ruled out factors that similarly altered toxicity caused by the GR dipeptide, supporting that they act on pathways not associated with toxicity of the GR protein. We also ruled out mechanisms underlying G4C2 transcription and RNA stability for eIF4B and eIF4H, as G4C2 transcript levels are unaltered by their depletion (see Fig. 5b). Although our investigations here focused on eIF4B and eIF4H, we note that we defined a number of other intriguing factors that act on either on G4C2- and/or GR-associated toxicity (see Table 1).
Model comparing potential G4C2 RAN-translation mechanisms and canonical translation. a. Ternary complex formation requires eIF5-mediated exchange of GDP to GTP on eIF2 complex (includes eIFs 2α, 2β, 2γ). eIF2α is highly regulated during stress and is reported to mediate G4C2 translation [12, 27]. eIF2β and eIF5 were identified as modifiers in this study. b. In normal translation, the formation of the 43S pre-initiation complex (PIC) involves the joining of a number of factors, including Ternary complex (described in a) and eIFs 1, 1A, 3, and 5. c. A minimal PIC complex may potentially mediate RAN-translation [1, 42, 76, 86]. d. mRNA transcripts are recognized by the eIF4F complex, includes eIFs 4E, 4G, 4A. All of these have been defined as G4C2 translation factors arguing that G4C2 RAN-translation is cap-dependent [12, 84]. eIF4E recognizes the 5-prime m7G cap on mRNAs [10, 77, 78]; notably, 4 of 6 eIF4E components were identified in our screen. eIF4A is recruited by eIF4E to mRNA transcripts (via the scaffold protein eIF4G). mRNA is then unwound by eIF4A, an activity that is significantly promoted by eIF4B or eIF4H, identified herein [24, 68, 70, 82, 91]. This action allows for the formation of the 48S scanning complex. e. In canonical translation, the 48S scanning complex moves down a transcript until identifying an AUG start codon. A CUG codon in the LDS sequence upstream of G4C2 may function as a start codon in the GA-reading frame [27, 84]. Frame-shifting could allow for translation of the GR and GP from this codon. Candidate RAN translation factors eIF5B, and potentially eIF5, mediate ribosome scanning, start codon recognition, and translation activation [10, 45, 61]. We note that mechanisms underlying RAN-translation are still relatively unknown. This model is based on current literature and canonical functions of translation factors
The fly model developed herein expresses the LDS-G4C2 transcript as an mRNA, containing a 5′-prime m7G cap and polyadenylated (poly(A)-) tail. In disease, the G4C2-RNA could exist in multiple forms [93], including: improperly spliced C9orf72-mRNA transcripts retaining the repeat [58], properly spliced intronic sequence kept stable by the repeat [30, 90], altered transcription initiation products [71], or aborted C9orf72-transcript products [29]. Interestingly, fly models that express expanded G4C2 within properly spliced introns do not show dipeptide expression nor toxicity, despite the formation of RNA foci [88]. Further, improperly spliced C9orf72-transcripts can be shuttled from the nucleus to the cytoplasm where they undergo RAN-translation [32]. Overall, these data support that G4C2 repeats retained in capped and polyadenylated C9orf72-mRNA are able to produce toxic dipeptides and are relevant to patients. Whether or not G4C2-associated RAN translation is dependent on the presence of this 5′-prime cap (and poly(A)-tail) is still debated [12, 27, 84].
Our investigations draw parallels between the canonical functions of translation factors [50] and RAN-translation (see Model, Fig. 7). However, there are multiple types of translation that may be pertinent to disease, including cap-independent mechanisms, such as IRES translation [28, 37, 76, 77]. Of the 11 candidate RAN-translation factors we identified, 4 have been reported to function in cap-independent translation: eIF4B [38, 75], eIF4H [91], eIF5B [23], eIF2β [44]. Interestingly, eIF4B or eIF4H significantly strengthen eIF4A-mediated unwinding of longer, more complex 5′ UTRs in transcripts [70, 74, 82, 91]. This activity is thought to mediate scanning of the 5′ UTR for translation start sites [79] and recruitment of ribosomal subunits [75, 92]. Overall, these data suggest a model for G4C2-associated RAN-translation where eIF4B/eIF4H and eIF4A mediate dipeptide production during this scanning process [28, 84]. Further, frame-shifting during scanning could result in the production of all three sense-strand associated dipeptides (GA, GR, GP) from a near-cognate alternative start codon, CUG, found upstream of the repeat in the GA-reading frame [84].
