Abstract
GJA1 (connexin43) has been predicted as the top key driver of an astrocyte enriched subnetwork associated with Alzheimer’s disease (AD). In this study, we comprehensively examined GJA1 expression across 29 transcriptomic and proteomic datasets from post-mortem AD and normal control brains. We demonstrated that GJA1 was strongly associated with AD amyloid and tau pathologies and cognitive functions. RNA sequencing analysis of Gja1−/− astrocytes validated that Gja1 regulated the subnetwork identified in AD, and many genes involved in Aβ metabolism. Astrocytes lacking Gja1 showed reduced Apoe protein levels as well as impaired Aβ phagocytosis. Consistent with this, wildtype neurons co-cultured with Gja1−/− astrocytes contained higher levels of Aβ species than those with wildtype astrocytes. Moreover, Gja1−/− astrocytes was more neuroprotective under Aβ stress. Our results underscore the importance of GJA1 in AD pathogenesis and its potential for further investigation as a promising pharmacological target in AD.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) reflects multifactorial genetic and environmental perturbations that, in turn, cause pleiotropic changes in molecular networks linking a host of biological processes. By employing an integrative network biology approach to analyze a large-scale genetic and gene expression dataset in late onset Alzheimer’s disease (LOAD), we previously conducted an unbiased identification and prioritization of gene networks associated with clinical and pathological progression of the disease [98]. In addition to a prominent driver of LOAD, an immune response subnetwork (module) governed by TYROBP, we identified other network modules within which gene coexpression patterns were significantly changed in LOAD subjects compared to controls. In the current study, we focused on the identification and characterization of causal regulators of a module that shows a dramatic loss of coordination in AD and that includes APOE, a well-known risk factor of AD [18, 82]. This module was highly enriched in astrocyte specific genes, with GJA1 predicted as its top regulator.
GJA1, also known as connexin43 (Cx43), is a member of the connexin family of proteins that is highly conserved among vertebrates. Members of the connexin family exist at the plasma membrane as hexameric complexes known as connexons, and function as connexin hemichannels allowing permeability to small molecules and ions [28]. Two connexons at the apposed cell surface of adjacent cells form a trans-dimer called a gap junction channel (GJC). Gap junctions are clusters of gap junction channels, and mediate efficient and rapid bidirectional inter-cellular transmission and transport of electrical and chemical signals [55, 80]. Cx43 also functions as a unitary channel (hemichannel) to participate in paracrine communication [17], but this activity is often associated with pathological conditions [21]. GJA1 and GJB6 (connexin30) are predominantly expressed in mature astrocytes [27, 93, 99], forming astrocytic networks facilitating propagation of calcium waves, potassium and glutamate buffering, and metabolic coupling [66, 70, 77, 78, 83, 89]. Astrocytic gap junctions are critical for neuronal function, as evidenced by the profound neurological phenotypes in Gja1/Gjb6 double knockout mice [56].
Numerous autosomal dominant GJA1 mutations have been reported to cause oculodentodigital dysplasia (ODDD), which is characterized by craniofacial and limb dysmorphisms [42]. Patients with ODDD frequently manifest neurological symptoms including paraparesis, white matter abnormalities, hearing and vision loss, and low IQ [29, 54]. GJA1 mutations associated with ODDD are mostly loss-of-function, although a few gain-of-function cases have also been identified, indicating that diverse functional perturbations of GJA1 can lead to ODDD [42].
Although mice deficient in astrocytic Gja1 are grossly normal, they show reduced synaptic plasticity [30], increased propagation of synaptic depolarization and neuronal inactivation (“spreading depression”) [84] and impaired in rotarod performance [26]. Mice deficient in both Gja1 and Gjb6 spontaneously develop pathological myelin degeneration without overt neuronal atrophy [56]. Loss or reduced expression of Gja1 leads to enlarged infarct size upon induced-ischemia, and the loss of only the C-terminal tail of Cx43 appeared to mediate the damaging effects [41, 44, 51, 64, 65, 85]. These studies indicated that Gja1 plays a neuroprotective function and restricts neuronal damage under oxidative and metabolic stress.
GJA1 has been previously investigated in the context of Alzheimer’s disease. GJA1 (Cx43) immunoreactivity was found to be enriched in astrocytes surrounding amyloid plaques in post-mortem AD brain [63] and a APP/PS1 mouse model [59] and Cx43 hemichannel activity was increased in the APP/PS1 mouse model [96]. Cx43 deficiency or pharmacological blockade of connexins in APP/PS1 mice appeared to reduce dystrophic neurites, mitochondrial oxidative stress, and cognitive impairment without altering amyloid pathology [73, 96, 97].
Here we corroborated the evidence of GJA1 dysregulation in AD by analyzing GJA1 expression in a large number of transcriptomic and proteomic datasets from pathologically and clinically characterized LOAD brain samples. We showed that a number of known AD risk factor genes were significantly correlated with Gja1 in multiple brain regions in AD. We constructed and validated GJA1 regulated gene networks in AD. We revealed that Gja1 regulated the expression of more than half of the known AD risk factor genes. We further demonstrated the effect of Gja1-deficiency on astrocyte function and the subsequent impact on co-cultured neurons.
Materials and methods
Data preprocessing
Three clinical cohorts of post-mortem brain samples from patients with Alzheimer’s disease (AD) symptoms of various severity, and normal controls were subject to RNA microarray and/or RNA-seq analysis to detect changes in mRNA expression caused by AD pathology. As summarized in (64), RNA microarray assays were performed on RNAs extracted from prefrontal cortex (PFC), visual cortex (VC), and cerebellum (CB) of post-mortem brain cortex tissues in the Harvard Brain Tissue Bank (HBTRC) [98]. Details of the research such as neuropathological phenotypic traits of AD subjects and normal controls, RNA microarray assays, data preprocessing and covariate adjustment were described in [98]. Both RNA microarray and RNA-seq assays were conducted using RNAs collected from post-mortem brains in the Mount Sinai Brain Bank (MSBB) cohort [31, 91]. Experimental design and data analysis of RNA microarrays on 19 distinct brain regions of post-mortem brain tissues in the MSBB cohort were presented by Wang et al. [91]. Single-end RNA-seq assays were performed on RNAs extracted from four selected regions (BM10, frontal pole (FP); BM22, superior temporal gyrus (STG); BM36, parahippocampal gyrus (PHG); BM44, inferior frontal gyrus (IFG)) of post-mortem brain tissues in the MSBB cohort. The raw sequence reads were aligned to human genome hg19 with the star aligner (v2.3.0e) [22]. Then the gene level expression was quantified by featureCounts (v1.4.4) [49] based on Ensemble gene model GRCh37.70. The gene level read counts data was normalized using the trimmed mean of M-values normalization (TMM) [76] method to adjust for sequencing library size difference. The normalized data was further adjusted for the covariates postmortem interval (PMI), race, RNA integrity number (RIN), gender, rate of exonic reads, and batch using a linear mixed model [33], where batch was treated as a random effect. The residuals from the regression model were used for downstream analysis. In addition, proteomics assays were performed on proteins extracted from the BM10 region of post-mortem brains in the MSBB cohort, and raw counts summarized at the gene/protein level were provided by the Genomics Core at the Icahn School of Medicine at Mount Sinai. These raw counts were further corrected by covariates (PMI + AOD + batch + gender) using a linear model described by Wang et al. [91], and the residuals after correction were used for further downstream analysis.
The Religious Orders Study and Memory and Aging Project (ROSMAP) cohort consists of ROS and MAP studies [7]. The detailed information regarding ROSMAP study was described in previous studies [8, 9]. Normalized RNA-seq expression data were downloaded from the AMP-AD Knowledge Portal at Synapse upon authentication by the AMP-AD Consortium (doi:https://doi.org/10.7303/syn3388564). Genes with at least 1 FPKM in at least 10% of the samples were selected and then the data was corrected for confounding factors including batch, PMI, gender and RIN via a linear model. The residuals after correction were used for further downstream analysis.
Bayesian causal network analysis
Bayesian causal network was constructed by integrating genome-wide gene expression, SNP genotype, and known transcription factor (TF)-target relationships. Briefly, we first computed expression quantitative trait loci (eQTLs) and then employed a formal statistical causal inference test (CIT) [62] to infer the causal probability between gene pairs associated with the same eQTL. The causal relationships inferred were used, together with TF-target relationships from the ENCODE project, as structural priors for building a causal gene regulatory network from the gene expression data through a Monte Carlo Markov Chain (MCMC) simulation based procedure [100]. We followed a network averaging strategy in which 1000 networks were generated from the MCMC procedure starting with different random structure, and links that shared by more than 30% of the networks were used to define a final consensus network structure. To ensure the consensus network is a directed acyclic graph, an iterative de-loop procedure was conducted, removing the most-weakly supported link of all links involved in any loop. Following Zhang et al. [98], we performed Key Driver Analysis (KDA) on the consensus Bayesian network to identify key hub genes which regulated many downstream nodes.
RNA sequencing and data processing
Total RNAs were sequenced via the Illumina HiSeq 2500 system with 100 nt paired-end read. Sequencing reads were aligned to mouse reference genome mm10 (GRCm38.75) STAR aligner [22] guided by UCSC gene model. Accepted mapped reads were summarized to gene levels using the featureCounts [49] program. Raw count data were normalized by the voom function in the R limma package [75], and then differential expression was called by the moderated t-test implemented in limma. Differentially expressed genes (DEGs) were defined to have at least 1.2-fold change in expression and BH-adjusted p < 0.05 for two comparisons including Gja1−/− astrocytes versus wildtype astrocytes and coculture of Gja1−/− astrocytes and neurons versus co-culture of wildtype astrocytes and neurons. The raw and processed data are available at doi: https://doi.org/10.7303/syn11711769.
Construction of GJA1 centric co-expression networks
We constructed GJA1 centric consensus co-expression networks from 8 cohort datasets from three cohorts including MSBB (4 brain regions), ROSMAP (1 brain region) and HBTRC (3 brain regions). In each dataset, the genes significantly correlated with GJA1 were identified based on BH-corrected p value < 0.05. From those significant correlations, a directional voting method was utilized to calculate the frequency of positive correlation as well as the frequency of negative correlations between GJA1 and each other gene. Gja1 centric networks were then defined as a function of frequency threshold n (=1, 2, …, 8}.
