Abstract
Background
The t(1;19)(q23;p13), which can result in the TCF3-PBX1 chimeric gene, is one of the most frequent translocations in B-acute lymphoblastic leukemia (B-ALL) and is observed in both adult and pediatric populations at an overall frequency of 6%. It can occur in a balanced or unbalanced form and as a sole abnormality is associated with an intermediate prognosis. Additionally, this translocation is observed in the context of hyperdiploid B-ALL, in which case it is associated with a poor prognosis. However, due to different translocation partner genes at chromosomes 1 and 19, distinct subtypes of hyperdiploid B-ALL with t(1;19)/der(19)t(1;19) are recognized based on the presence or absence of the TCF3-PBX1 fusion gene, but the cytogenetic and etiologic differences between the two remain understudied.
Findings
We report a case of an adult with a history of relapsed precursor B-ALL whose conventional cytogenetics showed an abnormal female karyotype with both hyperdiploidy and a t(1;19)(q23;p13). Fluorescence in situ hybridization (FISH) on previously G-banded metaphases using the LSI TCF3/PBX1 Dual Color, Dual Fusion Translocation Probe confirmed the presence of the TCF3-PBX1 gene fusion.
Conclusions
This particular pattern with a TCF3-PBX1 fusion within the context of a hyperdiploid karyotype is seen in B-ALL and is usually associated with a poor outcome. This case is one of only a few cases with both hyperdiploidy and a confirmed TCF3-PBX1 fusion, demonstrating the importance of using FISH for proper molecular classification of these cases in order to distinguish them from those with hyperdiploidy but no TCF3-PBX1 fusion gene. Such molecular studies may provide insight into the precise differences between TCF3-PBX1 positive and negative hyperdiploid B-ALL bearing the t(1;19)(q23;p13).
Similar content being viewed by others
Introduction
The t(1;19)(q23;p13) is one of the most frequent translocations in B-acute lymphoblastic leukemia (B-ALL), and is observed in both adult and pediatric populations at an overall frequency of 6% . This translocation can occur in a balanced – t (1;19)(q23;p13) – or unbalanced – der(19)t(1;19)(q23;p13) – form and can result in the fusion of TCF3 (transcription factor 3) found at 19p13 and PBX1 (pre-B cell leukemia homebox 1) found at 1q23 to form a chimeric gene whose protein product alters cell differentiation arrest, among other cellular processes [1]. Specifically, the fusion gene encodes a transcription factor bearing the transactivation domain of TCF3 and the DNA-binding domain of PBX1, which facilitates constitutive activation of genes bound by the protein encoded by PBX1 and other PBX proteins [2]. As a sole abnormality, t(1;19)/der(19)t(1;19) is associated with an intermediate prognosis in B-ALL, and hyperdiploidy is associated with a favorable prognosis [1]. However, more rarely, cases of t(1;19)/der(19)t(1;19) within the context of a hyperdiploid karyotype have been observed, only some of which express the TCF3-PBX1 fusion gene and are associated with a poor prognosis [3]. In addition to PBX1, other partner genes involved in rearrangements of TCF3, although at much lower frequencies, include ZNF384 (12p13; prognosis unknown), NOL1 (12p13; prognosis unknown), an unknown partner gene at 13q14 (prognosis unknown), HLF (17q22; extremely poor prognosis), and FB1/TFPT (19q13.4; prognosis unknown) [4-6]. The cytogenetic and etiologic differences between TCF3-PBX1 positive and negative B-ALL with hyperdiploidy and t(1;19)/der(19)t(1;19) remain understudied due to lack of molecular classification of the cases reported in the literature.
Case presentation
The patient was a forty-four year old woman with a history of relapsed precursor B-ALL, who was initially diagnosed in March 2013 with leukemic cells showing an immunophenotype positive for CD10, CD19, icCD22, CD38, icCD79a, CD138, TdT, HLA-DR and icIgM as well as a normal karyotype. Initial diagnosis was established at another institution at which point FISH analysis was not performed. After UK ALL 14 protocol consolidation therapy, she was considered to be in remission. In December 2013, a bone marrow biopsy showed evidence of relapse, and was comprised of approximately 85% blasts with a pre-B immunophenotype and a hyperdiploid, complex, poor-risk karyotype, further described in the results section. In January 2014, the patient underwent therapy with FLAG-Ida, resulting in a hypoplastic marrow with no significant residual blast population. Later in April 2014 she enrolled in a clinical trial with blinatumomab, which was eventually discontinued because the patient experienced multiple seizure episodes. A bone marrow biopsy showed extensive tumor necrosis with involvement by B-lymphoblasts representing over 90% of viable cells and comprising 5% of the total surface area. The immunophenotype was positive for CD10, CD19, PAX-5, CD79a and TdT (weak, rare), and negative for CD34 and CD20. The patient expired in May 2014 of relapsed B-lymphoblastic leukemia. Autopsy included a bone marrow biopsy, which revealed a hypercellular marrow of greater than 95% cellularity with sheets of lymphoblasts and extensive tumor necrosis.