In addition to their involvement in non-canonical translation and in stimulating eIF4A (previously reported as a RAN-translation factor [27, 84]), we chose to focus on eIF4B and eIF4H as they are RNA-binding proteins (RBPs) containing homologous RNA recognition motifs (RRMs) [66]. Screens for RBPs that interact with G4C2-RNA identified both eIF4B and eIF4H, supporting our data that they function in translation from G4C2 transcripts [14, 29, 72]. Both eIF4B and eIF4H can independently stimulate the helicase activity of eIF4A during translation [24, 31, 59, 68, 70, 74, 82, 91] while key differences between them are noted. Structural data supports that eIF4H is constitutively active while eIF4B contains a regulatory carboxyl domain containing multiple phosphorylation sites [52, 64]. This may explain why eIF4H is downregulated in C9+ ALS/FTD but not eIF4B (see Fig. 6), as eIF4B can be regulated by de/phosphorylation. However, no significant changes were observed for phospho-eIF4B at Ser422, an inhibitory modification [69, 77], in four C9+ derived cell lines versus five control cell lines. Extended analyses are needed to increase the sample size and to test for eIF4B phosphorylation at other marks [9, 89]. Overall, we hypothesize that eIF4H is downregulated as the result of compensatory mechanisms: cells may actively downregulate eIF4H to reduce expression of toxic GR dipeptide. Alternatively, as eIF4H had previously been reported to bind G4C2-RNA [14, 29, 72], the reduced eIF4H RNA levels could be the result of a more complex feedback loop: as eIF4H protein is sequestered by G4C2 RNA foci, cells may respond by downregulating eIF4H transcription as they may sense that there is plenty of this translation factor present. However, the eIF4H that is present may not be functioning normally. Localization studies in patient tissue are needed to determine if eIF4H is indeed sequestered into G4C2-foci. In either scenario, data supports that eIF4H plays an important role in C9+ disease while further investigations would help define its role in disease progression.
Studies in multiple model systems support that eIF4B and eIF4H loss does not inhibit global translation, including yeast [2, 15], flies [33], and mice [11]. Interestingly, yeast and flies (see Fig. 3b) with downregulated eIF4B or eIF4H are viable [2, 15, 33] and eIF4H+/− mice do not have notable deficits [11]; although eIF4B and eIF4H have been suggested to be important for brain development [9, 11, 20, 67]. The downstream consequences of eIF4H downregulation in C9+ ALS/FTD may be broader than simply altering RAN-translation as depletion of eIF4B and eIF4H in cultured cells has been shown to induce stress granule formation [54]. Interestingly, since eIF4H expression is reduced in C9+ ALS/FTD and C9+ derived cells (see Fig. 6), this raises a potential connection to mechanisms underlying TDP-43 pathology/toxicity [16, 21]. Further, our data in GR-expressing flies argues that the depletion of these factors downstream of GR-production can feed into pathways disrupted by this toxic dipeptide (see Fig. 4c-d) [35, 83, 85, 94].
In conclusion, in an unbiased, targeted screen we identified eIF4B and eIF4H as canonical translation factors that, when depleted in flies, disrupted toxicity caused by the expression of expanded G4C2 RNA. Interestingly, eIF4H was downregulated in C9+ ALS/FTD patients, indicating a distinct role in C9orf72-associated disease. These factors may represent unique G4C2 modifiers that couple RAN-translation to dysregulation of RNA metabolism in disease [16, 34, 95].
Methods and materials
Patient samples and clinical, genetic and pathological assessments
Participant information is summarized in Additional file 4: Table S4. Protocols were approved by the Mayo Clinic Institutional Review Board and Ethics Committee. All participants (or authorized family members) were provided written informed consent before information gathering, autopsies and postmortem analyses. Trained neurologists diagnosed patients with ALS and/or FTD after reviewing neurological and pathological information. The presence or absence of an expanded G4C2 within intron 1 of C9orf72 was done using a previously established protocol for repeat-primed polymerase chain reaction [17].
Drosophila work
Stocks were maintained on standard cornmeal-molasses medium. Fly lines used are detailed in Additional file 2: Table S2 and Additional file 5: Table S5. Fly lines obtained from Bloomington Drosophila Stock Center (BDSC) and Vienna Drosophila Resource Center (VDRC) are noted.
Fly RNAi efficacy
All control and RNAi lines are defined in Additional file 5: Table S5. RNAi efficacy was determined using Da-GAL4 (adults or larvae) as previously described [26, 51].
Characterization of LDS-(G4C2)n fly models
Transgenes were inserted into pUAST vectors and randomly inserted into w1118 fly genomes. The LDS-G4C2 model has a 5′ leader sequence (LDS) inserted immediately upstream of the G4C2 repeats, 114 bp of sequence upstream of the repeat in intron 1 of C9orf72 in patients, and a 3′ GFP tag in the GR reading frame. Repeat-length determination: Genomic DNA was extracted from individual fly lines and the transgenes present were amplified by PCR using primers designed to flank the repeat (Additional file 6: Table S6). Amplification was done using a KAPA HiFi HotStart kit (Kappa #KK2501) and PCR product sizes were quantified using agarose gels and a Bioanalyzer, previously described [26]. Control w1118 animals were included in experiments and showed no signal. RNA expression: Transgenes were expressed as previously described using HS-Gal4 [26] and RNA levels were assessed by qPCR, using primers designed to amplify the GFP tag (Additional file 6: Table S6). Control w1118 animals were included in experiments and showed no signal.
LOF external fly eye screen
Publicly available RNAi [57, 60] or mutant [6, 7, 80, 81] loss-of-function (LOF) fly lines targeting canonical translation factors were obtained from the Bloomington Drosophila Stock Center (BDSC). Additional UAS-RNAi lines targeting eIF4B and eIF4H1 were obtained from Vienna Drosophila Resource Center (VDRC) [18].