Construction of GJA1 signaling map
Starting from GJA1 as the root, we searched through the khaki module based Bayesian network for further three layers of genetic neighborhood, and trimmed away those single leaf nodes while keeping the remaining nodes. Then, we used the individual nodes or its first neighborhood in the remaining network to query the reactome (https://reactome.org/) and MSigDB database for gene ontology enrichment. A GJA1 signal map was constructed by replacing each individual nodes in the network plot with a relevant gene ontology entry acquired from the query.
Quantitative PCR
Quantitative PCR was performed as previously described [38]. Briefly, total RNA was isolated using Qiagen RNeasy kit, and 1–2 μg of total RNA was used to synthesize cDNA using Ecodry (Clontech, 639,543, Mountain view, CA). Universal probe library in combination with the primers listed (Additional file 1: Table S10) was used to perform qPCR with KAPA PROBE FAST qPCR master mix (KAPA Biosystems, KK4703, Wilmington, MA). Acquired Ct values were loaded onto qbase Plus software package (Biogazelle, Belgium) for data quality control and normalization. Actb, Rpl13a, and Rplp0 were used as normalizer genes.
Cell culture and reagents
Generation of Gja1−/− astrocytes were previously described [66]. WT and Gja1−/− primary astrocytes were prepared by dissecting and dissociating forebrains of P1–3 pups and culturing for 2 weeks in DMEM containing 10% fetal bovine serum and penicillin/streptomycin in T75 flask. Contaminating microglia were reduced by vigorous shaking flasks at 500 rpm for 20 min and replacement of medium containing floating cells. Mouse TNFα (14–8321-62) and IL-1β (14–8012-62) were purchased from eBiosciences and used at concentration of 10 ng/ml. Carbenoxolone (C4790), lanthanum (575275), quinine (Q1125) were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Neuron astrocyte cocultures
For RNA-seq in astrocytes and neuron/astrocyte cocultures, cortical neuron cultures were prepared from E15.5 wildtype C57BL6/J embryos as previously described [14]. 1 × 105 neurons were seeded in poly-D-lysine coated 6 well plates with or without 1 × 105 wildtype or Gja1−/− astrocytes in neurobasal medium (Life Technologies, 21,103–049, Grand Island, NY, USA) supplemented with B27 (Life Technologies, 17,504–044), and cultured for 10 days. At div10, half of medium was replaced with neurobasal containing 20 μM Aβ 1–42 oligomer for four days, and total RNA was harvested using RNA-Bee (AMSBIO, Cambridge, MA, USA) following the manufacturer’s instruction. Four replicate wells were used for each condition.
For MEA assays, E15.5 cortical neuron cultures were prepared from wildtype C57BL6/J as above. MEA wells were coated with poly-D-lysine and one day prior to plating 1 × 105 neurons, 5 × 104 wildtype or Gja1−/− astrocytes were plated in neurobasal medium supplemented with B27. At days in vitro (div) 16, neuron/astrocyte cocultures were treated with 10 μM Aβ1–42 oligomers until div 20.
For neuronal death and viability assays, 2 × 105 cortical neurons were plated onto PDL coated 12 well plate and 1 × 105 wildtype or Gja1−/− astrocytes are plated in Transwells (Costar, 3460, Kennebunk, ME USA). At div7, Transwells with astrocytes were inserted to neuronal cultures and incubated with the indicated Aβ1–42 for 4 or 7 days. LDH assay was performed as in (Kajiwara, 2014) using LDH Cytotoxicity Detection Kit (Clontech, Mountain View, CA, USA). MTT assay was performed as described in (Bruban 2015).
Apoe and Aβ ELISA
Astrocytes were seeded in 6 well plates at the density of 100,000 cells per well in serum free DMEM medium for 48 h with vehicle, 2 or 20 μM Aβ1–42 oligomers. Mouse Apoe secreted in conditioned medium was quantitated using mouse Apoe ELISA kit (Mybiosource, MBS705227, San Diego, CA, USA) following manufacture’s instruction. Astrocytes attached to the plate were used to prepare cell lysates as described below. Mouse amyloidβ 40 and 42 species were quantitatively determined using Human/Rat Amyloid ELISA kits (294–64,701 and 294–64,501, Wako Chemicals, Richmond, VA, USA).
Immunoblotting
Lysates were prepared from astrocytes underwent above procedure in buffer containing phosphate buffered saline, protease inhibitor cocktail (Roche, Indianapolis, IN, USA), and 1x loading buffer. Lysates were resolved by denaturing SDS-PAGE gel and transferred to PVDF membrane. The membranes were blotted with following primary antibody: Cx43 (1:500, 3512, Cell Signaling Technology); Apoe (1:1000, sc-6384, Santa Cruz Biotechnology); Actin (1:1000, A5060, Sigma-Aldrich), and specific signals were detected with HRP-conjugated secondary antibodies followed by SuperSignal West Dura (34,075, ThermoFisher Scientific).
Multielectrode array recordings
Recordings of neuronal activity was performed on Axion 768 channel system (The Maestro, Axion Biosystems, Atlanta, GA, USA) using 48-well plate, which records or stimulates neuronal activity from up to 16 electrodes per well. Manufacture’s recommended settings were used for detection of spikes, bursts and network bursts. Briefly, spikes were detected using Adaptive Threshold Crossing with 6x Standard Deviation; bursts were defined as minimum of 5 spikes with 100 ms of maximum inter-spike interval; and network bursts were defined as minimum of 10 spikes with 100 ms of maximum inter-spike interval and 20 ms of synchrony window. Spontaneous activities were recorded at days in vitro (div) 10, 14, 18, and 20 as follow. A MEA plate was set on heated stage at 37C for at least 5 min prior to initiation of recordings, which lasted for 10 min. All the recordings and analysis were performed on Axion Integrated Studio (Axion Biosystems). Stimulation was performed div 14 and 20 using the same spike and burst settings as in spontaneous recordings. A stimulation given was comprised of 3 repeated cycles of voltage-controlled (500 mV/− 500 mV), biphasic (500 μs each), and positive-first stimulus followed by 3 ms of floating duration [88], and was given to all electrodes. Stimulation was given 5 times with 5 s interval and stimulation data were analyzed using NeuralMetricTool (Axion Biosystems). Results are given as means of 5 stimulations.
Statistics
All of the statistical significance levels reported in this study were corrected for multiple testing unless otherwise specified. Statistical analysis was performed using either R programming language or SPSS version 20 or higher (IBM corporation, Somers, NY, USA). Amyloid uptake assays were analyzed by Student’s t-test. Spontaneous neural recordings were analyzed by Man-Whitney, or Kruskal-Wallis non-parametric analysis.
Results
Figure 1 shows the workflow of the integrative network analysis and functional validation experiments performed in this study. We first examined the relationships between GJA1 mRNA expression and clinical and pathological traits in 29 AD transcriptomic datasets. We also investigated the role of GJA1 in gene networks underlying AD. We then performed in vitro experiments to study the role of GJA1 in regulating AD gene networks and AD related phenotypes using primary astrocytes purified and cultured from wildtype and astrocyte specific Gja1−/− mice. Gja1’s target gene signatures identified from the RNA-seq data from the in vitro experiments were then projected onto the GJA1 centric networks to validate these networks’ structures.
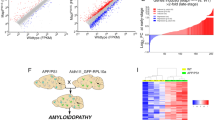
Overview of the integrative network analyses and validation experiments performed in the study. a. GJA1 mRNA expression changes and its correlations with clinical and pathological traits were systematically investigated in 29 datasets. GJA1-centric coexpression and regulatory networks. b. Workflow of in vitro functional validation study. Wildtype and Gja1−/− astrocytes with or without wildtype neurons were used to prepare RNA for sequencing and to perform various functional validations (c). d. GJA1’s gene signatures between wildtype and Gja1−/− astrocytes and between coculture of wildtype astrocytes and wildtype neurons and coculture of Gja1−/− astrocytes and wildtype neurons were identified from RNA-sequencing data from the experiments in b. e. GJA1’s gene signatures were used to validate the network structures predicted from the transcriptomic datasets in human AD brains. Functional relevance of GJA1’s gene signatures was also investigated
GJA1 is a key regulator of an astrocyte specific gene subnetwork dysregulated in LOAD
Several studies have used co-expression network analysis to find modules of co-regulated genes in AD [36, 60, 61]. Our previous study developed a novel network approach capable of integrating clinical and neuropathological data with large-scale genetic and gene expression [98]. This network biology approach led to a novel multiscale network model of LOAD, which identified a number of coexpressed gene modules that were strongly associated with AD pathological traits or underwent dramatic disruption of high-order gene-gene interactions [98]. One such module, referred to as the khaki module in the original construction of this network, was of particular interest since it included APOE, the top AD risk factor gene. Moreover, the average interaction strength among its member genes in LOAD was reduced by 71% compared to that in normal control at a false discovery rate (FDR) < 2%, suggesting a huge loss of coordination among this group of genes in AD. The khaki module was enriched for the genes in Gamma-aminobutyrate (GABA) biosynthesis and metabolism (24 fold enrichment (FE), Fisher’s exact test (FET) p = 0.046) and harbored 12 (ALDOC, APOE, AQP4, ATP1A2, CSPG3, CST3, EDG1, EMX2, GJA1, PPAP2B, PRDX6 and SPARCL1) of 46 known astrocyte marker genes, a 15-fold enrichment over what would be expected by chance (FET p = 6.55E-9). The module was also enriched for the expression of the common variants identified as genome-wide significant by AD genome wide association studies (GWAS) (3-FE, FET p = 1.92E-11). Bayesian causal network analysis showed that GJA1 was the top driver of the module followed by FXYD1, STON2 and CST3 [98]. The key drivers of the corresponding causal network of the module were the nodes that had a large number of downstream nodes [90, 98]. These results indicate that GJA1 is a potential regulator of molecular networks in AD. In the next subsection, we will investigate the association between Gja1 mRNA expression and AD. All of the statistical significance levels reported were corrected for multiple testing unless otherwise specified.