Material and methods
Chromosome analysis was performed using standard cytogenetic techniques on the bone marrow of this patient. The karyotypes were prepared using the Applied Imaging CytoVision software (Applied Imaging, Genetix, Santa Clara, CA) and described according to the ISCN 2013 nomenclature [7].
Fluorescence in situ hybridization (FISH) was performed on interphase nuclei using the Vysis MYC-IGH Dual Color, Dual Fusion Probe, Vysis LSI BCR,ABL ES Dual Color Translocation Probe, and Vysis LSI MLL Dual Color, Break Apart Rearrangement Probe from Abbott Molecular (Des Plaines, Illinois 60018). Additionally, FISH was performed with the LSI TCF3/PBX1 Dual Color, Dual Fusion Translocation Probe on previously G-banded metaphases.
Findings
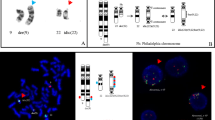
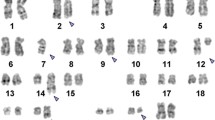
Only three metaphase cells were available for chromosome analysis due to a poor mitotic index. These cells revealed an abnormal female karyotype with numerical and structural abnormalities including extra copies of chromosomes 1, 8, 11, 20, 22, a (1;19) translocation, an unbalanced rearrangement of the long arm of chromosome 13 leading to 13q-, and a marker chromosome of unknown origin. This karyotype was described as (Figure 1):
FISH on interphase nuclei confirmed the additional copies of chromosome 8 in 73.8% of nuclei (79/107), chromosome 22 in 80% of nuclei (44/55), as well as chromosome 11 in 4.7% of nuclei (4/85) examined. The FISH results (Figure 2) were described as:
-
nuc ish(MYCx5,IGH)x2)[79/107]
-
nuc ish(BCRx3,ABL1x2)[44/55]
-
nuc ish(MLLx3)[4/85]
To further characterize and confirm the previous conventional cytogenetics findings [t(1;19) which fuses TCF3 (green signal) on 19p13 with PBX1 (red signal) at 1q23], FISH studies on previously G-banded metaphases were performed, and detected two fusion [t(1;19)] signals and an additional copy of red signal (+1q) with the TCF3-PBX1 probe indicative of translocation between TCF3 and PBX1, as well as an additional copy of the 1q23 locus, which is consistent with the karyotype results found previously. Gain of chromosome 1q is often seen in association with disease progression or advanced disease. Based on these studies the karyotype was described as (Figure 3):
Discussion
The t(1;19)(q23;p13)/der(19)t(1;19)(q23;p13) is one of the most common translocations seen in B-ALL cases and is typically found as a sole abnormality. It creates a fusion of TCF3 on 19p13 with PBX1 at 1q23, can be present in balanced or unbalanced form, and is usually associated with an intermediate prognosis [1]. Hunger et al. noted in an early study that 95% of t(1;19)/der(19)t(1;19)-positive cases of B-ALL with <50 chromosomes expressed the TCF3-PBX1 fusion transcript, whereas only 25% of cases with >50 chromosomes did. Furthermore, immunophenotypic differences between TCF3-PBX1 positive and TCF3-PBX1 negative cases were observed, which suggested etiologic differences between the two subtypes [3].
In a recent study conducted by Paulsson et al., 42 cases with both t(1;19)/der(19)t(1;19) and high hyperdiploidy (HeH; 51–67 chromosomes) from both published literature and the LRCG database were analyzed, revealing similar numerical chromosomal gains in both translocation-HeH (t-HeH) and classic-HeH (c-HeH) cases, most commonly involving chromosomes 21, 4, 6, 10, 18, 14, X, and 17, in decreasing frequency [8]. Furthermore, none of these cases were found to have a stemline balanced or unbalanced t(1;19), whereas 11% had hyperdiploid stemlines, suggesting that numerical chromosomal gains resulting in HeH are primary cytogenetic aberrations and occur prior to t(1;19)/der(19)t(1;19) [8]. This may result in clinical similarities between t(1;19)/der(19)t(1;19)-HeH ALL and c-HeH ALL, as the two may share a similar cytogenetic progression and etiology. It was also found that the majority of t(1;19)/der(19)t(1;19)-HeH cases tested by molecular methods were negative for the presence of the TCF3-PBX1 fusion gene [8]. Additionally, previous studies have found that greater than 90% of t(1;19) positive, TCF3-PBX1 fusion negative cases have an unbalanced form of the rearrangement [4]. In Paulsson et al’s study, only 18% of the cases had a balanced rearrangement [8], while 40% of TCF3-PBX1 positive cases overall have been found to have a balanced rearrangement [1], ultimately suggesting etiologically distinct subtypes of B-ALL with both hyperdiploidy and t(1;19)/der(19)t(1;19) based on the presence of the TCF3-PBX1 fusion gene by FISH and/or polymerase chain reaction (PCR) [8].