External eye imaging
LOF males were crossed to recombinant females: UAS-LDS-(G4C2)EXP, GMR-GAL4 (III) (26 °C). Multiple w− and w+ controls were setup with every experiment to assess any natural variability, including a UAS-Luc RNAi (BDSC # 31603) and w1118; UAS-DSRED. External eyes for 1-2d progeny were imaged on a Leica Z16 APO microscope as described [26]. Any changes to the ommatidia organization, eye size, pigmentation, and ability to eclose from pupae were noted. Resulting phenotype was categorized into one of six groups: suppressors, mild suppressors, no effect, mild enhancers, enhancers, and lethal enhancers (Additional file 1: Figure S2).
External eye fluorescence imaging
LOF males were crossed to recombinant females: UAS-LDS-(G4C2)EXP, GMR-GAL4 (III) (26 °C) and 1-2d progeny were imaged on a Leica DM6000B and quantified as previously described [26]. w− or w+ controls were used for accurate comparisons depending on the background of the LOF lines and the final genotypes of animals. Any changes to the GR-GFP levels using LUT Z-stacked images were noted. Resulting phenotype was categorized into one of six groups: suppressors, mild suppressors, no effect, mild enhancers, enhancers, and lethal enhancers (Additional file 1: Figure S2). Researchers were blinded to the LOF targets during screening. Modifiers of LDS-(G4C2)EXP toxicity and/or GR-GFP levels were further assessed in GMR-GAL4 > UAS-(GR)36 animals. Modifier crosses were repeated 3+ independent times to confirm reproducibility of results.
Quality control experiments
Defining unspecific LOF lines were performed as previously described [26].
Fibroblast cells
Cultured using standard protocols in DMEM complete media: 15% FBS (Sigma), 1x MEM Amino Acids (ThermoSci # 11130051), 1% pen/strep, DMEM (high glucose, plus sodium pyruvate). Cells were maintained in a 5% CO2 incubator at 37 °C.
Western immunoblots (WB)
Fly tissue: Triplicate samples of 5–10 heads per genotype were homogenized using disposable pellet/pestles tissue grinders (Kimble Chase #749520–0000) and motor (Kimble Chase #749540–0000). For βgal: heads were directly homogenized into 1X NuPAGE LDS sample buffer. For GR/GFP: heads were homogenized into RIPA buffer (50 mM Tris-HCL (pH 7.5), 150 mM NaCl, 1% NP-40, 50 mM NaF, 0.5% DOC), plus protease inhibitors (Sigma # 05892970001), 1 mM PMSF, and 1 mM DTT. Fibroblast cells: cells were lysed in RIPA buffer plus protease inhibitors, 1 mM PMSF, and 1 mM DTT, and phosphatase inhibitors (Sigma # 04906845001) for 30 min at 4 °C. All RIPA lysates: quantified by Bradford; 20 μg of protein was run per lane. All WBs: run using a standard protocol with Invitrogen’s XCell SureLock blot system, 4–12% Bis-Tris NuPAGE gels and a wet transfer with PVDF membrane, except βgal which was transferred with an iBlot dry transfer system (program 2, 8 min) and nitrocellulose membrane. Antibodies: anti-βgalactosidase (Promega #Z3781, 1:2000), anti-αTubulin (DSHB #AA4.3, 1:2000), anti-GFPJL8 (Takara #632380, 1:10,000), anti-GR (gift from V. M-Y. Lee #2316, 1:1000). H. sapiens antibodies: anti-eIF4B (Cell Signaling #3592, 1:1000), anti-eIF4H (Cell Signaling #3469,1:1000), anti-phospho-eIF4B (Cell Signaling #3591,1:1000), anti-GAPDH (Sigma #G8795, 1:5000). Secondary antibodies: Mouse-HRP (Jackson Immunoresearch Labs #115–035-146, 1:5000), Rabbit-HRP (Jackson Immunoresearch Labs #111–035-144, 1:5000) [8]. Blots were analyzed using Amersham ECL Prime Detection Reagent and imaged on an Amersham Imager 600.
Quantitative real-time PCR (qPCR)
All primers are defined in Additional file 6: Table S6. For both fly and human qPCRs, protocols are previously described with the following changes [26]. Flies: For Da-GAL4 assays, triplicate samples of 5 whole animals were processed per condition. For GMR-GAL4 assays, triplicate samples of 20 fly heads (1-2d) were processed per condition. Humans: Total RNA was extracted from frozen postmortem tissue from the cerebellum using the RNAeasy Plus Mini Kit (QIAGEN), previously described [62]. RNA integrity (RIN) was verified on an Agilent 2100 bioanalyzer. RIN values ranged from 6.7 to 10, with most of the samples falling between 9.1 and 9.8.