GJA1 expression is associated with AD clinical and pathophysiological traits
To gain insight into the role of GJA1 in cognitive functions and AD pathogenesis, we first extensively investigated how GJA1 expression at the mRNA level was correlated with AD neuropathological traits in 29 gene expression datasets from three AD cohort studies of aging and dementia that included organ donation at death: the Mount Sinai/JJ Peters VA Medical Center Brain Bank (MSBB; Additional file 1: Table S1) [91], the Religious Orders Study and the Rush Memory and Aging Project (ROSMAP) [8, 9] and in the Harvard Brain Tissue Resource Center Alzheimer’s Disease study (HBTRC) [98]. We chose six different clinical and pathological criteria to evaluate the clinical relevance of GJA1 on AD pathology and cognitive functions: the MiniMental State Examination (MMSE) score [25, 32], the sum of NFT density estimates for all cortical regions examined (NTrSum), Mean Plaque density (PLQ_Mn) for the estimation of average plaque density, Braak stage score for quantitative assessment of neurofibrillary tangles [11], the Consortium to Establish a Registry for Alzheimer’s disease (CERAD: 1 for definite AD, 2 for probable AD, 3 possible AD, 4 for normal control) score for quantitative measure of neuritic plaques, and clinical dementia rating score (CDR ranging between 0 and 5 with 0 for normal control and 5 for severe dementia).
In the microarray data in the ROSMAP cohort, GJA1 expression was significantly correlated with CERAD score (r = − 0.15, p = 3.3E-3) and the MiniMental State Examination (MMSE) score (r = − 0.14, p = 6.4E-3). Similar results were observed in the ROSMAP RNA-seq dataset (Fig. 2a, and Additional file 1: Table S2), suggesting that the mRNA expression of GJA1 is associated with AD pathogenesis and dementia. The MSBB AD cohort includes microarray and RNA-seq data from a battery of distinct brain cortical regions and thus provides an excellent opportunity to investigate regional differences in the correlation between GJA1 expression and AD neuropathological traits [91]. Among the 19 brain cortex regions investigated in the MSBB AD microarray data, GJA1 expression in six cortex regions including BM10 (frontal pole), BM20 (inferior temporal gyrus), BM21 (middle temporal gyrus), BM32 (anterior cingulate), BM36 (parahippocampal gyrus) and BM46 (dorsolateral prefrontal cortex) was significantly correlated with at least three AD neuropathological traits (Fig. 2b, and Additional file 1: Table S2). Overall, GJA1 expression in these six cortex regions displayed a significant positive correlation with Braak stage score, PLQ_Mn, NTrSum and CDR. The MSBB AD RNA-seq data revealed a consistent pattern of correlation between GJA1 expression and AD clinic traits across the cortical regions studied (Fig. 2c-e; Additional file 1: Table S2). Notably, in BM10, BM36 and BM44 cortex regions, the microarray and RNA-seq data converged to show a consistent correlation between GJA1 expression and AD neuropathological traits (Additional file 1: Table S2). Thus, in the MSBB cohort, the association between GJA1 expression and AD neuropathological traits was cortex-specific. At the protein level, GJA1 in the brain cortex BM10 region was significantly correlated with CERAD (r = − 0.32, p = 1.26E-07), PLQ_Mn (r = 0.37, p = 4.08E-10) (Fig. 2f) and CDR (r = 0.35, p = 5.69E-9) (Additional file 1: Table S2). Also the total soluble amyloidβ (Aβ) levels had a significant positive correlation with GJA1 protein levels in the BM10 region (r = 0.18, p = 0.0036).
GJA1 mRNA/protein expression is correlated with AD pathogenesis, dementia and known AD risk factor genes. GJA1 mRNA expression is correlated with the MMSE score in the prefrontal cortex region in the ROSMAP cohort (a), the NTrSum score in the cortex region frontal pole (BM10) the MSBB cohort (b), the Braak score in the parahippocampal gyrus (BM36) in the MSBB cohort (c), and the CDR score in BM36 in the MSBB cohort (d), and CERAD in BM36 in the MSBB cohort (e). The GJA1 protein level is also correlated with the plaque mean density (PLQ_Mn) in the frontal pole (BM10) in the MSBB cohort (f). Inset was the correlation coefficient along its p value between individual clinical traits and GJA1 mRNA/protein level. Severity of AD symptoms was classified based on each individual AD neuropathological and cognitive traits according to the criteria established in the ROSMAP clinic codebook [91, 92]. g. Gja1 mRNA expression is correlated with a vast majority of the ADGWAS genes in the MSBB (BM10, BM22, BM36 and BM44) and ROSMAP RNA-seq datasets. Colors represent Correlation coefficients: red, high; blue, low; yellow, in between. * and ** stand for significance at 5 and 0.5% level, respectively while ns indicates insignificant correlation and na stands for “not applicable”. Shown in the heatmap are only the ADGWAS genes that passed RNA-seq data preprocessing, normalization and annotation
We showed previously in the HBTRC cohort that Gja1 had a significant correlation with Braak score (r = 0.61 and 0.52 for dorsolateral prefrontal cortex and cerebellum cortex regions, respectively) [98].
We further checked how GJA1 mRNA expression was correlated with 30 known AD risk factor genes. As shown in Fig. 2g, a majority of those AD genes were significantly correlated to GJA1 in the five RNA-seq datasets.
We also examined GJA1 differential expression between various subgroups of AD severity with respect to each individual AD neuropathological or functional cognitive trait using pairwise Student’s t-test. Consistent with the correlation analysis, Gja1 expression increased significantly as the disease deteriorated (Additional file 1: Table S3).
Because APOE is one of the major AD risk factors, and age and sex are critical clinic covariables in AD neuropathology, we investigated whether age, sex and APOE genotypes had any impact on the association of GJA1 expression with AD clinical and pathological traits. We stratified this cohort by age of death (AOD) (AOD > 85 versus AOD < 85), sex (female versus male), and APOE genotypes (E23, E34 and E33). As demonstrated in Additional file 1: Table S4 and Additional file 2: Figure S1, GJA1 expression is more significantly associated with clinical dementia rating (CDR) in the group with AOD > 85 than that with AOD < 85, in females than males, and in the group with APOE E33 than that with E34 or E23, across four brain regions in the MSBB cohort, suggesting that age, sex and APOE genotypes impact the association of GJA1 expression with clinical and pathological traits.
We further assessed if GJA1 mRNA expression was correlated with the variants of the known AD risk genes using the RNA-seq data from the brain region BM36 in the MSBB cohort. Among the 28 ADGWAS genes that had identifiable variants in the present study, 18 had at least one variant that possessed a significant correlation with GJA1 (Additional file 2: Figure S2). For example, the transcript/isoform ENST00000532146 of the risk factor CELF1 is significantly correlated with GJA1 expression in BM10 and BM44, but not BM22 and BM36 while ENST00000534614 and ENST00000539254 are correlated with GJA1 expression only in BM36 and BM22, respectively (Additional file 2: Figure S2). These results suggested that variation in transcripts’ abundance might be an important factor for determining the co-regulation between GJA1 and AD risk factors.
In summary, both correlation and differential expression analyses revealed that GJA1 was associated with amyloid and tau pathologies of AD as well as cognitive functions suggesting that GJA1 may play an important role in AD.
Transcriptomic changes caused by Gja1 deficiency in mouse astrocytes
To validate the role for GJA1 in orchestrating the astrocytic transcriptome, we purified and cultured primary astrocytes from wildtype and astrocyte specific Gja1−/− mice, and identified differentially expressed genes (DEGs) by RNA-seq in Gja1−/− vs wildtype primary astrocytic cultures in the absence or presence of wildtype primary cortical neurons. Each group had four replicates. All cultures were treated with 10 μM Aβ1–42 oligomer from div 10 through 14, when total RNAs were harvested. We identified 2891 upregulated (termed AST(up)) and 2605 downregulated (termed AST(dn)) DEGs upon the ablation of the Gja1 gene as compared to wildtype primary astrocytes (Fig. 3a). We identified 573 upregulated genes (termed AST + NEU(up)) and 1391 downregulated genes (termed AST + NEU(dn)) in Gja1−/− vs. wildtype primary astrocytes co-cultured with primary neurons (Fig. 3a). AST(up) shared 328 genes (11.3%) with AST + NEU(up) and 208 genes with AST + NEU(dn) while AST(dn) shared 672 and 60 genes with AST-NEU(dn) and AST-NEU(up), respectively (Fig. 3a). These DEG signatures were enriched for a variety of biological pathways including translational processes, immune response, cell-cell communication, extracellular matrix, microtubule cytoskeleton, synaptic transmission, lipid biosynthesis, steroid biosynthesis, cholesterol biosynthesis and cell-matrix adhesion (Fig. 3a-b, Additional file 1: Table S5).
The impact of GJA1 deficiency on AD relevant gene ontologies and AD risk networks. a. Venn diagram for genes overlapping among various differentially expressed genes (DEG) signatures. AST(dn) and AST(up) were down- and up- regulated DEG signatures in Gja1−/− vs. wildtype astrocyte cultures, while AST + NEU(dn) and AST + NEU(up) were down- and up- regulated DEG signatures in Gja1−/− vs. wildtype astrocytes co-cultured with neurons. Top gene ontology for each individual DEGs were shown above each cluster. b. Heatmap of Gene enrichment significance (−log10(p)) for the DEG signatures. Top 10 enriched gene ontologies were shown for each individual DEGs. c. GJA1−/− signatures including AST(all), AST(up), AST(dn), AST + NEU(all), AST + NEU(up) and AST + NEU(dn) are enriched for three AD genetic gene sets including the one identified by the International Genomics of Alzheimer’s Project (IGAP) [1], the Abeta (Aβ) genetic network [16], the AD risk gene list (ADGWAS) [43]
AST(up) was significantly enriched for the genes in an Aβ network signature [16] (2.03 FE, FET p = 1.08 E-10), the AD genome-wide significant risk factor gene signature (ADGWAS) (3.47 FE, FET p = 6.58E-6) (Fig. 2c) while AST + NEU(dn) was enriched for the genes in the Aβ network signature (1.68 FE, FET p = 1.1E-3) (Fig. 3c and Additional file 1: Table S6). These results suggested that Gja1 is a critical regulator of the AD GWAS genes and may play an important role in Aβ metabolism.
