Abstract
Background
Curbing the potential negative impact of antibiotic resistance, one of our era's growing global public health crises, requires regular monitoring of the resistance situations, including the reservoir of resistance genes. Wild birds, a possible bioindicator of antibiotic resistance, have been suggested to play a role in the dissemination of antibiotic-resistant bacteria. Therefore, this study was conducted with the objective of determining the phenotypic and genotypic antibiotic resistance profiles of 100 Escherichia coli isolates of gull and pigeon origin by using the Kirby-Bauer disk diffusion method and PCR. Furthermore, the genetic relationships of the isolates were determined by RAPD-PCR.
Results
Phenotypic antibiotic susceptibility testing revealed that 63% (63/100) and 29% (29/100) of E. coli isolates were resistant to at least one antibiotic and multidrug-resistant (MDR), respectively. With the exception of cephalothin, to which the E. coli isolates were 100% susceptible, tetracycline (52%), kanamycin (38%), streptomycin (37%), ampicillin (28%), chloramphenicol (21%), trimethoprim/sulfamethoxazole (19%), gentamicin (13%), enrofloxacin (12%) and ciprofloxacin (12%) resistances were detected at varying degrees. Among the investigated resistance genes, tet(B) (66%), tet(A) (63%), aphA1 (48%), sul3 (34%), sul2 (26%), strA/strB (24%) and sul1 (16%) were detected. Regarding the genetic diversity of the isolates, the RAPD-PCR-based dendrograms divided both pigeon and gull isolates into five different clusters based on a 70% similarity threshold. Dendrogram analysis revealed 47–100% similarities among pigeon-origin strains and 40–100% similarities among gull-origin E.coli strains.
Conclusions
This study revealed that gulls and pigeons carry MDR E. coli isolates, which may pose a risk to animal and human health by contaminating the environment with their feces. However, a large-scale epidemiological study investigating the genetic relationship of the strains from a "one health" point of view is warranted to determine the possible transmission patterns of antibiotic-resistant bacteria between wild birds, the environment, humans, and other hosts.
Video Abstract
Similar content being viewed by others
Background
Wild animals, including wild birds, are considered part of our ecosystem’s biodiversity, contributing to the continuity of the normal ecosystem on Earth. Assessing their possible epidemiological role as a carrier of infection contributes to the assessment of ecosystem health status as well as diseases that can affect other hosts, including humans [1]. It has been proposed that wild birds may act as reservoirs of pathogenic microorganisms [1,2,3], suggesting that the diseases carried by wild birds may affect the health of other potential hosts [4,5,6].
Escherichia coli (E. coli), Campylobacter, and Salmonella, common zoonotic bacterial pathogens that cause severe human infections [7], are among the most frequently reported pathogens associated with wild birds [4, 8]. Even though E. coli is a commensal pathogen in the intestines of humans and different animal species, including avian species, it is one of the most common animal and human pathogens responsible for significant infections [9]. Moreover, this bacterium is known to transfer its resistance genes to other bacteria besides acquiring resistance genes from other bacteria [10]. Hence, this bacterium is used as a model bacteria for detecting antibiotic resistance in veterinary and human medicine [11].
Currently, antimicrobial-resistant microorganisms have become one of the biggest health challenges in public health and veterinary medicine [12,13,14]. This growing threat of antibiotic resistance can significantly impact both sectors by limiting antibiotic options [15]. One critical factor that aggravates this situation is the improper use of antibiotics [16]. Several genes conferring resistance to drugs, which are highly important in human and animal health, have been reported in E. coli isolated from wild birds from different parts of the world [17,18,19], showing the commonness of antibiotic-resistant pathogens originating from other hosts (e.g., animals and humans) in the environment [20]. Therefore, wild birds, which are not directly exposed to antibiotic agents, could become infected by resistant bacteria through contact with a contaminated environment (for example, contaminated water/food) [21, 22]. In support of this argument, molecular studies have reported the genetic similarities of E. coli strains obtained from wild birds, landfills, wastewater, and humans [23, 24]. The carriage of drug-resistant E. coli isolates by wild birds suggests that wild birds may serve as a reservoir in the transmission of this pathogen to domestic animals and also cause environmental contamination [1, 24].
In general, several studies have highlighted the possible role of wild birds as the carriers/reservoirs of antibiotic-resistant E. coli pathotypes [3, 22, 25,26,27,28] although the situation in developing countries is not adequately elucidated yet. In Turkey, despite the availability of some studies [5, 29, 30] on isolation and antibiotic susceptibility testing, antibiotic resistance patterns and the resistance genes' distribution in wild bird populations have not been clearly elucidated. However, addressing the growing challenge of antibiotic resistance requires a detailed understanding of potential sources and vectors of resistance genes [31]. Hence, determining antibiotic resistance patterns and genes that cause resistance in bacteria obtained from hosts like wild birds for which sufficient data are unavailable is of great importance in designing effective prevention strategies against antibiotic resistance challenges. Therefore, this study aimed to determine the phenotypic and genotypic antibiotic resistance profile of E. coli isolated from wild bird species in close contact with humans (gulls and pigeons). Moreover, the isolates’ genetic diversity was analyzed by using RAPD-PCR.