Statistical analysis and data availability
GraphPad Prism 8.00 software was used to develop all graphs and for all statistical analyses. P-values < 0.05 were considered significant. All relevant data are included within the manuscript and supplementary data. Additional inquiries can be directed to the corresponding author, including reagent requests. No statistical methods were used to predetermine sample sizes and data distributions were assumed to be normal, similar to previous work [8, 19, 26, 39, 40, 51, 53, 62, 63]. Fly and fibroblast data: A two-tailed unpaired student t-test or one-way ANOVA with Tukey’s multiple comparisons test was performed when appropriate. Researchers were blinded to the genotype of all samples to maintain unbiased scoring. Human qPCRs: Nonparametric, one-way ANOVAs with Dunn’s multiple comparisons test were performed as data distribution was not normal.
Abbreviations
- ALS:
-
Amyotrophic Lateral Sclerosis
- C9- ALS/FTD:
-
ALS/FTD cases lacking the G4C2 expansion in C9orf72
- C9+ ALS/FTD:
-
ALS/FTD cases harboring the G4C2 expansion in C9orf72
- DPR:
-
Dipeptide repeats
- FTD:
-
Frontotemporal Dementia
- G4C2:
-
Expanded (GGGGCC)30+ mutation found within C9orf72
- GFP:
-
Green fluorescent protein
- qPCR:
-
quantitative reverse transcribed polymerase chain reaction
- RAN-translation:
-
Repeat Associated Non-AUG translation; also RANT
- RBP:
-
RNA binding protein
- RRM:
-
RNA recognition motif
- WT:
-
Wild-type
References
Akulich KA, Andreev DE, Terenin IM, Smirnova VV, Anisimova AS, Makeeva DS, Arkhipova VI, Stolboushkina EA, Garber MB, Prokofjeva MM et al (2016) Four translation initiation pathways employed by the leaderless mRNA in eukaryotes. Sci Rep 6:37905
Altmann M, Müller PP, Wittmer B, Ruchti F, Lanker S, Trachsel H (1993) A Saccharomyces cerevisiae homologue of mammalian translation initiation factor 4B contributes to RNA helicase activity. EMBO J 12:3997–4003
Andreou AZ, Klostermeier D (2013) The DEAD-box helicase eIF4A. RNA Biol 10:19–32
Ash PEA, Bieniek KF, Gendron TF, Caulfield T, Lin W-L, DeJesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, Rademakers R et al (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77:639–646
Balendra R, Isaacs AM (2018) C9orf72 -mediated ALS and FTD: multiple pathways to disease. Nat Rev Neurol 14:544
Bellen HJ, Levis RW, He Y, Carlson JW, Evans-Holm M, Bae E, Kim J, Metaxakis A, Savakis C, Schulze KL et al (2011) The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics 188:731–743
Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, Hiesinger PR, Schulze KL, Rubin GM et al (2004) The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167:761–781
Berson A et al (2017) TDP-43 promotes neurodegeneration by impairing chromatin remodeling. Curr. Biol 27:3579–3590 e6
Bettegazzi B, Bellani S, Roncon P, Guarnieri FC, Bertero A, Codazzi F, Valtorta F, Simonato M, Grohovaz F, Zacchetti D (2017) eIF4B phosphorylation at Ser504 links synaptic activity with protein translation in physiology and pathology. Sci Rep 7:10563
Browning KS, Bailey-Serres J (2015) Mechanism of cytoplasmic mRNA translation. Arab Book Am Soc Plant Biol 13
Capossela S, Muzio L, Bertolo A, Bianchi V, Dati G, Chaabane L, Godi C, Politi LS, Biffo S, D’Adamo P et al (2012) Growth defects and impaired cognitive–behavioral abilities in mice with knockout for Eif4h, a gene located in the mouse homolog of the Williams-Beuren syndrome critical region. Am J Pathol 180:1121–1135
Cheng W, Wang S, Mestre AA, Fu C, Makarem A, Xian F, Hayes LR, Lopez-Gonzalez R, Drenner K, Jiang J et al (2018) C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2α phosphorylation. Nat Commun 9:51
Chung C-Y, Berson A, Kennerdell JR, Sartoris A, Unger T, Porta S, Kim H-J, Smith ER, Shilatifard A, Van Deerlin V et al (2018) Aberrant activation of non-coding RNA targets of transcriptional elongation complexes contributes to TDP-43 toxicity. Nat Commun 9
Cooper-Knock J, Walsh MJ, Higginbottom A, Robin Highley J, Dickman MJ, Edbauer D, Ince PG, Wharton SB, Wilson SA, Kirby J et al (2014) Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain 137:2040–2051
Coppolecchia R, Buser P, Stotz A, Linder P (1993) A new yeast translation initiation factor suppresses a mutation in the eIF-4A RNA helicase. EMBO J 12:4005–4011
Coyne AN, Zaepfel BL, Zarnescu DC (2017) Failure to deliver and translate—new insights into RNA dysregulation in ALS. Front Cell Neurosci 11
DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Gilmer HF, Adamson J et al (2011) Expanded GGGGCC hexanucleotide repeat in non-coding region of C9ORF72 causes chromosome 9p-linked frontotemporal dementia and amyotrophic lateral sclerosis. Neuron 72:245–256