We further intersected the GJA1 KO DEG signatures with the signatures from other inflammatory diseases to better understand of the immune response component in GJA1 regulated gene expression. We considered 2 well-established inflammatory gene signatures, an inflammatome signature of 2461 genes from eleven rodent inflammatory disease models [90] and the human macrophage and immune response enriched module of 2483 genes causally linked to obesity and diabetes [23]. These two inflammatory signatures share 758 genes [termed the core disease-related inflammatory gene (CDIG)]. About half of the CDIGs fall into AST(up) (accounting for about 12.6% of AST(up), 6.57 FE, FET p = 1.85E-208) and the intersection includes well known inflammatory markers (e.g., CD44, CD53, FCER1G, HCK, TYROBP and TREM2), inflammation complement component members (C1QA, C1QB, and C1QC), CXC chemokines (CXCL10, CXCL3 and CXCL6) and TNF Receptors (TNFRSF11B and TNFRSF13B). The CDIGs are also significantly enriched in AST + NEU(up) (accounting for about 15% of AST + NEU(up), 7.84 FE, FET p = 1.06E-49). These results suggest that inflammation is a critical component in GJA1-regulated gene expression.
We then investigated expression changes of astrocyte- and neuron-specific genes in various gene signatures using the recently identified brain cell type specific signatures [57]. Astrocyte- and neuron-specific marker gene signatures are enriched in the down- and up-regulated gene signatures in the neuron and Gja1−/− astrocyte co-cultures versus the neuron and wildtype astrocyte co-cultures, respectively (Additional file 1: Table S9A). The co-culture systems with and without Gja1−/− upregulated a significant portion of the neuron-specific marker genes when compared with the respective astrocyte alone models (with or without Gja1−/−) while down-regulating many astrocyte marker genes (Additional file 1: Table S9B). The gene lists from the abovementioned intersection analyses can be found in (Additional file 1: Tables S9C and S9D).
Gja1 deficiency induced transcriptomic changes highly overlap the GJA1 centric gene networks in AD
We further examined the molecular mechanisms of GJA1 in AD pathogenesis and cognitive function. We first examined if these Gja1−/− DEG signatures were enriched in the Khaki module where GJA1 resided based on our previous study [98]. As shown in Additional file 1: Table S7, AST + NEU(dn), AST(dn) and AST(up) were all significantly enriched in the khaki module with FET p = 3.74E-54 (9.0 FE), 3.64E-28 (4.4 FE), 1.28E-5 (2.1 FE), respectively. On the other hand, the overall signatures AST(all) and AST + NEU(all) also significantly overlapped the module with FET p = 3.03E-32 (3.2FE) and 1.9E-47 (6.7 FE), respectively .
To gain more insights into the signaling circuit of the GJA1 regulation in AD, we constructed the Bayesian causal network for the khaki module and projected the Gja1−/− DEG signatures onto this network in order to delineate the underlying causal relationship among the molecular constituents of this module. As shown in Fig. 4a and Additional file 1: Table S7, 13 of the 19 predicted key drivers except for GJA1 in the khaki module were regulated by GJA1 (4.94 FE, FET p = 4.70E-05). Strikingly, 11 out of the 13 GJA1 regulated key driver genes were within the GJA1’s downstream network neighborhood (Additional file 2: Figure S3A). To better gain insights into the functions of the Bayesian causal network in Fig. 4a, we built up a GIA1 signaling pathway map. As shown in Additional file 2: Figure S3B, GIA1 regulates a diversity of pathways such as regulation of gap junction activity, innate immune system, TGFβ signaling, DNA repair and lipid metabolism. These results suggested that Gja1 deficiency significantly impacted the khaki module and thus strongly validated our previous prediction of GJA1 as a driver of this important AD related subnetwork.
Network analysis on GJA1 genetic networks. a. BN (Bayesian Network) based on astrocyte (khiki) module. Gja1−/− DEG signatures were projected onto BN to detect overlapped gene signatures, which were classified into eight categories: up-regulated only in AST (in red upright triangle), down-regulated only in AST (in blue downside triangle), only up-regulated in AST + NEU (red square), only down-regulated in AST + NEU (blue square), up-regulated in both AST and AST + NEU (in pink diamond), up-regulated in AST but down-regulated in AST + NEU (in pink hexagon), down-regulated in both AST and AST + NEU (in gray diamond), and down-regulated in AST but up-regulated in AST + NEU (in gray hexagon)). In enlarged label were genes that were validated experimentally. Gja1 was in black. In turquoise circle were genes non-overlapped. AST, Gja1−/− astrocyte culture while AST + NEU, Gja1−/− astrocyte culture in the presence of co-cultured neurons. b. GJA1 centered genetic networks inferred by projecting Gja1−/− DEG signatures onto Gja1 correlation concensus network (CGCCS(6)). Overlapped gene nodes were denoted as in A
To more comprehensively determine GJA1’s target genes in AD, we further systematically identified the genes correlated with GJA1 in a number of human AD cohorts. Genes significantly correlated with GJA1 (Benjamini-Hochberg (BH)-corrected p-value < 0.05) were first identified in each of eight datasets including four from the MSBB RNA-seq cohort, one from the ROSMAP RNA-seq data and three from the HBTRC cohort. Note that only the data from the AD subjects were used for the correlation analysis. A majority voting method was utilized to calculate the frequency of each correlation across the eight datasets (Additional file 1: Table S8). We defined consensus GJA1-centered correlation signature (CGCCS) as a function of frequency threshold n, i.e., CGCCS(n) = {g | frequency(r(g, GJA1)) ≥ n}, where r(g, GJA1) represents a significant correlation between a gene g and GJA1 at FDR < 0.05, and n = 1, 2, …, 8. This process led to GJA1 centered correlation networks, also denoted as CGCCS(n).
Additional file 2: Figure S4 shows the enrichment of the GJA1 centered correlation networks for the previously identified DEGs in AST and AST + NEU. CGCCS (4) was most significantly enriched for the AST(all) and AST + NEU(all) DEG signatures with FET p = 1.0E-330 (3.2 FE) and 7.5E-311 (3.9 FE), respectively (Black lines, Additional file 2: Figure S4). CGCCS (6) is enriched for the DEG signature in AST with FET p = 6.2E-309 (3.8 FE). Figure 4b shows the network CGCCS (6) which includes 201 up-regulated (red nodes, termed GJA1_centered_AST(up)) and 307 down-regulated (blue nodes, termed GJA1_centered_AST(dn)) genes upon the knockout of Gja1 in astrocytes. About 30% of the genes in each of the two GJA1_centered_signatures belonged to the khaki module (FET p = 1.52E-90, 32.37 FE). This GJA1-centered correlation network included eight driver genes (AGT, BMPR1B, CST3, FXYD1, NTSR2, SLC15A2, SPON1, and STON2) in the khaki module [98]. Similar results were derived for the AST + NEU(up) and AST + NEU(down) signatures. Therefore, GJA1 impacts the expression of many key network drivers in the astrocytic subnetwork, suggesting its central role in this AD-related gene network. Furthermore, the analysis of the genes specific to the A1/A2 astrocytic activation in our DEG from Gja1−/− astrocytes [50] revealed that most of pan-, A1-, and A2-specific genes were generally upregulated (Additional file 2: Figure S5).
Inflammatory cytokines downregulate expression of Gja1 and the astrocytic subnetwork module
Previously it has been shown that LOAD-relevant inflammatory cytokines such as TNFα and IL-1β downregulated expression of Gja1 in astrocytes [19, 74]. Wildtype astrocytes were treated with TNFα, or IL-1β, or both for 7 days, confirming that Cx43 (Gja1 protein) was profoundly and synergistically reduced by both cytokines (Fig. 5a-b). Paradoxically, IL-1β significantly increased, but TNFα significantly decreased, Apoe protein levels (Fig. 5b). Following cytokine treatment, we tested Gja1, Apoe and eight key network driver genes found to be differentially expressed in Gja1−/− astrocytes by RNA-seq analysis, as a proxy to capture the network changes. qPCR analysis revealed that Gja1, Apoe and the other astrocytic subnetwork drivers were similarly downregulated by these cytokines (Fig. 5c).
Inflammatory cytokines downregulates Gja1, Apoe, and other network genes. a. Wildtype primary mouse astrocytes were treated with either IL-1β (10 ng/ml) or TNFα (10 ng/ml) or in combination for 7 days and levels of Cx43, Apoe expression were analyzed by immunoblot. Representative results from 4 (Gja1) and 3 (Apoe) independent experiments are shown. b. Protein levels of Cx43 (left) and Apoe (right) were quantitatively analyzed for these technical replicates after normalization to Actin. ANOVA followed by Bonferroni post-hoc tests are indicated by asterisks. * p < 0.05, ** p < 0.01, *** p < 0.001. c. Quantitative gene expression analysis in wildtype astrocytes with treatments similar to a) was performed to analyze the other key drivers of the GJA1-centered network. Results representative of two independent experiments are shown. Statistical analysis was performed as in B, and only the comparison between control and IL-1β/TNFα is shown
Gja1 channel activity increases the expression of the astrocytic subnetwork module
Since these cytokines inhibit GJC and potentiate hemichannel activities [74], we asked whether inhibition of GJC and hemichannel activities regulates Gja1 and other astrocytic subnetwork drivers. Treatment of wildtype astrocytes with carbenoxolone (CBX, inhibitor of GJC and hemichannel [2, 95]) or lanthanum (La3+, inhibitor of hemichannel [2]) led to significant reduction of Gja1 and Apoe protein levels (Fig. 6a-b). Interestingly, the CBX treatment had broader effects on the reduction of the driver genes (Fig. 6c), while La3+ had generally milder and more selective effects, suggesting that GJC and hemichannel activities contribute to the regulation of distinct sets of the genes.