In the present study, we report a case of hyperdiploid B-ALL with a balanced t(1;19) bearing the TCF3-PBX1 fusion gene confirmed by metaphase FISH, which has only been previously reported in a small number of cases and represents a distinct subtype of B-ALL based on the presence of the confirmed fusion gene in conjunction with hyperdiploidy and t(1;19)/der(19)t(1;19). Interestingly, the numerical gains present in our case, of chromosomes 1, 8, 11, 20, and 22, are not consistent with the most common gains found in t-HeH/t(1;19)-positive B-ALL by Paulsson et al. [8]. In that study, the majority of cases were found to have unbalanced rearrangements and out of those that had molecular evidence, most did not bear the TCF3-PBX1 fusion [8]. The presence of different numerical gains between our case and those of Paulsson et al. further supports the fact that the TCF3-PBX1 positive and negative variants of t(1;19)/hyperdiploid B-ALL represent distinct subtypes of the disease. Furthermore, we queried the Mitelman Database of Chromosome Aberrations in Cancer for reported cases of ALL with t(1;19)/der(19)t(1;19), hyperdiploidy, and molecular evidence (FISH and/or PCR) of TCF3-PBX1 fusion, and only identified 5 cases that were positive for the fusion (Table 1). When compiling the karyotypes of these cases and the present case, we noted that four out of six cases had additional copies of chromosome 8, which was interestingly not found to be one of the most common numerical gains in t-HeH/t(1;19)-positive B-ALL [8].
Recent molecular insights into the TCF3-PBX1 fusion protein have revealed its involvement in complex signaling pathways. In particular, deregulation of JunD and NFX1-regulated transcriptional processes has been noted to be a significant effect of the fusion protein [9]. Additionally, PAX5 (19p13.2) haploinsufficiency, detectable both by conventional and molecular cytogenetics, is associated with TCF3-PBX1 in B-ALL. Specifically, FISH using both TCF3 split signal probes in conjunction with PAX5 locus-specific deletion probes suggests that PAX5 is a secondary event in the oncogenesis of TCF3-PBX1-positive B-ALL, and may be associated with clonal evolution of the malignancy [10]. Furthermore, studies have revealed that vascular endothelial growth factor-C (VEGF-C), encoded by VEGFC (4q34.3), is involved and perhaps essential to proliferation of TCF3-PBX1 positive leukemic B cells [11]. Finally, treatment with hyperfractionated cyclophosphamide, vincristine, doxorubicin, and dexamethasone alternating with methotrexate and high-dose cytarabine (hyper-CVAD) has shown a favorable outcome in adults with t(1;19)-positive ALL [12].
Hyperdiploidy in B-ALL normally conveys a favorable prognosis, but in the present study, the particular pattern of a t(1;19)(q23;p13.3) with TCF3-PBX1 fusion within the context of a complex karyotype (>3 abnormalities) and hyperdiploidy due to extra copies of chromosomes 8, 11 and 22 (confirmed by FISH) plus the presence of a marker chromosome of unknown origin is associated with an unfavorable prognosis in B-ALL [3]. It is one of only a few published cases with hyperdiploidy, t(1;19)/der(19)t(1;19), and a confirmed TCF3-PBX1 fusion in B-ALL, demonstrating the importance of using FISH and PCR for proper cytogenetic and molecular classification in order to distinguish the present scenario from hyperdiploid B-ALL with t(1;19)/der(19)t(1;19), but lacking the TCF3-PBX1 fusion. The latter represents a different subtype of B-ALL that may be primarily driven by chromosomal gains or other fusion genes rather than the t(1;19)/der(19)t(1;19) resulting in the TCF3-PBX1 fusion and should not be confused with the entity presented in this report. Further investigation of the cytogenetic and molecular etiologies of these subtypes of B-ALL is warranted to determine their implications in the diagnosis and prognosis of the malignancy.
References
Heim S, Mitelman F. Cancer Cytogenetics. 3rd ed. Hoboken, New Jersey: Wiley-Blackwell Publishers; 2009.
Troussard X, Rimokh R, Valensi F, Leboeuf D, Fenneteau O, Guitard AM, et al. Heterogeneity of t(1;19)(q23;p13) acute leukaemias. French Haematological Cytology Group. Br J Haematol. 1995;89(3):516–26.