Dietzl G, Chen D, Schnorrer F, Su K-C, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S et al (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448:151
Elden AC, Kim H-J, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM et al (2010) Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466:1069–1075
Eom T, Muslimov IA, Tsokas P, Berardi V, Zhong J, Sacktor TC, Tiedge H (2014) Neuronal BC RNAs cooperate with eIF4B to mediate activity-dependent translational control. J Cell Biol 207:237–252
Fernandes, N., Eshleman, N. and Buchan, J.R. (2018) Stress granules and ALS: a case of causation or correlation? In Sattler,R., Donnelly,C.J. (eds), RNA Metabolism in Neurodegenerative Diseases. Springer International Publishing, Cham, Vol. 20, pp. 173–212
Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee K-H, Badders N, Valentine M, Miller BL, Wong PC et al (2015) GGGGCC repeat expansion in C9ORF72 compromises nucleocytoplasmic transport. Nature 525:129–133
Galmozzi E, Aghemo A, Colombo M (2012) Eukaryotic initiation factor 5B: a new player for the anti-hepatitis C virus effect of ribavirin? Med Hypotheses 79:471–473
García-García C, Frieda KL, Feoktistova K, Fraser CS, Block SM (2015) Factor-dependent processivity in human eIF4A DEAD-box helicase. Science 348:1486–1488
Gendron TF, Bieniek KF, Zhang Y-J, Jansen-West K, Ash PEA, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J et al (2013) Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. (Berl.) 126:829–844
Goodman LD, Prudencio M, Kramer NJ, Martinez-Ramirez LF, Srinivasan AR, Lan M, Parisi MJ, Zhu Y, Chew J, Cook CN, et al. (2019) Toxic expanded GGGGCC repeat transcription is mediated by the PAF1 complex in C9orf72-associated FTD. Nature Neuroscience. in press
Green KM, Glineburg MR, Kearse MG, Flores BN, Linsalata AE, Fedak SJ, Goldstrohm AC, Barmada SJ, Todd PK (2017) RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nat Commun 8
Green KM, Linsalata AE, Todd PK (2016) RAN translation—what makes it run? Brain Res 1647:30–42
Haeusler AR, Donnelly CJ, Periz G, Simko EAJ, Shaw PG, Kim M-S, Maragakis NJ, Troncoso JC, Pandey A, Sattler R et al (2014) C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507:195–200
Haeusler AR, Donnelly CJ, Rothstein JD (2016) The expanding biology of the C9orf72 nucleotide repeat expansion in neurodegenerative disease. Nat Rev Neurosci 17:383–395
Harms U, Andreou AZ, Gubaev A, Klostermeier D (2014) eIF4B, eIF4G and RNA regulate eIF4A activity in translation initiation by modulating the eIF4A conformational cycle. Nucleic Acids Res 42:7911–7922
Hautbergue GM, Castelli LM, Ferraiuolo L, Sanchez-Martinez A, Cooper-Knock J, Higginbottom A, Lin Y-H, Bauer CS, Dodd JE, Myszczynska MA et al (2017) SRSF1-dependent nuclear export inhibition of C9ORF72 repeat transcripts prevents neurodegeneration and associated motor deficits. Nat Commun 8
Hernández G, Vázquez-Pianzola P, Zurbriggen A, Altmann M, Sierra JM, Rivera-Pomar R (2004) Two functionally redundant isoforms of Drosophila melanogaster eukaryotic initiation factor 4B are involved in cap-dependent translation, cell survival, and proliferation. Eur J Biochem 271:2923–2936
Ito D, Hatano M, Suzuki N (2017) RNA binding proteins and the pathological cascade in ALS/FTD neurodegeneration. Sci. Transl. Med 9:–eaah5436
Kanekura K, Yagi T, Cammack AJ, Mahadevan J, Kuroda M, Harms MB, Miller TM, Urano F (2016) Poly-dipeptides encoded by the C9ORF72 repeats block global protein translation. Hum Mol Genet 25:1803–1813
Kearse MG, Green KM, Krans A, Rodriguez CM, Linsalata AE, Goldstrohm AC, Todd PK (2016) CGG repeat-associated non-AUG translation utilizes a cap-dependent scanning mechanism of initiation to produce toxic proteins. Mol Cell 62:314–322
Kearse MG, Wilusz JE (2017) Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev 31:1717–1731
Khan MA, Goss DJ (2012) Poly(a)-binding protein increases the binding affinity and kinetic rates of interaction of viral protein linked to genome with translation initiation factors eIFiso4F and eIFiso4F·4B complex. Biochemistry 51:1388–1395
Kim H-J, Raphael AR, LaDow ES, McGurk L, Weber R, Trojanowski JQ, Lee VM-Y, Finkbeiner S, Gitler AD, Bonini NM (2014) Therapeutic modulation of eIF2α-phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat Genet 46:152–160
Kramer NJ, Carlomagno Y, Zhang Y-J, Almeida S, Cook CN, Gendron TF, Prudencio M, Blitterswijk MV, Belzil V, Couthouis J et al (2016) Spt4 selectively regulates the expression of C9orf72 sense and antisense mutant transcripts. Science 353:708–712