GJA1 inhibitors downregulate Gja1 and Apoe, and other network genes. a. Wildtype primary mouse astrocytes were treated with 200 μM carbenoxolone (CBX, gap junction inhibitor) or Lanthanum (La3+, hemichannel inhibitor) for 3 days and levels of Cx43, Apoe were analyzed by immunoblot. b. Quantitative analysis of Cx43 and Apoe protein levels were normalized to Actin. ANOVA followed by Bonferroni post-hoc tests are indicated by asterisks. ** p < 0.01, *** p < 0.001. c. Quantitative gene expression analysis in wildtype astrocytes treated similar to A was performed to analyze GJA1 network drivers. Statistical analysis was performed as in b, and only the comparisons to control are shown. Results representative of two independent experiments are shown for all experiments
We next tested whether Gja1 channel activation can alter the astrocytic subnetwork. Gja1 hemichannel activity can be increased by quinine [81]. Treatment of wildtype astrocytes with quinine significantly upregulated Gja1 and Apoe protein and mRNA levels, along with transcriptional upregulation of a subset of the driver genes (Additional file 2: Figure S6). These data collectively supports Gja1, and specifically its channel activity, as an important regulator of astrocytic gene coexpression network including Apoe.
Increased neuronal survival following Aβ treatment when co-cultured with Gja1−/− astrocytes
We assessed the role for Gja1 in neuronal death and viability in a transwell co-culture system, in which cortical neurons were grown on the bottom of the plate, while astrocytes were cultured on the transwell insert. This allowed physical separation of the two cell types, while maintaining chemical continuity. Incubation of wildtype neurons and wildtype astrocytes with the indicated concentration of Aβ1–42 oligomers resulted in increased neuronal death [t (4) = − 2.941, p = 0.030 at 2 μM; t (4) = − 5.857, p = 0.004 at 20 μM] and decreased viability [t (6) = − 5.395, p = 0.002 at 2 μM; t (6) = 4.671, p = 0.003 at 20 μM] at day 7 (but not at day 4 (Fig. 7a)), in a dose dependent manner (Fig. 7b). Wildtype neurons cultured with Gja1−/− astrocytes were significantly resistant to death and loss of viability (Fig. 7b), consistent with a previous study showing that Gja1−/− deficient astrocytes conferred neuroprotection [69].
Gja1 contributes to neuronal death and reduces viability by Aβ1–42 treatment. Cortical neurons were plated onto 12 well plate and wildtype or Gja1−/− astrocytes grown on transwells were inserted to the neuron cultures. Indicated concentration of Aβ1–42 oligomers are applied for 4 days (a) or 7 days (b) and neuronal death and viability was determined by LDH and MTT assays, respectively. Student’s t-tests were used for all the statistical analysis. * p < 0.05, ** p < 0.01, *** p < 0.001. For all experiments, representative of at least two independent experiments is shown
Reduced Apoe secretion and synthesis in Gja1−/− astrocytes
APOE is a well-known risk gene for LOAD and primarily produced by human and mouse astrocytes as well as microglia and neurons [52]. In addition, APOE was in the coexpression network module governed by GJA1. Therefore, we tested how Apoe levels were affected in the absence of Gja1 in mouse primary astrocytes. Quantitative PCR revealed significantly reduced Apoe expression in Gja1−/− astrocytes (t (6) = − 5.330, p = 0.002) (Fig. 8a). Consistently, we found lower Apoe protein levels in Gja1−/− astrocytes by immunoblot analysis of the lysates, with or without Aβ1–42 oligomer treatment (Fig. 8b). We analyzed Apoe in the conditioned medium from these astrocytes and found that secreted Apoe in the conditioned medium was consistently reduced in Gja1−/− astrocytes compared to wildtype, with or without Aβ1–42 oligomer treatment (Fig. 8c). Two way ANOVA revealed that there were significant main effects by genotype and treatment (F(1, 18)=121.618, p < 0.0005, F(2, 18)=9.364, p = 0.0016, respectively) but there was no interaction of genotype and treatment (F(2, 18) = 0.305, p = 0.741].
Gja1 deficiency causes reduction of Apoe production and promotes Aβ production in neuron/astrocyte cocultures. a. Quantitative PCR analysis of Apoe in wildtype (WT) and Gja1−/− astrocytes. b. Apoe, Cx43, and actin levels in wildtype and Gja1−/− astrocytes treated with the indicated concentration of Aβ1–42 oligomers for 48 h were determined by immunoblot analysis of cell lysates. c. Apoe secreted into the conditioned medium of astrocytes in B was quantitatively determined by specific ELISA in quadruplicate wells. d and e. Amyloidβ secreted to the conditioned medium of wildtype cortical neurons cocultured with either wildtype or Gja1−/− astrocytes was quantitatively determined by Aβ40 and Aβ42 specific ELISA
Neurons cocultured with Gja1−/− astrocytes produce higher levels of Aβ species
Since our RNA-seq analysis revealed that Gja1 regulated genes were highly enriched in Aβ network, we therefore analyzed Aβ production in neuron/astrocyte cocultures. Indeed, we observed higher levels of both Aβ40 and Aβ42 species in the conditioned medium from neurons co-cultured with Gja1−/− astrocytes at all time points examined [Aβ40; t(4) = 17.684, p < 0.0005 (div6), t(4) = 3.447, p = 0.026 (div13), t(4) = 11.112, p < 0.0005 (div16), t(4) = 11.112, p < 0.0005 (div20): Aβ42; t(4) = 8.422, p < 0.0005 (div6), t(4) = 2.047, p = 0.110 (div13), t(4) = 11.245, p < 0.0005 (div16), t(4) = 8.016, p < 0.001 (div20)] (Fig. 8d-e). Because astrocytes have been shown to take up Aβ, preferentially oligomers [67], we investigated the role for Gja1 in controlling Aβ clearance. When we tested whether wildtype and Gja1−/− astrocytes were proficient in taking up exogenously applied fluorescently labeled Aβ1–42 oligomers, we found that the number of Gja1−/− astrocytes with cell-associated Aβ1–42 oligomers was significantly reduced compared to wildtype astrocytes [t(20) = − 6.670, p < 0.0005] (Additional file 2: Figure S7).
Reduced neuronal activity in response to Aβ treatment when co-cultured with Gja1−/− astrocytes
In order to understand the impact of the network perturbation by loss of astrocytic Gja1−/− on neuronal functions, we co-cultured wildtype cortical neurons with either wildtype or Gja1−/− astrocytes on multielectrode arrays and compared spontaneous and stimulated neuronal activities before and after Aβ1–42 oligomer treatment (Fig. 9a). Although few significant differences were detected prior to Aβ treatment at 16 days in vitro (div), at 18 and/or 20 div (2 and 4 days after 10 μM Aβ1–42 treatment) most metrics of spontaneous activity (i.e. number of spikes, bursts, network bursts, and spikes per network burst) were significantly reduced in Gja1−/− astrocyte-cocultured neurons (Fig. 9b). The reduced spontaneous neuronal activity at div 20 suggested that co-culture with Gja1−/− astrocytes made neurons less responsive to Aβ-induced changes. While stimulated neuronal activity at div 14 was not affected by co-culture with Gja1−/− astrocytes (Fig. 10a), it was reduced at 20 div (number of spikes, spikes per burst, mean firing rate, and burst duration) (Fig. 10b-c).
Gja1 deficiency reduces spontaneous neuronal activity in cortical neuron/astrocyte cocultures as determined by multielectrode array. a. Experimental scheme is indicated. One day prior to cortical neuron preparation and plating, wildtype or Gja1−/− astrocytes were plated onto 48-well MEA array plate in quadruplicate wells. Spontaneous activities of the cocultures were recorded at day 10, 14, 18 and 20 on Axion Maestro. At day 16, cocultures were treated with 10 μM Aβ1–42 oligomers. b. Boxplots of the indicated metrics from the MEA recordings are shown. Each circles represent mean values of individual well, and the horizontal bars within the box indicates means of quadruplicates. Mann-Whitney U tests were used for statistical analysis. * p < 0.05
Gja1 deficiency reduces stimulated neuronal activity in cortical neuron/astrocyte cocultures as determined by multielectrode array. a and b. Cocultures were stimulated by 3 cycles of voltage-controlled biphasic stimulus from all electrodes within the wells at 14 days (a) and 20 days (b) in vitro. At least 5 times of repeated stimulations were recorded and analyzed. c. Two representative raster plots as recorded on day 20 are shown for wildtype neurons cocultured with wildtype astrocytes (upper panel) or with Gja1−/− astrocytes (lower panel). Y-axis represents 16 electrodes and X-axis represents time after stimulation indicated by red arrowhead. Black horizontal scale bar indicates 100 milliseconds (ms). Each vertical bar represents a detected spike
Discussion
Functional interaction between neurons and astrocytes is an appealing research area that has expanded vigorously in recent years. The involvement of astrocytes in the pathogenesis of neurodegenerative diseases such as AD is just beginning to be appreciated. Astrocyte-specific GJA1 expression has been shown to be upregulated in the postmortem AD brains [6, 34]. Consistent with but beyond these studies, we predicted and validated not only upregualtion of GJA1 expression in AD but also its regulatory role in driving a large astrocyte-specific molecular network underlying AD. Indeed, our functional analysis using Gja1−/− astrocytes revealed that the Gja1 network contributed to neuronal death and decreased their viability under Aβ1–42 treatment and, which may explain synapse and neuron loss in the Alzheimer brains [20]. This is consistent with recent studies showing that GJA1 play critical roles in AD-relevant phenotypes in AD mouse models [40, 73, 96, 97]. On the other hand, a number of other studies indicate that Gja1 isneuroprotective [41, 44, 51, 64, 65, 85]. Indeed, our data showed that the Gja1 network appeared to be involved in phagocytosis of Aβ, and maintenance of neuronal activities under Aβ stress. Taken together with the previous findings of the increase of Cx43 protein and its channel activity specifically in astrocytes surrounding plaques [59, 63, 96], we suggest that the upregulation of GJA1 in AD brains may start as a neuroprotective response to amyloid plaques. It is interesting to note that previous clinical studies have consistently demonstrated an increased neuronal activity in the hippocampus and cortex in non-demented individuals at high risk for AD including APOE4 carriers, minor cognitive impairment (MCI) or at presymptomatic stage of familial Alzheimer’s disease [10, 72], and individuals with early Alzheimer’s disease are prone to developing seizures [87]. Furthermore, the hippocampal hyperactivation is a viable therapeutic target for AD [4]. We speculate that the role of GJA1 in supporting neuronal activity in the AD brain might explain partly the observation of elevated brain activity in the prodromal stage of AD.