Hunger SP, Sun T, Boswell AF, Carroll AJ, McGavran L. Hyperdiploidy and E2A-PBX1 fusion in an adult with t(1;19) + acute lymphoblastic leukemia: case report and review of the literature. Genes Chromosomes Cancer. 1997;20(4):392–8.
Barber KE, Harrison CJ, Broadfield ZJ, Stewart AR, Wright SL, Martineau M, et al. Molecular cytogenetic characterization of TCF3 (E2A)/19p13.3 rearrangements in B-cell precursor acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2007;46(5):478–86.
Boomer T, Varella-Garcia M, McGavran L, Meltesen L, Olsen AS, Hunger SP. Detection of E2A translocations in leukemias via fluorescence in situ hybridization. Leukemia. 2001;15(1):95–102.
Brambillasca F, Mosna G, Colombo M, Rivolta A, Caslini C, Minuzzo M, et al. Identification of a novel molecular partner of the E2A gene in childhood leukemia. Leukemia. 1999;13(3):369–75.
Shaffer LG, McGowan-Joran J, Schmid MS. ISCN 2013: An International System of Human Cytogenetic Nomenclature. Unionville, CT,USA: S. Karger Publications, Inc; 2013.
Paulsson K, Harrison CJ, Andersen MK, Chilton L, Nordgren A, Moorman AV, et al. Distinct patterns of gained chromosomes in high hyperdiploid acute lymphoblastic leukemia with t(1;19)(q23;p13), t(9;22)(q34;q22) or MLL rearrangements. Leukemia. 2013;27(4):974–7.
Hajingabo LJ, Daakour S, Martin M, Grausenburger R, Panzer-Grümayer R, Dequiedt F, et al. Predicting interactome network perturbations in human cancer: application to gene fusions in acute lymphoblastic leukemia. Mol Biol Cell. 2014;25(24):3973–85. Doi: 10.1091/mbc.E14-06-1038. Epub 2014 Oct 1.
Familiades J, Bousquet M, Lafage-Pochitaloff M, Béné MC, Beldjord K, De Vos J, et al. PAX5 mutations occur frequently in adult B-cell progenitor acute lymphoblastic leukemia and PAX5 haploinsufficiency is associated with BCR-ABL1 and TCF3-PBX1 fusion genes: a GRAALL study. Leukemia. 2009;23(11):1989–98. Doi: 10.1038/leu.2009.135. Epub 2009 Jul 9.
Shirasaki R, Tashiro H, Oka Y, Sugao T, Yamamoto T, Yoshimi M, et al. Vascular endothelial growth factor-C and its receptor type-3 expressed in acute lymphocytic leukemia cases with t(1;19). Int J Hematol. 2011;94(2):203–8. Doi: 10.1007/s12185-011-0889-5. Epub 2011 Jul 6.
Garg R, Kantarjian H, Thomas D, Faderl S, Ravandi F, Lovshe D, et al. Adults with acute lymphoblastic leukemia and translocation (1;19) abnormality have a favorable outcome with hyperfractionated cyclophosphamide, vincristine, doxorubicin, and dexamethasone alternating with methotrexate and high-dose cytarabine chemotherapy. Cancer. 2009;115(10):2147–54. doi:10.1002/cncr.24266.
Rowe D, Devaraj PE, Irving JA, Hogarth L, Hall AG, Turner GE. A case of mature B-cell ALL with coexistence of t(1;19) and t(14;18) and expression of the E2A/PBX1 fusion gene. Br J Haematol. 1996;94(1):133–5.
Foa R, Vitale A, Mancini M, Cuneo A, Mecucci C, Elia L, et al. E2A-PBX1 fusion in adult acute lymphoblastic leukaemia: biological andclinical features. Br J Haematol. 2003;120(3):484–7.
Acknowledgments
To UCLA Clinical Cytogenetics Laboratory.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CAT and DS had equal contribution to this manuscript and led drafting, conducted survey of relevant literature, and edited and revised all drafts. LY wrote the initial draft. SP revised the manuscript and added various comments. LY conducted the bench work analysis. MK provided the clinical presentation of the patient. NR edited the manuscript. All authors read and approved the final manuscript.
Carlos A Tirado and David Shabsovich contributed equally to this work.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Tirado, C.A., Shabsovich, D., Yeh, L. et al. A (1;19) translocation involving TCF3-PBX1 fusion within the context of a hyperdiploid karyotype in adult B-ALL: a case report and review of the literature. Biomark Res 3, 4 (2015). https://doi.org/10.1186/s40364-015-0029-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40364-015-0029-0