Kramer NJ, Carlomagno Y, Zhang Y-J, Almeida S, Cook CN, Gendron TF, Prudencio M, Van Blitterswijk M, Belzil V, Couthouis J et al (2016) Spt4 selectively regulates the expression of C9orf72 sense and antisense mutant transcripts associated with c9FTD/ALS. Science 353:708–712
Leppek K, Das R, Barna M (2018) Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol 19:158–174
Levis R, Hazelrigg T, Rubin GM (1985) Effects of genomic position on the expression of transduced copies of the white gene of Drosophila. Science 229:558–561
Liberman N, Gandin V, Svitkin YV, David M, Virgili G, Jaramillo M, Holcik M, Nagar B, Kimchi A, Sonenberg N (2015) DAP5 associates with eIF2β and eIF4AI to promote internal ribosome entry site driven translation. Nucleic Acids Res 43:3764–3775
Lin KY, Nag N, Pestova TV, Marintchev A (2018) Human eIF5 and eIF1A compete for binding to eIF5B. Biochemistry 57:5910–5920
Liu EY, Russ J, Wu K, Neal D, Suh E, McNally AG, Irwin DJ, Van Deerlin VM, Lee EB (2014) C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD. Acta Neuropathol. (Berl.) 128:525–541
Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, Weng S-M, Haass C, Kretzschmar HA, Edbauer D et al (2013) Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol. (Berl.) 126:859–879
Mackenzie IRA, Frick P, Grässer FA, Gendron TF, Petrucelli L, Cashman NR, Edbauer D, Kremmer E, Prudlo J, Troost D et al (2015) Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathol (Berl) 130:845–861
Mann DM, Rollinson S, Robinson A, Bennion Callister J, Thompson JC, Snowden JS, Gendron T, Petrucelli L, Masuda-Suzukake M, Hasegawa M et al (2013) Dipeptide repeat proteins are present in the p62 positive inclusions in patients with frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol. Commun. 1:68
Marygold SJ, Attrill H, Lasko P (2017) The translation factors of Drosophila melanogaster. Fly (Austin) 11:65–74
McGurk L, Bonini NM (2012) Protein interacting with C kinase (PICK1) is a suppressor of spinocerebellar ataxia 3-associated neurodegeneration in Drosophila. Hum Mol Genet 21:76
Méthot N, Song MS, Sonenberg N (1996) A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol Cell Biol 16:5328–5334
Mizielinska S, Grönke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IOC, Pietrzyk J et al (2014) C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345:1192–1194
Mokas S, Mills JR, Garreau C, Fournier M-J, Robert F, Arya P, Kaufman RJ, Pelletier J, Mazroui R (2009) Uncoupling stress granule assembly and translation initiation inhibition. Mol Biol Cell 20:2673–2683
Mordes DA, Prudencio M, Goodman LD, Klim JR, Moccia R, Limone F, Pietilainen O, Chowdhary K, Dickson DW, Rademakers R et al (2018) Dipeptide repeat proteins activate a heat shock response found in C9ORF72-ALS/FTLD patients. Acta Neuropathol Commun 6
Mori K, Weng S-M, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C et al (2013) The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339:1335–1338
Ni J-Q, Zhou R, Czech B, Liu L-P, Holderbaum L, Yang-Zhou D, Shim H-S, Tao R, Handler D, Karpowicz P et al (2011) A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods 8:405
Niblock M, Smith BN, Lee Y-B, Sardone V, Topp S, Troakes C, Al-Sarraj S, Leblond CS, Dion PA, Rouleau GA et al (2016) Retention of hexanucleotide repeat-containing intron in C9orf72 mRNA: implications for the pathogenesis of ALS/FTD. Acta Neuropathol. Commun. 4
Nielsen KH, Behrens MA, He Y, Oliveira CLP, Sottrup Jensen L, Hoffmann SV, Pedersen JS, Andersen GR (2011) Synergistic activation of eIF4A by eIF4B and eIF4G. Nucleic Acids Res 39:2678–2689
Perkins LA, Holderbaum L, Tao R, Hu Y, Sopko R, McCall K, Yang-Zhou D, Flockhart I, Binari R, Shim H-S et al (2015) The transgenic RNAi project at Harvard Medical School: resources and validation. Genetics 201:843–852
Pisareva VP, Pisarev AV (2014) eIF5 and eIF5B together stimulate 48S initiation complex formation during ribosomal scanning. Nucleic Acids Res 42:12052–12069
Prudencio M, Belzil VV, Batra R, Ross CA, Gendron TF, Pregent LJ, Murray ME, Overstreet KK, Piazza-Johnston AE, Desaro P et al (2015) Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat Neurosci 18:1175–1182
Prudencio M, Gonzales PK, Cook CN, Gendron TF, Daughrity LM, Song Y, Ebbert MTW, van Blitterswijk M, Zhang Y-J, Jansen-West K et al (2017) Repetitive element transcripts are elevated in the brain of C9orf72 ALS/FTLD patients. Hum Mol Genet 26:3421–3431
Raught B, Peiretti F, Gingras A-C, Livingstone M, Shahbazian D, Mayeur GL, Polakiewicz RD, Sonenberg N, Hershey JW (2004) Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J 23:1761–1769
Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L et al (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72:257–268
Richter-Cook NJ, Dever TE, Hensold JO, Merrick WC (1998) Purification and characterization of a new EUKARYOTIC protein translation factor EUKARYOTIC INITIATION FACTOR 4H. J Biol Chem 273:7579–7587
Rode S, Ohm H, Anhäuser L, Wagner M, Rosing M, Deng X, Sin O, Leidel SA, Storkebaum E, Rentmeister A et al (2018) Differential requirement for translation initiation factor pathways during ecdysone-dependent neuronal remodeling in Drosophila. Cell Rep 24:2287–2299 e4
Rogers GW, Richter NJ, Lima WF, Merrick WC (2001) Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem 276:30914–30922
Roux PP, Topisirovic I (2018) Signaling pathways involved in the regulation of mRNA translation. Mol Cell Biol 38
Rozovsky N, Butterworth AC, Moore MJ (2008) Interactions between eIF4AI and its accessory factors eIF4B and eIF4H. RNA 14:2136–2148
Sareen D, O’Rourke JG, Meera P, Muhammad AKMG, Grant S, Simpkinson M, Bell S, Carmona S, Ornelas L, Sahabian A et al (2013) Targeting RNA foci in iPSC-derived motor neurons from ALS patients with C9ORF72 repeat expansion. Sci. Transl. Med. 5:208ra149
Satoh J, Yamamoto Y, Kitano S, Takitani M, Asahina N, Kino Y (2014) Molecular network analysis suggests a logical hypothesis for the pathological role of C9orf72 in amyotrophic lateral sclerosis/frontotemporal dementia. J Cent Nerv Syst Dis 6:69–78
Sellier C, Buijsen RAM, He F, Natla S, Jung L, Tropel P, Gaucherot A, Jacobs H, Meziane H, Vincent A et al (2017) Translation of expanded CGG repeats into FMRpolyG is pathogenic and may contribute to fragile X tremor Ataxia syndrome. Neuron 93:331–347
Sen ND, Zhou F, Harris MS, Ingolia NT, Hinnebusch AG (2016) eIF4B stimulates translation of long mRNAs with structured 5′ UTRs and low closed-loop potential but weak dependence on eIF4G. Proc Natl Acad Sci U S A 113:10464–10472
Sharma SD, Kraft JJ, Miller WA, Goss DJ (2015) Recruitment of the 40S ribosome subunit to the 3′-untranslated region (UTR) of a viral mRNA, via the eIF4 complex, facilitates cap-independent translation. J Biol Chem 290:11268–11281
Shatsky IN, Terenin IM, Smirnova VV, Andreev DE (2018) Cap-independent translation: What’s in a name? Trends Biochem Sci. https://doi.org/10.1016/j.tibs.2018.04.011
Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745
Spilka R, Ernst C, Mehta AK, Haybaeck J (2013) Eukaryotic translation initiation factors in cancer development and progression. Cancer Lett 340:9–21
Spirin AS (2009) How does a scanning ribosomal particle move along the 5′-untranslated region of eukaryotic mRNA? Brownian ratchet model. Biochemistry 48:10688–10692
Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, Misra S, Rubin GM (1999) The Berkeley Drosophila genome project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153:135–177
Spradling AC, Stern DM, Kiss I, Roote J, Laverty T, Rubin GM (1995) Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc Natl Acad Sci U S A 92:10824–10830
Sun Y, Atas E, Lindqvist L, Sonenberg N, Pelletier J, Meller A (2012) The eukaryotic initiation factor eIF4H facilitates loop-binding, repetitive RNA unwinding by the eIF4A DEAD-box helicase. Nucleic Acids Res 40:6199–6207
Suzuki, H., Shibagaki, Y., Hattori, S. and Matsuoka, M. (2018) The proline–arginine repeat protein linked to C9-ALS/FTD causes neuronal toxicity by inhibiting the DEAD-box RNA helicase-mediated ribosome biogenesis. Cell Death Dis., 9
Tabet R, Schaeffer L, Freyermuth F, Jambeau M, Workman M, Lee C-Z, Lin C-C, Jiang J, Jansen-West K, Abou-Hamdan H et al (2018) CUG initiation and frameshifting enable production of dipeptide repeat proteins from ALS/FTD C9ORF72 transcripts. Nat Commun 9:152
Tao Z, Wang H, Xia Q, Li K, Li K, Jiang X, Xu G, Wang G, Ying Z (2015) Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum Mol Genet 24:2426–2441
Terenin IM, Smirnova VV, Andreev DE, Dmitriev SE, Shatsky IN (2017) A researcher’s guide to the galaxy of IRESs. Cell Mol Life Sci 74:1431–1455
Todd PK, Oh SY, Krans A, He F, Sellier C, Frazer M, Renoux AJ, Chen K, Scaglione KM, Basrur V et al (2013) CGG repeat-associated translation mediates neurodegeneration in fragile X tremor Ataxia syndrome. Neuron 78:440–455
Tran H, Almeida S, Moore J, Gendron TF, Chalasani U, Lu Y, Du X, Nickerson JA, Petrucelli L, Weng Z et al (2015) Differential toxicity of nuclear RNA foci versus dipeptide repeat proteins in a Drosophila model of C9ORF72 FTD/ALS. Neuron 87:1207–1214