In the present study, we demonstrated GJA1 as a master regulator, which regulates not an AD-related astrocyte-specific gene subnetwork [98] but also an array of AD risk genes including the most important AD gene APOE (Fig. 4a-b). More broadly speaking, in in vitro cultured astrocytes, Gja1 deficiency caused significant change in expression of 5496 genes involved a diverse of biological pathways and processes (Fig. 3b). Similarly, in in vitro astrocyte and neuron cocultures, thousands of genes were regulated by Gja1 (−/−). Of note, more than half of the known AD risk factor genes (Fig. 2g) and hundreds of inflammatome genes in chronic inflammation diseases [23] were the targets of GJA1. Upregulation of GJA1 expression in AD brains of human subjects and in vitro or in vivo perturbation of Gja1 (e.g., Gja1 deficiency in primary cultured astrocytes in the present study) lead to a cascade of biological and pathological reactions and processes including subsequent alteration in cells’ biological functions and responses to their environments, which are manifested as a complex and intertwined GJA1 centered molecular network. Interestingly, our data pointed to the key role of Cx43 gap junction channel and hemichannel activities in regulating gene networks, and indicated that the network can be fine-tuned by pharmacologically modulating Cx43 channel activity (Fig. 6 and Additional file 2: Figures S5 and S8). Cx43 could be a useful therapeutic target in diseases such as LOAD, ODDD, and other neuropsychiatric disorders [71]. In support for such a hypothesis, recent studies showed that Gja1-deficiency or a connexin inhibitor (Boldine) treatment in APP/PS1 mice had beneficial effects [73, 96, 97], underscoring that Cx43 is a critical mediator of AD pathophysiology and an important therapeutic target for LOAD.
One of the important novel discoveries in the present study was the Gja1 mediated production of ApoE in astrocytes. APOE is mainly produced in astrocytes, but also in neurons and microglia when under stress [35]. APOE is the major LOAD risk factor, critical to AD neuropathology and neurocognitions [15, 39]. We therefore focused on characterizing Apoe in Gja1−/− astrocytes. Manipulation of Gja1 by genetic disruption, cytokines, CBX, and quinine in astrocytes as well as neuron/astrocyte/microglia culture system consistently resulted in the regulation of Apoe in the same direction as Gja1. Abnormal production of APOE, in particular, the risk-associated isoform APOE4, caused either gain of toxic functions, when over-expressed, such as neuronal toxicity, Aβ aggregation and tangle formation or loss of physiological functions, when under-expressed, such as Aβ clearance, synaptic function and neurogenesis. Consistent with this notion, we observed drastic reduction in uptake and retention of Aβ oligomers by Gja1(−/−) astrocytes (Additional file 2: Figure S7 A-B), with concomitant increase in concentration of both Aβ1–40 and Aβ1–42 species in in vitro culture medium (Fig. 8d-e). Since APOE is known to be regulated by transcription factors such as LXR and RXR [48], and taken together with downregulation of NR1H3 (LXRA) expression in our RNA-seq data, we speculate that NR1H3 regulation by GJA1 might have mediated the APOE expression. Further investigation is required to dissect the mechanism underlying GJA1-mediated expression of APOE and its biological significance, which would lead to the development of novel therapeutic strategies.
A recent study identified the transcriptomic basis of the two distinct polarization states of A1 neurotoxic and A2 neuroprotective astrocytes [50]. Interestingly, our RNA-seq analysis revealed that most of pan-reactive, A1 and A2 specific transcripts were significantly upregulated in Gja1−/− astrocytes (Additional file 2: Figure S5). This pattern of expression is most similar to those induced by combination of IL-1β and TNFα or C1q (Fig. 1a in [50]), and consistent with this, our data showed that IL-1β and TNFα caused downregulation of Cx43 and other key drivers, mimicking the loss of Gja1. This pattern of expression may indicate that Gja1 deficiency induces non-specific activation of either A1 or A2 polarized-astrocytes, or A1/A2 overlapping polarization. Although these two possibilities need to be clarified by transcriptomic analysis at a single cell level, these mixed transcriptomic signatures suggest how Gja1 may contribute to opposing neuroprotective (Figs. 8, 9 and 10) and neurotoxic (Fig. 7) phenotypes, as demonstrated here as well as in previous studies [41, 44, 51, 64, 65, 69, 85, 96, 97].
Based upon the results from this study and the discussion above, we developed a working hypothesis of GJA1 dysregulation in AD (Fig. 11), in which upregulation of GJA1, a master regulator of astrocytic gene expression, concomitant with amyloid accumulation during Alzheimer pathogenesis ultimately drives AD clinical and pathological traits as evidenced by highly significant correlation with AD progression and AD GWAS genes (Fig. 2a-g). According to our model, GJA1 induction initially serves to support neuronal functions by elevating gene expression signatures for the glial neurosupportive functions such as Abeta network (Fig. 3c), Apoe (Fig. 8a-c), vascular and neurodevelopment, extracellular matrix, and cytoskeletal and cell adhesion (Fig. 3a-b) as well as suppressing inflammatory response (Fig. 3b). These findings were supported by amyloid phagocytosis (Fig. 8d-e, Additional file 2: Figure S5) and multielectrode array (Figs. 9 and 10) assays showing that the amyloid uptake, Apoe expression, and neuronal activity are attenuated with Gja1−/− astrocytes. This is consistent with several key clinical and pathological observations in LOAD. For example, neuronal activity is found to be enhanced at the early stage of Alzheimer’s disease subjects [4, 10, 72, 87] and evidence of cell cycle activation in AD neurons [3, 53, 58, 79, 86, 94], changes in extracellular matrix [12, 46], and neuronal cytoskeletal components [5, 13, 45, 91] have been reported. However, we speculate that chronic GJA1 gain of function could lead to detrimental effects on neurons in a long run, as evidenced by our observation, in which prolonged neuron/astrocyte cocultures were better protected in the absence of Gja1 in astrocytes (Fig. 7a-b) possibly due to excessive neuronal activity and glutamate-induced excitotoxicity [24, 37, 47, 68].
A working hypothesis of GJA1 dysregulation in AD. Progressive upregulation of GJA1 in the brains during LOAD pathogenesis (in response to amyloid accumulation) serves to enhance the coordinated gene network function to orchestrate neuroprotective response by astrocytes including Aβ production and clearance (CST3, CLU, CTSB, ECE2, MEGF10, PSENEN and GSAP), APOE production (APOE, ABCA1, CAV1 and SPON1), and supporting neuronal activity (DLG4, NLGN3, GRINA and GRIN2D). However, the enhanced neuronal activity due to chronic and prolonged GJA1 upregulation triggers neuronal wear and eventual death
In summary, this study elucidated the previously unrecognized role for GJA1 and its channel activity in regulating a key AD-related gene regulatory network comprised of APOE and other astrocyte-specific genes. We demonstrated that the disruption of this astrocytic gene coexpression network by knocking out Gja1 can contribute to LOAD-related pathology. This study represents the first step towards a comprehensive functional characterization of one of the most significantly dysregulated modules in our previous integrative network analysis of LOAD brains. The strong validation of the predictive power of our multiscale gene network approach paves a way to dissect the complex pathogenic mechanisms of LOAD.