Material and method
Bacterial isolates
The bacterial isolates used in this study were obtained from culture collections of the Department of Veterinary Microbiology, Ondokuz Mayis University, Turkey. The isolates were comprised of 100 E. coli isolates of gulls (50) and pigeons (50) origin, which were stored at − 20 °C. The isolates obtained from culture collections were isolated from fecal sample of healthy wild birds collected in different times from the Black Sea Region of Turkey. MacConkey agar, eosin methylene blue (EMB), and tryptic soy agar (TSA) were used for the revival and purification of the isolates. E. coli isolates were first inoculated on MacConkey agar (Oxoid, UK), and then a single colony was taken from MacConkey agar and inoculated on EMB (Oxoid, UK) agar. Similarly, a colony grown on the EMB agar was taken and sub-cultured on TSA (Oxoid, UK) to be used for various purposes (e.g., DNA extraction). All media were incubated at 37 °C for 24 h to allow bacterial growth.
DNA extraction
In order to obtain the bacterial DNA, a few pure cultured colonies grown on TSA were selected and suspended in an Eppendorf tube containing 500 µl of sterile distilled water. The mixture was then vortexed and boiled at 100 °C for 10 min and centrifuged at 10,000 rpm for 10 min. The supernatant obtained from the centrifugation process was stored at − 20 °C to be used as the template DNA [3, 32].
E. coli genotypic confirmation
PCR-based genotypic confirmation of E. coli isolates was performed by modifying and optimizing the protocol previously described by Abd El-Razik et al. [33]. For this purpose, 16S rRNA primer pair (Eco 2083 (F): 5'-GCTTGACACTGAACATTGAG-3′; Eco 2745 (R): 5'-GCACTTATCTCTTCCGCATT-3′) were used, and 662 bp bands were considered positive. A PCR mixture comprised of a total volume of 25 μl mixtures which include 10 × PCR buffer (2.5 µl), MgCl2 (3 µl), primer (0.25 µl for each primer), deoxynucleotide triphosphate (dNTP) (0.5 µl), Taq polymerase (0.3 µl), 13.2 μl of sterile distilled water, and template DNA (5 μl). The mixture prepared according to the determined protocol was subjected to amplification conditions consisting of initial denaturation for 2 min at 95 °C, followed by 35 cycles of denaturation (45 s at 94 °C), annealing (45 s at 57 °C), and extension (45 s at 72 °C), and final extension for 10 min at 72 °C. After mixing the amplified PCR product with the loading dye, 10 µL of the mixture was taken using a micropipette and loaded into the wells of agarose gel (1.5%) containing 2 µg/ml ethidium bromide. The mixture was then subjected to gel electrophoresis at 150 V for 60 min and visualized using an ultraviolet (UV) transilluminator. To determine the amplicons’ length and whether the band with targeted amplicon sizes (662 bp) was formed or not, DNA Marker (Thermo Scientific, SM0241, 100 bp DNA Ladder) was used. E. coli ATCC® 25,922 strain was used as a positive control, whereas a mixture without target DNA was used as a negative control.
Antibiotic susceptibility test
The Kirby–Bauer standard disk diffusion method was used to determine the resistance patterns of isolates against commonly used antibiotics following the protocols specified in CLSI guidelines [34,35,36]. Resistance/susceptibility to ampicillin (AMP; 10 µg), trimethoprim/sulfamethoxazole (SXT; 1.25/23.75 µg), cephalothin (KF; 30 µg), tetracycline (TE; 30 µg), chloramphenicol (C;30 µg), streptomycin (S;10 µg), kanamycin (K;30 µg), enrofloxacin (ENR;5 µg), gentamicin (CN; 10 µg), and ciprofloxacin (CIP;5 µg) was investigated. The selection of antibiotics was done in a way that represent major group of antibiotics (beta-lactams, aminoglycoside, phenicol, fluroquinolones, and folate antagonist) taking in to account their common usage and also their public and animal health importance.
The bacterial suspensions used for determining the resistance pattern were prepared by suspending the colonies grown on TSA in sterile physiological saline and adjusting the turbidity to the 0.5 McFarland standard. The prepared suspensions were spread on Mueller–Hinton agar (MHA) within 15 min. Following this, antibiotic discs were aseptically placed on the MHA surface with the help of sterile forceps and incubated overnight at 37 °C under aerobic conditions. Interpretation of the results was performed according to CLSI criteria by measuring the diameters of the inhibition zones and recorded as resistant (R), moderate (I), or susceptible (S) [34,35,36]. E. coli ATCC® 25,922 strain was used as the quality control strain. Bacterial isolates found resistant to at least three antibiotics in different classes (≥ 3 antibiotic groups) were considered as MDR isolates [15]. However, in analyzing the isolates’ resistance profile, only isolates that showed phenotypic resistance to a particular antibiotic were considered (intermediate resistance isolates were not taken into account).