van Gorp AGM, van der Vos KE, Brenkman AB, Bremer A, van den Broek N, Zwartkruis F, Hershey JW, Burgering BMT, Calkhoven CF, Coffer PJ (2009) AGC kinases regulate phosphorylation and activation of eukaryotic translation initiation factor 4B. Oncogene 28:95–106
Vatovec S, Kovanda A, Rogelj B (2014) Unconventional features of C9ORF72 expanded repeat in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Neurobiol. Aging 35:2421.e1–2421.e12
Vaysse C, Philippe C, Martineau Y, Quelen C, Hieblot C, Renaud C, Nicaise Y, Desquesnes A, Pannese M, Filleron T et al (2015) Key contribution of eIF4H-mediated translational control in tumor promotion. Oncotarget 6:39924–39940
Walker SE, Zhou F, Mitchell SF, Larson VS, Valasek L, Hinnebusch AG, Lorsch JR (2013) Yeast eIF4B binds to the head of the 40S ribosomal subunit and promotes mRNA recruitment through its N-terminal and internal repeat domains. RNA 19:191–207
Yuva-Aydemir Y, Almeida S, Gao F-B (2018) Insights into C9ORF72-related ALS/FTD from Drosophila and iPSC models. Trends Neurosci. https://doi.org/10.1016/j.tins.2018.04.002
Zhang Y-J, Gendron TF, Ebbert MTW, O’Raw AD, Yue M, Jansen-West K, Zhang X, Prudencio M, Chew J, Cook CN et al (2018) Poly (GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat Med 24:1136–1142
Zhao M, Kim JR, van Bruggen R, Park J (2018) RNA-binding proteins in amyotrophic lateral sclerosis. Mol Cells 41:818–829
Zu T, Pattamatta A, Ranum LPW (2018) Repeat-associated non-ATG translation in neurological diseases. Cold Spring Harb Perspect Biol 10:a033019
Sonobe Y, Ghadge G, Masaki K, Sendoel A, Fuchs E, Roos RP (2018) Translation of dipeptide repeat proteins from the C9ORF72 expanded repeat is associated with cellular stress. Neurobiology of Disease 116:155-165
Westergard T, McAvoy K, Russell K, Wen X, Pang Y, Morris B, Pasinelli P, Trotti D, and Haeusler A (2019). Repeat‐associated non‐AUG translation in C9orf72‐ALS/FTD is driven by neuronal excitation and stress. EMBO Mol Med: e9423.
Acknowledgements
We thank Shizuka B. Yamada and Aaron Gitler at Stanford University for post-screening investigations. Also, thank you to Edward B. Lee, Thomas A. Jongens, Zhaolan (Joe) Zhou and members of the Bonini laboratory for helpful comments. Further thanks to Peter Todd for discussions on unpublished translation factor data. Undergraduate and post-baccalaureate students Luis F. Martinez-Ramirez, Kimberley Newman, Benjamin Gallo, and Ken Yanagisawa provided minimal technical support. We thank the Transgenic RNAi Project (TRiP) at Harvard Medical School (NIH/NIGMS R01-GM084947) and the Vienna Drosophila Research Center for developing transgenic RNAi fly stocks used in this study. We thank the Center for Neurodegenerative Disease Research (CNDR) at the University of Pennsylvania for the antibody targeting GR. This work was supported by the Systems and Integrative Biology NIH/NIGMS training grant T32-GM07517 (to LDG), Amyotrophic Lateral Sclerosis Association (to MP and LP), NIH/NINDS R35-NS097273 (to LP), NIH/NINDS P01-NS084974 (to LP), NIH/NINDS P01-NS099114 (LP)], Mayo Clinic Foundation (to LP), Robert Packard Center for ALS Research at Johns Hopkins (to LP), Target ALS Foundation (to LP), NIH/NINDS R01-NS078283 (to NMB), and NIH/NINDS R35-NS09727 (to NMB).
Author information
Authors and Affiliations
Contributions
This work was done by LDG under the mentorship of NMB. MP performed patient studies and analyses under the mentorship of LP. ARS performed western blots on fibroblast cells. OR setup crosses and collected samples for DsRed fluorescence imaging. VYML provided a GR-antibody. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Figure S1. Extended LDS-G4C2 fly line characterization. Figure S2. Extended translation factor screen data. Figure S3. eIF4B and eIF4H1 RNAi-2 data. Figure S4. Full western immunoblot images. (DOCX 1150 kb)
Additional file 2:
Table S2. Translation Factor screen details. (PDF 157 kb)
Additional file 3:
Table S3. Fibroblast lines. (PDF 56 kb)
Additional file 4:
Table S4. Patient samples. (PDF 34 kb)
Additional file 5:
Table S5. Fly lines used. (PDF 80 kb)
Additional file 6:
Table S6. primers. (PDF 74 kb)
Additional file 7:
Table S1. Full genotypes in figures. (PDF 121 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Goodman, L.D., Prudencio, M., Srinivasan, A.R. et al. eIF4B and eIF4H mediate GR production from expanded G4C2 in a Drosophila model for C9orf72-associated ALS. acta neuropathol commun 7, 62 (2019). https://doi.org/10.1186/s40478-019-0711-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40478-019-0711-9