Abstract
Background
Linear bone growth is achieved by the division of chondrocytes at the growth plate and is regulated by endocrine and paracrine factors such as growth hormone. Mutations that negatively affect chondrogenesis can be a contributor to short stature. One such mutation can occur in the ACAN gene, causing short stature and advanced bone age. Similarly, mutations in growth hormone receptors (GHR) can lead to Laron syndrome (LS), one of the several disorders that are collectively called growth hormone insensitivity syndrome (GHI). Another example is Floating-Harbor syndrome (FHS), a rare autosomal dominant due to mutations in the SRCAP gene that can also result in short stature.
Case presentation
We report the case of a 6-year-old female with concomitant mutations in the three genes mentioned above. The mutations reported here were found on genetic studies and are usually benign, causing a variant of undetermined significance. However, our patient’s phenotype could only be explained by the compounded effects of pathogenic mutations of these genes. Some of the same mutations were also found in the patient’s father and her paternal grandfather. Both also presented with short stature, though not to the same degree as our patient. While these mutations are often reported to be insignificant, they gave rise to severe short stature and a specific phenotype in the patient when presented together. We think that even though the GHI spectrum is inherited through an autosomal recessive pattern, the sum of these heterozygous mutations resulted in severe short stature despite the limited GHI seen in our patient, the father, and the grandfather, through a rare ACAN and SRCAP mutation that, to our knowledge, has not been previously reported as a pathogenic mutation in the literature.
Conclusion
We investigated the possible synergistic effects of these variations on exacerbation or masking of the signs and symptoms of GHI with the hope of providing a better understanding of these genes and their function through our rare case.
Similar content being viewed by others
Background
Linear bone growth is achieved by chondrocyte division at the growth plate. Chondrocyte division is regulated by endocrine and paracrine factors such as growth hormone [1]. Mutations that negatively affect chondrogenesis can potentially be a contributor to short stature [1]. One such mutation occurs in the ACAN gene that encodes Aggrecan, which is a critical proteoglycan component of the extracellular matrix of the growth plate cartilage [2]. Most ACAN mutations are inherited through an autosomal dominant pattern and differ in phenotype, but all of them are characterized by short stature and advanced bone age [3,4,5].
Similarly, mutations in the growth hormone receptor (GHR) can lead to Laron syndrome (LS), one of the several disorders that are collectively called growth hormone insensitivity syndrome (GHI) [6,7,8,9,10]. Studies into heterozygous mutations have found that such mutations may indeed result in shorter-than-average stature, but not to what would be considered a pathological extent; as well, there have been limited studies to suggest that heterozygous mutations may result in GHI [11, 12]. While LS is an autosomal recessive condition, our investigation indicates that if there are simultaneous heterozygous mutations in different genes that normally contribute to linear bone growth, the occurrence of short stature with highly variable phenotypes is quite possible [10, 12,13,14,15].
Here, we report a case with three concomitant mutations in the aforementioned genes. Most of these mutations are of undetermined significance due to a lack of available research into these mutations. On the other hand, there is controversy as to the pathogenicity of the same variant of the GHR gene of our patient. However, our patient presented with a phenotype that could be explained by the compounded pathogenic effect of these mutations. Our aim in this report is to provide a better understanding of these mutations and to establish that they are likely pathogenic.
Case presentation
Clinical data
A 6-year-old girl presented at our clinic with short stature. She was born healthy at 38 weeks by vaginal delivery to non-consanguineous parents. At birth, she was 48 cm in height and weighed 2655 g with a head circumference of 34.5 cm. The pregnancy was uncomplicated. The subject had an Apgar score of 8 to 9. The patient’s father’s height was 155 cm, and her mother’s height was 153 cm. These values were respectively 2.1 and 0.6 standard deviations below average in their country of origin, Iran, when matched by age and gender.