References
(2015) Convergent genetic and expression data implicate immunity in Alzheimer's disease. Alzheimers Dement 11:658–671. https://doi.org/10.1016/j.jalz.2014.05.1757
Anselmi F et al (2008) ATP release through connexin hemichannels and gap junction transfer of second messengers propagate Ca2+ signals across the inner ear. Proc Natl Acad Sci U S A 105:18770–18775. https://doi.org/10.1073/pnas.0800793105
Arendt T, Rodel L, Gartner U, Holzer M (1996) Expression of the cyclin-dependent kinase inhibitor p16 in Alzheimer's disease. Neuroreport 7:3047–3049
Bakker A et al (2012) Reduction of hippocampal hyperactivity improves cognition in amnestic mild cognitive impairment. Neuron 74:467–474. https://doi.org/10.1016/j.neuron.2012.03.023
Bamburg JR, Bloom GS (2009) Cytoskeletal pathologies of Alzheimer disease. Cell Motil Cytoskeleton 66:635–649. https://doi.org/10.1002/cm.20388
Begcevic I et al (2013) Semiquantitative proteomic analysis of human hippocampal tissues from Alzheimer's disease and age-matched control brains. Clin Proteomics 10:5. https://doi.org/10.1186/1559-0275-10-5
Bennett DA (2006) Postmortem indices linking risk factors to cognition: results from the Religious Order Study and the Memory and Aging Project. Alzheimer Dis Assoc Disord 20:S63–S68
Bennett DA, Schneider JA, Arvanitakis Z, Wilson RS (2012) Overview and findings from the religious orders study. Current Alzheimer Research 9:628–645
Bennett DA et al (2005) The rush memory and aging project: study design and baseline characteristics of the study cohort. Neuroepidemiology 25:163–175
Bookheimer SY et al (2000) Patterns of brain activation in people at risk for Alzheimer's disease. N Engl J Med 343:450–456. https://doi.org/10.1056/NEJM200008173430701
Braak H, Braak E (1991) Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol 82:239–259
Brandan E, Inestrosa NC (1993) Extracellular matrix components and amyloid in neuritic plaques of Alzheimer's disease. Gen Pharmacol 24:1063–1068
Brion JP (1992) The pathology of the neuronal cytoskeleton in Alzheimer's disease. Biochim Biophys Acta 1160:134–142
Bruban J et al (2015) Presenilin 1 is necessary for neuronal, but not glial, EGFR expression and neuroprotection via gamma-secretase-independent transcriptional mechanisms. FASEB J 29:3702–3712. https://doi.org/10.1096/fj.15-270645
Bu G (2009) Apolipoprotein E and its receptors in Alzheimer's disease: pathways, pathogenesis and therapy. Nat Rev Neurosci 10:333–344. https://doi.org/10.1038/nrn2620
Campion D, Pottier C, Nicolas G, Le Guennec K, Rovelet-Lecrux A (2016) Alzheimer disease: modeling an Abeta-centered biological network. Mol Psychiatry 21:861–871. https://doi.org/10.1038/mp.2016.38
Chever O, Lee CY, Rouach N (2014) Astroglial connexin43 hemichannels tune basal excitatory synaptic transmission. J Neurosci 34:11228–11232. https://doi.org/10.1523/JNEUROSCI.0015-14.2014
Corder EH et al (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 261:921–923
De Vuyst E et al (2007) Connexin hemichannels and gap junction channels are differentially influenced by lipopolysaccharide and basic fibroblast growth factor. Mol Biol Cell 18:34–46. https://doi.org/10.1091/mbc.E06-03-0182
DeKosky ST, Scheff SW (1990) Synapse loss in frontal cortex biopsies in Alzheimer's disease: correlation with cognitive severity. Ann Neurol 27:457–464. https://doi.org/10.1002/ana.410270502
D'Hondt C, Iyyathurai J, Himpens B, Leybaert L, Bultynck G (2014) Cx43-hemichannel function and regulation in physiology and pathophysiology: insights from the bovine corneal endothelial cell system and beyond. Front Physiol 5:348. https://doi.org/10.3389/fphys.2014.00348
Dobin A et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. https://doi.org/10.1093/bioinformatics/bts635
Emilsson V et al (2008) Genetics of gene expression and its effect on disease. Nature 452:423–428
Esposito Z et al (2013) Amyloid beta, glutamate, excitotoxicity in Alzheimer's disease: are we on the right track? CNS Neurosci Ther 19:549–555. https://doi.org/10.1111/cns.12095
Folstein MF, Folstein SE, McHugh PR (1975) "Mini-mental state". A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 12:189–198
Frisch C et al (2003) Mice with astrocyte-directed inactivation of connexin43 exhibit increased exploratory behaviour, impaired motor capacities, and changes in brain acetylcholine levels. Eur J Neurosci 18:2313–2318
Giaume C et al (1991) Gap junctions in cultured astrocytes: single-channel currents and characterization of channel-forming protein. Neuron 6:133–143
Goodenough DA, Paul DL (2003) Beyond the gap: functions of unpaired connexon channels. Nat Rev Mol Cell Biol 4:285–294. https://doi.org/10.1038/nrm1072
Gutmann DH, Zackai EH, McDonald-McGinn DM, Fischbeck KH, Kamholz J (1991) Oculodentodigital dysplasia syndrome associated with abnormal cerebral white matter. Am J Med Genet 41:18–20. https://doi.org/10.1002/ajmg.1320410106
Han Y et al (2014) Astrocyte-restricted disruption of connexin-43 impairs neuronal plasticity in mouse barrel cortex. Eur J Neurosci 39:35–45. https://doi.org/10.1111/ejn.12394
Haroutunian V, Katsel P, Schmeidler J (2009) Transcriptional vulnerability of brain regions in Alzheimer's disease and dementia. Neurobiol Aging 30:561–573. https://doi.org/10.1016/j.neurobiolaging.2007.07.021
Harrell LE, Marson D, Chatterjee A, Parrish JA (2000) The severe mini-mental state examination: a new neuropsychologic instrument for the bedside assessment of severely impaired patients with Alzheimer disease. Alzheimer Dis Assoc Disord 14:168–175
Hoffman GE, Schadt EE (2016) variancePartition: interpreting drivers of variation in complex gene expression studies. BMC Bioinformatics 17:483. https://doi.org/10.1186/s12859-016-1323-z
Hokama M et al (2014) Altered expression of diabetes-related genes in Alzheimer's disease brains: the Hisayama study. Cereb Cortex 24:2476–2488. https://doi.org/10.1093/cercor/bht101
Huang Y, Mucke L (2012) Alzheimer mechanisms and therapeutic strategies. Cell 148:1204–1222. https://doi.org/10.1016/j.cell.2012.02.040
Humphries CE et al (2015) Integrated whole transcriptome and DNA methylation analysis identifies gene networks specific to late-onset Alzheimer's disease. J Alzheimers Dis 44:977–987. https://doi.org/10.3233/JAD-141989
Hynd MR, Scott HL, Dodd PR (2004) Glutamate-mediated excitotoxicity and neurodegeneration in Alzheimer's disease. Neurochem Int 45:583–595. https://doi.org/10.1016/j.neuint.2004.03.007
Kajiwara Y et al (2016) The human-specific CASP4 gene contributes to Alzheimer-related synaptic and behavioral deficits. Hum Mol Genet. https://doi.org/10.1093/hmg/ddw265
Kim J, Basak JM, Holtzman DM (2009) The role of apolipoprotein E in Alzheimer's disease. Neuron 63:287–303. https://doi.org/10.1016/j.neuron.2009.06.026
Koulakoff A, Mei X, Orellana JA, Saez JC, Giaume C (2012) Glial connexin expression and function in the context of Alzheimer's disease. Biochim Biophys Acta 1818:2048–2057. https://doi.org/10.1016/j.bbamem.2011.10.001
Kozoriz MG et al (2010) The connexin43 C-terminal region mediates neuroprotection during stroke. J Neuropathol Exp Neurol 69:196–206. https://doi.org/10.1097/NEN.0b013e3181cd44df
Laird DW (2014) Syndromic and non-syndromic disease-linked Cx43 mutations. FEBS Lett 588:1339–1348. https://doi.org/10.1016/j.febslet.2013.12.022
Lambert JC et al (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 45:1452–1458. https://doi.org/10.1038/ng.2802
Le HT et al (2014) Gap junction intercellular communication mediated by connexin43 in astrocytes is essential for their resistance to oxidative stress. J Biol Chem 289:1345–1354. https://doi.org/10.1074/jbc.M113.508390
Lee VM (1995) Disruption of the cytoskeleton in Alzheimer's disease. Curr Opin Neurobiol 5:663
Lepelletier FX, Mann DM, Robinson AC, Pinteaux E, Boutin H (2017) Early changes in extracellular matrix in Alzheimer's disease. Neuropathol Appl Neurobiol 43:167–182. https://doi.org/10.1111/nan.12295
Lewerenz J, Maher P (2015) Chronic glutamate toxicity in neurodegenerative diseases-what is the evidence? Front Neurosci 9:469. https://doi.org/10.3389/fnins.2015.00469
Liang Y et al (2004) A liver X receptor and retinoid X receptor heterodimer mediates apolipoprotein E expression, secretion and cholesterol homeostasis in astrocytes. J Neurochem 88:623–634
Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. https://doi.org/10.1093/bioinformatics/btt656
Liddelow SA et al (2017) Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541:481–487. https://doi.org/10.1038/nature21029
Lin JH et al (2008) A central role of connexin 43 in hypoxic preconditioning. J Neurosci 28:681–695. https://doi.org/10.1523/JNEUROSCI.3827-07.2008
Liu CC, Liu CC, Kanekiyo T, Xu H, Bu G (2013) Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol 9:106–118. https://doi.org/10.1038/nrneurol.2012.263
Liu WK, Williams RT, Hall FL, Dickson DW, Yen SH (1995) Detection of a Cdc2-related kinase associated with Alzheimer paired helical filaments. Am J Pathol 146:228–238
Loddenkemper T, Grote K, Evers S, Oelerich M, Stogbauer F (2002) Neurological manifestations of the oculodentodigital dysplasia syndrome. J Neurol 249:584–595. https://doi.org/10.1007/s004150200068
Lohman AW, Isakson BE (2014) Differentiating connexin hemichannels and pannexin channels in cellular ATP release. FEBS Lett 588:1379–1388. https://doi.org/10.1016/j.febslet.2014.02.004
Lutz SE et al (2009) Deletion of astrocyte connexins 43 and 30 leads to a dysmyelinating phenotype and hippocampal CA1 vacuolation. J Neurosci 29:7743–7752. https://doi.org/10.1523/JNEUROSCI.0341-09.2009
McKenzie AT et al (2018) Brain cell type specific gene expression and co-expression network architectures. Sci Rep 8:8868. https://doi.org/10.1038/s41598-018-27293-5
McShea A, Harris PL, Webster KR, Wahl AF, Smith MA (1997) Abnormal expression of the cell cycle regulators P16 and CDK4 in Alzheimer's disease. Am J Pathol 150:1933–1939
Mei X, Ezan P, Giaume C, Koulakoff A (2010) Astroglial connexin immunoreactivity is specifically altered at beta-amyloid plaques in beta-amyloid precursor protein/presenilin1 mice. Neuroscience 171:92–105. https://doi.org/10.1016/j.neuroscience.2010.08.001
Miller JA, Oldham MC, Geschwind DH (2008) A systems level analysis of transcriptional changes in Alzheimer's disease and Normal aging. J Neurosci 28:1410–1420
Miller JA, Woltjer RL, Goodenbour JM, Horvath S, Geschwind DH (2013) Genes and pathways underlying regional and cell type changes in Alzheimer's disease. Genome Medicine 5:48