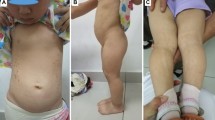
On initial evaluation, the patient was of disproportionately short stature, 96 cm tall (− 3.5 SDS), and weighing 21.6 kg. Examination of the subject revealed facial features of long eyelashes, a wide columella, and a short philtrum, with no signs of early-onset puberty, and no evidence of congenital anomalies. Given the exam, an underlying genetic cause was suspected, but we aimed to rule out other possible causes through a complete medical evaluation that yielded the results presented below.
Methods
Our investigations were performed in two phases: we firstly assessed the subject with a physical examination, blood tests, imaging, and genetic testing, and secondly, based on the results, genetic analyses of the subject’s parents were performed.
Blood tests were ordered to assess thyroid function, as well as insulin-like growth factor 1 (IGF1), growth hormone (GH), prolactin levels, and urinary AM cortisol; to rule out autoimmune-related malabsorptive diseases, anti-tissue transglutaminase antibody (anti-tTG) IgG, and IgA levels were evaluated. A GH stimulation test by clonidine was also performed. A urinalysis was conducted, as well as a renal ultrasound, and bone density was assessed. An X-ray was performed of her left hand/wrist to determine bone age. The national growth reference provided by Iran’s Ministry of Health and Medical Education was used in this study.
Subsequently, we ordered conventional G-banding karyotyping to screen for large duplications and deletions, balanced translocations, inversions, and ploidy changes. Then, for a more detailed analysis, we followed this with whole-exome enrichment, performed by the Agilent SureSelect V6 target enrichment kit on Illumina Hiseq 4000 platform with an average depth of 206X performed by Macrogen, South Korea. Nearly all exons and flanking 10 bp (base pairs) were detected and analyzed (detected variants included single-point mutations and small indels (within 20 bp)). The analytical sensitivity and specificity of the next-generation sequencing (NGS) method used in this assay for detection variants mentioned above was more than 95%, and nearly all exons were detected and analyzed.
Based on the results of our analysis of the subject’s genetic testing, we performed a polymerase chain reaction (PCR) followed by Sanger sequencing on the parents to further investigate the validity of the detected variants. This procedure was repeated on available relatives of the subject’s father (his father and his sister). All bioinformatics analysis of sequencing results was performed using international databases (American College of Medical Genetics, The Human Gene Mutation Database and ClinVar) and standard bioinformatics software.
After all evaluations had been completed, growth hormone therapy was initiated. The GH dose was increased to a maximum amount of 67 μg/kg per day for a course of 6 months of treatment.
Clinical results
Analysis of the blood tests from the subject showed results within normal limits (WNL), although IGF1 levels were found to be at borderline low-normal limits (2.5th percentile) and GH levels were found to be mildly deficient (0.96 ng/ml), while GH stimulation was WNL. Urinalysis, prolactin, AM cortisol level, and bone density were all WNL, and the renal ultrasound was unremarkable. X-ray results were reviewed to determine bone age; her left hand/wrist X-ray showed results consistent with an age of 7 years when she was, in fact, 6 years old. Conventional G-banding karyotype showed no chromosomal abnormalities. Anti-tTG, IgG, and IgA were found to be WNL, suggesting no evidence of autoimmune-related malabsorptive disease.
Once all tests were concluded, GH therapy was initiated for the subject given her unsatisfactory growth velocity. With GH therapy, her growth velocity increased about 1.75 cm above the growth velocity prior to treatment. These results thus far point to treatment failure and partial insensitivity to GH.
Molecular results
Results from whole exome enrichment showed that the subject was heterozygous for the GHR gene (NM_001242399.2 exon6: c.556C>T) that led to amino acid changes p.R186C, heterozygous for the ACAN gene (NM_013227.3 exon17: c.7418G>A) that led to amino acid changes p.R2473Q, heterozygous for the SRCAP gene (NM_006662.2 exon25: c.4259C>T) that led to amino acid changes p.S1420F, and heterozygous for the AGBL1 gene (NM_152336.2 exon22: c.2969G>C) that led to amino acid changes p.C990S. These results are summarized in Table 1.