Millstein J, Zhang B, Zhu J, Schadt EE (2009) Disentangling molecular relationships with a causal inference test. BMC Genet 10:23. https://doi.org/10.1186/1471-2156-10-23
Nagy JI, Li W, Hertzberg EL, Marotta CA (1996) Elevated connexin43 immunoreactivity at sites of amyloid plaques in Alzheimer's disease. Brain Res 717:173–178
Nakase T, Fushiki S, Naus CC (2003) Astrocytic gap junctions composed of connexin 43 reduce apoptotic neuronal damage in cerebral ischemia. Stroke 34:1987–1993. https://doi.org/10.1161/01.STR.0000079814.72027.34
Nakase T, Sohl G, Theis M, Willecke K, Naus CC (2004) Increased apoptosis and inflammation after focal brain ischemia in mice lacking connexin43 in astrocytes. Am J Pathol 164:2067–2075. https://doi.org/10.1016/S0002-9440(10)63765-0
Naus CC et al (1997) Altered gap junctional communication, intercellular signaling, and growth in cultured astrocytes deficient in connexin43. J Neurosci Res 49:528–540
Nielsen HM et al (2010) Astrocytic a beta 1-42 uptake is determined by a beta-aggregation state and the presence of amyloid-associated proteins. Glia 58:1235–1246. https://doi.org/10.1002/glia.21004
Ong WY, Tanaka K, Dawe GS, Ittner LM, Farooqui AA (2013) Slow excitotoxicity in Alzheimer's disease. J Alzheimers Dis 35:643–668. https://doi.org/10.3233/JAD-121990
Orellana JA et al (2011) Amyloid beta-induced death in neurons involves glial and neuronal hemichannels. J Neurosci 31:4962–4977. https://doi.org/10.1523/JNEUROSCI.6417-10.2011
Pannasch U et al (2011) Astroglial networks scale synaptic activity and plasticity. Proc Natl Acad Sci U S A 108:8467–8472. https://doi.org/10.1073/pnas.1016650108
Parikshak NN, Gandal MJ, Geschwind DH (2015) Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat Rev Genet 16:441–458. https://doi.org/10.1038/nrg3934
Quiroz YT et al (2010) Hippocampal hyperactivation in presymptomatic familial Alzheimer's disease. Ann Neurol 68:865–875. https://doi.org/10.1002/ana.22105
Ren R, Zhang L, Wang M (2018) Specific deletion connexin43 in astrocyte ameliorates cognitive dysfunction in APP/PS1 mice. Life Sci 208:175–191. https://doi.org/10.1016/j.lfs.2018.07.033
Retamal MA et al (2007) Cx43 hemichannels and gap junction channels in astrocytes are regulated oppositely by proinflammatory cytokines released from activated microglia. J Neurosci 27. https://doi.org/10.1523/JNEUROSCI.2042-07.2007
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47, doi:https://doi.org/10.1093/nar/gkv007 (2015)
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. https://doi.org/10.1093/bioinformatics/btp616
Rouach N, Koulakoff A, Abudara V, Willecke K, Giaume C (2008) Astroglial metabolic networks sustain hippocampal synaptic transmission. Science 322:1551–1555. https://doi.org/10.1126/science.1164022
Scemes E, Giaume C (2006) Astrocyte calcium waves: what they are and what they do. Glia 54:716–725. https://doi.org/10.1002/glia.20374
Smith TW, Lippa CF (1995) Ki-67 immunoreactivity in Alzheimer's disease and other neurodegenerative disorders. J Neuropathol Exp Neurol 54:297–303
Sohl G, Maxeiner S, Willecke K (2005) Expression and functions of neuronal gap junctions. Nat Rev Neurosci 6:191–200. https://doi.org/10.1038/nrn1627
Stout CE, Costantin JL, Naus CC, Charles AC (2002) Intercellular calcium signaling in astrocytes via ATP release through connexin hemichannels. 277:10482–10488. https://doi.org/10.1074/jbc.M109902200
Strittmatter WJ et al (1993) Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A 90:1977–1981
Tabernero, A., Giaume, C. & Medina, J. M. Endothelin-1 regulates glucose utilization in cultured astrocytes by controlling intercellular communication through gap junctions. Glia 16, 187–195, doi:https://doi.org/10.1002/(SICI)1098-1136(199603)16:3<187::AID-GLIA1>3.0.CO;2-# [pii]. https://doi.org/10.1002/(SICI)1098-1136(199603)16:3<187::AID-GLIA1>3.0.CO;2-# (1996)
Theis M et al (2003) Accelerated hippocampal spreading depression and enhanced locomotory activity in mice with astrocyte-directed inactivation of connexin43. J Neurosci 23:766–776
Theodoric N, Bechberger JF, Naus CC, Sin WC (2012) Role of gap junction protein connexin43 in astrogliosis induced by brain injury. PLoS One 7:e47311. https://doi.org/10.1371/journal.pone.0047311
Vincent I, Rosado M, Davies P (1996) Mitotic mechanisms in Alzheimer's disease? J Cell Biol 132:413–425
Vossel KA et al (2013) Seizures and epileptiform activity in the early stages of Alzheimer disease. JAMA Neurol 70:1158–1166. https://doi.org/10.1001/jamaneurol.2013.136
Wagenaar DA, Pine J, Potter SM (2004) Effective parameters for stimulation of dissociated cultures using multi-electrode arrays. J Neurosci Methods 138:27–37. https://doi.org/10.1016/j.jneumeth.2004.03.005
Wallraff A et al (2006) The impact of astrocytic gap junctional coupling on potassium buffering in the hippocampus. J Neurosci 26:5438–5447. https://doi.org/10.1523/JNEUROSCI.0037-06.2006
Wang IM et al (2012) Systems analysis of eleven rodent disease models reveals an inflammatome signature and key drivers. Mol Syst Biol 8:594. https://doi.org/10.1038/msb.2012.24
Wang M et al (2016) Integrative network analysis of nineteen brain regions identifies molecular signatures and networks underlying selective regional vulnerability to Alzheimer's disease. Genome Med 8:104. https://doi.org/10.1186/s13073-016-0355-3
Wilson RS, Arnold SE, Schneider JA, Li Y, Bennett DA (2007) Chronic distress, age-related neuropathology, and late-life dementia. Psychosom Med 69:47–53. https://doi.org/10.1097/01.psy.0000250264.25017.21
Yamamoto T, Ochalski A, Hertzberg EL, Nagy JI (1990) On the organization of astrocytic gap junctions in rat brain as suggested by LM and EM immunohistochemistry of connexin43 expression. J Comp Neurol 302:853–883. https://doi.org/10.1002/cne.903020414
Yang Y, Mufson EJ, Herrup K (2003) Neuronal cell death is preceded by cell cycle events at all stages of Alzheimer's disease. J Neurosci 23:2557–2563
Ye ZC, Oberheim N, Kettenmann H, Ransom BR (2009) Pharmacological "cross-inhibition" of connexin hemichannels and swelling activated anion channels. Glia 57:258–269. https://doi.org/10.1002/glia.20754
Yi C et al (2016) Astroglial connexin43 contributes to neuronal suffering in a mouse model of Alzheimer's disease. Cell Death Differ 23:1691–1701. https://doi.org/10.1038/cdd.2016.63
Yi C et al (2017) Inhibition of glial hemichannels by boldine treatment reduces neuronal suffering in a murine model of Alzheimer's disease. Glia 65:1607–1625. https://doi.org/10.1002/glia.23182
Zhang B et al (2013) Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell 153:707–720. https://doi.org/10.1016/j.cell.2013.03.030
Zhang Y et al (2016) Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron 89:37–53. https://doi.org/10.1016/j.neuron.2015.11.013
Zhu J et al (2007) Increasing the power to detect causal associations by combining genotypic and expression data in segregating populations. PLoS Comput Biol 3:e69. https://doi.org/10.1371/journal.pcbi.0030069
Acknowledgments
This study was supported in parts by grants from the National Institutes of Health/National Institute on Aging (R01AG046170, RF1AG054014, RF1AG057440, R01AG057907). This study has been made possible through the support of Brain Canada and the financial support of Health Canada, Michael Smith Foundation for Health Research, Genome BC and the Pacific Alzheimer’s Research Foundation to CCN and WCS. CCN holds a Canada Research Chair.
Availability of data and materials
The RNA-seq and other Omics data from the MSBB cohort used in this study are available at https://doi.org/10.7303/syn3159438, https://doi.org/10.7303/syn7392158, https://doi.org/10.7303/syn3157743, https://doi.org/10.7303/syn4645334, https://doi.org/10.7303/syn10901600 and https://doi.org/10.7303/syn5759470. The RNA-seq data from the Gja1 experiments are available at https://doi.org/10.7303/syn11711769.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Tables S1. Clinical and pathological traits, brain regions analyzed by transcriptome profiling, and sample classification with respect to the severity stage of each trait. Table S2. Correlations between GJA1 expression and individual phenotypical traits. Table S3. Differential expression analysis. TableS4. Covariables' impacts on GJA1 association with AD traits. Table S5. Gene ontology analysis. Table S6. Intersection of GJA1 gene signatures and AD gene signatures. Table S7. Intersection of GJA1 gene signatures and the khiki module. Table S8. GJA1_centered genetic network analysis. Table S9A. GJA1 DEG signatures (AST+NEU) overlapped with brain cell type specific gene signatures. Table S9B. AST+NEU vs AST in Gja1 (-/-) or (+/+) DEG signatures overlapped with brain cell type specific gene signatures. Table S9C. differentially expressed genes in neurons upon GJA1 KO or not in astrocyte and neuron coculture. Table S9D. Difference of change degree in AST+NEU_vs_AST_Gja1(-/-) vs AST+NEU_vs_AST_Gja1(+/+). Table S10. List of primer sets and Universal Probe Library (UPL) probe for qRT-PCR. (XLSX 234 kb)
Additional file 2:
Figure S1. Correlations between GIA1 and individual clinic traits stratified by AOD (age of death), gender and APOE genotypes in BM36 region. Figure S2. Correlation between Gja1 and individual transcripts of ADGWAS genes. Figure S3. GJA1-centric Bayesian causal network and GJA1 signaling pathway map. Figure S4. Enrichment of Gia1-/- gene signatures in GJA1 centric correlation networks. Figure S5. Regulation of A1/A2 astrocyte marker genes in Gja1-/- astrocytes. Figure S6. Quinine (GJA1 channel agonist) upregulates Gja1 and Apoe, and other network genes. Figure S7. A. Wildtype and Gja1-/- astrocytes were treated with fluorescently labeled Aβ1-42 oligomers for 24 hours and the number of astrocytes in association with Aβ1-42 oligomers were quantitatively estimated by counting total cells (DAPI+) and Aβ positive cells. B. Representative images of phase contrast, Hilyte Fluor488-labeled Aβ, and DAPI staining from wildtype (upper panels) and Gja1-/- (lower panels) astrocytes were shown. These images were taken by bright field microscope and the representatives of 2 independent experiments with similar results are shown. Figure S8. Complex regulation of Gja1 key drivers in neuron/astrocyte/microglia cocultures by carbenoxolone and quinine. (DOCX 1.4 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kajiwara, Y., Wang, E., Wang, M. et al. GJA1 (connexin43) is a key regulator of Alzheimer’s disease pathogenesis. acta neuropathol commun 6, 144 (2018). https://doi.org/10.1186/s40478-018-0642-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40478-018-0642-x