To determine the origin of her mutations and to rule out any other variation in her family, WES was performed on the subject’s parents. Her father was heterozygous for the GHR gene (NM_001242399.2 exon6: c.556C>T) that led to amino acid changes p.R186C. Her mother was heterozygous for the AGBL1 gene (NM_152336.2 exon22: c.2969G>C) that led to amino acid changes p.C990S. These results are summarized in Table 2.
PCR amplification followed by Sanger sequencing for the variants c.556C>T p.R186C in GHR gene, c.2969G>C p.C990S in AGBL1 gene, and c.7418G>A p.R2473Q in ACAN gene validated the variants detected via WES. The results are shown in Table 3.
To evaluate the effects for the variant c.556C>T p.R186C in the GHR gene, we ordered PCR followed by Sanger sequencing for the available family members of the patient’s father (his father and his sister). His father was heterozygous for the variant, and his height was 157 cm (− 1.8 SDS), nearly the same as his son; however, his sister, who was of average height at 165 cm (+ 1.1 SDS), did not inherit the variant.
Based on these results, we concluded that the patient’s father’s familial short stature could be due to his GHR mutation resulting in GHI spectrum manifestation. Although we posit that heterozygous mutations in the GHR gene are seen resulting in GHI despite having an autosomal recessive pattern of inheritance, we are not yet aware of how this is possible; so far, only the signs and symptoms concurrent with the mutations stand as evidence of their detrimental effects on height.
Discussion
It is evident that our patient’s mutations in ACAN and SRCAP are sporadic and new, as they did not exist in her parents. Prior to GH therapy, the subject’s GH level was low while a GH stimulation test showed normal results; with GH therapy, the subject’s growth velocity did not increase to the degree expected, suggesting a partial insensitivity to GH.
As mentioned above, mutations in the GHR gene can result in GHI. The subject’s partial GH insensitivity is likely explained by her GHR mutation. Our patient had a heterozygous missense GHR mutation which previously has been reported as a pathogenic variant in The Human Gene Mutation Database (HGMD) and other studies [16], but the ClinVar Database does not support its pathogenicity [9]. Given that her father shares this mutation, it is highly likely that his short stature is also at least in part due to a similar GH insensitivity.
That said, the severity of our patient’s short stature is most likely the result of not only her GHR mutation, but the additive effects of her mutations in ACAN or SRCAP or indeed both these genes.
As explained earlier, pathogenic ACAN mutations can result in short stature and advanced bone age similar to those seen in our patient. However, to our knowledge, her specific mutation has not been reported as pathogenic in previous publications. Since her phenotype resembles those seen in pathogenic mutations of the ACAN gene, here we would like to suggest that her specific variation is likely pathogenic and a contributor to her advanced bone age and short stature.
Mutations in the SRCAP gene can result in Floating-Harbor syndrome (FHS), a rare autosomal dominant mutation in the SRCAP gene that can also result in short stature and other phenotypes like a triangular face, low hanging columella, short philtrum, wide mouth, and an elongated nose with a narrow bridge [17]. To our knowledge, the pathogenicity of our patient’s specific SRCAP mutation was not reported previously, and its significance is unknown; however, her long eyelashes, her seemingly wide columella, and her slightly short philtrum all resemble those futures seen in FHS, and could be the result of this mutation [18]. Since other work-up did not explain her mildly low levels of GH and IGF1, here we would like to report her SRCAP mutation as the cause; other studies have reported similar findings [19]. Mutations of the AGBL1 gene are associated with late-onset of Fuchs’ corneal dystrophy, an autosomal dominant condition that results in corneal epithelial destruction [20]. In our patient, her mutation is likely pathogenic based on the American College of Medical Genetics and Genome (ACMG) and as reported in other studies [20]. Therefore, we referred our patient to an ophthalmologist to be further evaluated for this condition and to avoid corneal epithelial destruction and loss of eyesight.
The frequency in the general population, pathogenicity, signs, and symptoms of the aforementioned variants are summarized in Table 4.
Conclusions
To our knowledge, the concomitance of these mutations as they relate to the resultant phenotype in our patient is unique and lead to a new hypothesis: Based on our clinical investigations and our review of the latest relevant literature, the highly unusual phenotype seen in our patient can most likely be attributed to her particular variations.
One explanation for her phenotype is that these variants are pathogenic and of the autosomal dominant pattern of inheritance, despite being categorized in the VUS section. The evidence that we have to support our claim are the signs and symptoms of these variants that are observed only in pathogenic mutations of the genes, mentioned above. Regardless of the pattern of inheritance of these variations, the attributed signs and symptoms were mild, so we could assume that if they do cause short stature, it would be to a mild degree, in correlation with other signs and symptoms. That said, with short stature being one of the signs of all of these mutations, and given that its severity does not correlate directly with the other signs and symptoms, we can assume that ultimately the impact on stature is a result of the synergistic effects of all the abovementioned variations. One possible mechanism is that, along with her partial GHI, the mutation in her SRCAP gene caused reduced plasma levels of GH and IGF1, resulting in synergistic effects with the GHI. Likewise, the synergistic effects of ACAN and SRCAP mutations coinciding with GHI could explain our patient’s lack of response to treatment, in spite of her heterozygous GHR mutation. Thus, we can presume that while these particular variations may have an insignificant effect individually on a person’s height, their usual phenotypes could be altered due to the synergistic effects they impart on one another, influencing a patient’s height dramatically. Also, many patients are labeled as having idiopathic or familial short stature (the father of our patient is an example) because no genetic studies are conducted and they do not show signs or symptoms that could lead the physician to the correct diagnosis. Therefore, we think that many of these mutations are not as rare as previously believed, but frequently go undiagnosed due to their variable phenotypes. We suggest that a complete genetic study and WES may be beneficial in certain patients who are suspected to carry the aforementioned genes prior to assigning a diagnosis of idiopathic or familial short stature.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author in response to reasonable requests and with the permission of the patient’s legal guardians.
Abbreviations
- Anti-tTG:
-
Anti-tissue transglutaminase antibody
- GHR:
-
Growth hormone receptor
- GHI:
-
Growth hormone insensitivity syndrome
- LS:
-
Laron syndrome
- FHS:
-
Floating-Harbor syndrome
- SDS:
-
Standard deviations
- IGF1:
-
Insulin-like growth factor 1
- HGMD:
-
Human Gene Mutation Database
- ACMG:
-
American College of Medical Genetics and Genome
- genomAD:
-
Genome Aggregation Database
- PCR:
-
Polymerase chain reaction
- NGS:
-
Next-generation sequencing
- het:
-
Heterozygous
References
Baron J et al (2015) Short and tall stature: a new paradigm emerges. Nat Rev Endocrinol 11:735–746
Roughley PJ, Mort JS (2014) The role of aggrecan in normal and osteoarthritic cartilage. J Exp Orthop 1:8
Dateki SJCPE (2017) ACAN mutations as a cause of familial short stature. Clin Pediatr Endocrinol 26:119–125
Quintos JB, Guo MH, Dauber A (2015) Idiopathic short stature due to novel heterozygous mutation of the aggrecan gene. J Pediatr Endocrinol Metab 28:927–932
Nilsson O et al (2014) Short stature, accelerated bone maturation, and early growth cessation due to heterozygous aggrecan mutations. J Clin Endocrinol Metab 99:E1510–E1518
Fang P et al (2007) Primary growth hormone (GH) insensitivity and insulin-like growth factor deficiency caused by novel compound heterozygous mutations of the GH receptor gene: genetic and functional studies of simple and compound heterozygous states. J Clin Endocrinol Metab 92:2223–2231
Gent J, Van Kerkhof P, Roza M, Bu G, Strous G (2002) Ligand-independent growth hormone receptor dimerization occurs in the endoplasmic reticulum and is required for ubiquitin system-dependent endocytosis. Proc Natl Acad Sci USA 99:9858–9863
Rosenfeld RG (1994) Rosenbloom, A.L. & Guevara-Aguirre, J.J.E.r. Growth hormone (GH) insensitivity due to primary GH receptor deficiency. Endocr Rev 15:369–390
Goddard AD et al (1995) Mutations of the growth hormone receptor in children with idiopathic short stature. N Engl J Med 333:1093–1098
Kaji H et al (1997) Novel compound heterozygous mutations of growth hormone (GH) receptor gene in a patient with GH insensitivity syndrome. J Clin Endocrinol Metab 82:3705–3709
Kou K, Lajara R, Rotwein P (1993) Amino acid substitutions in the intracellular part of the growth hormone receptor in a patient with the Laron syndrome. J Clin Endocrinol Metab 76:54–59
Kaji H, Ohashi S, Abe H, Chihara K (1994) Regulation of the growth hormone (GH) receptor and GH-binding protein mRNA. Proc Soc Exp Biol Med 206:257–262
Chujo S et al (1996) No correlation of growth hormone receptor gene mutation P561T with body height. Eur J Endocrinol 134:560–562
Moia S et al (2017) Compound heterozygosity for two GHR missense mutations in a patient affected by Laron Syndrome: a case report. Ital J Pediatr 43:94
Berg MA et al (1993) Diverse growth hormone receptor gene mutations in Laron syndrome. Am J Hum Genet 52:998–1005
Amselem S et al (1993) Spectrum of growth hormone receptor mutations and associated haplotypes in Laron syndrome. Human Mol Genet 2:355–359
Hood RL et al (2012) Mutations in SRCAP, encoding SNF2-related CREBBP activator protein, cause Floating-Harbor syndrome. Am J Human Genet 90:308–313
Wieczorek D, Wüsthof A, Harms E, Meinecke P (2001) Floating-Harbor syndrome in two unrelated girls: mild short stature in one patient and effective growth hormone therapy in the other. Am J Med Genet 104:47–52
Cannavo S et al (2002) Abnormalities of GH secretion in a young girl with Floating-Harbor syndrome. J Endocrinol Invest 25:58–64
Riazuddin SA, Vasanth S, Katsanis N, Gottsch JD (2013) Mutations in AGBL1 cause dominant late-onset Fuchs corneal dystrophy and alter protein-protein interaction with TCF4. Am J Hum Genet 93:758–764
Acknowledgements
We thank all the participants for their cooperation in this project.
Funding
Not applicable (no funds were received).
Author information
Authors and Affiliations
Contributions
S. MPN visited and carried out the treatment of the patient. N. MKH analyzed genetic findings, aided in the genetic study, drafted the initial manuscript, reviewed and revised the manuscript, enrolled the patient in the study, collected the data, and drafted the final manuscript. B.HZ and T.V carried data management, drafted, and revised the final manuscript. All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Due to the rarity of mutations and the condition of our patient, written informed consent was obtained from her parents to use her data for research purposes.
This study was approved by the University and Clinic’s Research and Ethics Committees.
An informed written consent to participate in our study was obtained from the patient’s parents and the members of the father’s family members for laboratory work-up and DNA analysis.
Consent for publication
A separate informed written consent was obtained from the patient’s parents and the father’s family members regarding their own and their child’s clinical details, research findings, and data for publication purposes.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mohammadian Khonsari, N., Mohammad Poor Nami, S., Hakak-Zargar, B. et al. Mutations of uncertain significance in heterozygous variants as a possible cause of severe short stature: a case report. Mol Cell Pediatr 7, 11 (2020). https://doi.org/10.1186/s40348-020-00104-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40348-020-00104-6