Antibiotic resistance genes determination
Simplex and multiplex PCR (mPCR) was employed to determine antibiotic resistance genes using specific primer sets. For this purpose, the antibiotic resistance genes conferring resistance to quinolones, tetracyclines, sulfonamides, and aminoglycosides were investigated by modifying and optimizing the PCR protocols described in previous studies (Tables 1 and 2) [37,38,39]. Quinolone [qnr(A), qnr(B) and qnr(S)], tetracyclines resistance genes [tet(A), tet(B) and tet(C)], aminoglycosides (strA/strB, aphA1, aphA2, aadB, and aac(3) IV), and sulfonamides (sul1, sul2 and sul3) resistance genes were investigated. qnr genes’ positive controls were kindly provided by Assoc. Prof. Dr. Yeliz Tanriverdi Çayci (Ondokuz Mayis University, Faculty of Medicine, Department of Medical Microbiology). For other resistance genes, E. coli strains known to have the genes under consideration, obtained from Ondokuz Mayıs University Veterinary Microbiology Laboratory, were used.
For determining sulfonamide resistance, mPCR was performed in a total volume of 25 µl PCR reaction mixture consisting of PCR components presented in Table 1 and sterile distilled water. Other genes were investigated using simplex PCR, and PCR amplifications were performed using the concentration of PCR components presented in Table 1, which were adjusted to a total of 25 µl with sterile distilled water. The amplification conditions for sul1, sul2, and sul3 consist of pre-denaturation (15 min at 95 °C) followed by 30 cycles of denaturation (for 60 s at 95 °C), annealing (60 s at 66 °C), and elongation (60 s at 72 °C), and final elongation for 10 min at 72 °C. The amplification conditions for the plasmid-mediated quinolone resistance (PMQR) genes (qnrA, qnrB, and qnrS) were as follows: 5 min of pre-denaturation at 95 °C, 35 cycles of denaturation at 95 °C for 60 s, 60 s of annealing at the specific primer temperature presented in Table 2, and extension at 72 °C for 1 min and 5 min of final elongation at 72 °C. tet(A), tet(B), tet(C), strA/strB, aac(3)IV, aphA1, aphA2, and aadB were subjected to PCR amplification consisting of pre-denaturation at 94 °C for 15 min, 30 cycles of denaturation at 94 °C for 60 s, annealing for 60 s at the specific primer temperature indicated in Table 2, and elongation at 72 °C for 60 s, and the final elongation at 72 °C for 10 min. Amplification products were subjected to gel electrophoresis (1.5% agarose) containing 2 μg/ml ethidium bromide and visualized using a UV transilluminator.
Genotyping of the isolates by RAPD-PCR
Determination of the E. coli isolates’ phylogenetic relatedness was performed by using “enterobacterial repetitive intergenic consensus-2 (ERIC-2)” primer (5′-AAGTAAGTGACTGGGGTGAGCG-3′) as previously described by Versalovic et al. [40]. The PCR amplification reaction mixture consists of 10 × PCR buffer (2.5 µl), MgCl2 (2.5 µl), dNTP (0.5 µl), ERIC-2 primer (0.8 µl), Taq DNA polymerase (0.2 µl), and template DNA (5 µl), which were adjusted to a total of volume 30 µl reaction mixture with DNase/RNase-free distilled water. The amplification conditions for RAPD-PCR consisted of 5 min initial denaturation at 94 °C, 40 cycles of 1 min denaturation at 94 °C, 60 s of annealing at 36 °C, and 3 min extension at 72 °C, and a final extension cycle at 72 °C for 7 min. For visualizing the amplicons, a 1.5% agarose gel was prepared in a 1XTBE buffer, and the amplified products were subjected to gel electrophoresis containing 2 μg/ml ethidium bromide for 80 min at 140 V. The bands formed after electrophoresis were visualized using a UV transilluminator, and the bands formed were recorded. The bands were then analyzed with the image analysis program (Quantity one, BioRad) using the UPGMA (“unweighted pair group method with arithmetic mean”) method and dendrograms were drawn.
Statistical analysis
Descriptive analysis of data obtained from laboratory results was performed using Microsoft Excel® and SPSS® version 26. A chi-square test (χ2) was used to determine the difference between gulls and pigeons in terms of carrying E. coli isolates that were resistant to at least one antibiotic, two or more antibiotics, multidrug-resistant, and also antibiotic-resistant genes carriage. Moreover, the chi-square test was used to determine the associations between specific antibiotic resistance gene carriage and phenotypic antibiotic resistance expression. The difference/relationship was considered statistically significant when the p-value was less than 0.05.
Results
Genotypic confirmation of E. coli
PCR-based genotypic confirmation of the isolates revealed 662 bp bands; all were confirmed to be E. coli.
Antibiotic susceptibility test
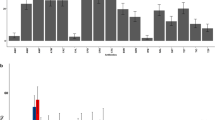
Disk diffusion-based phenotypic antibiotic susceptibility test revealed that 63% (63/100) of E. coli isolates were resistant to at least one antibiotic (≥ 1 antibiotic), and 29% (29/100) were resistant to three or more antibiotic groups (MDR). Out of 100 E. coli isolates, 52 (52%), 38 (38%), 37 (37%), 28 (28%), 21 (21%), 19 (19%), and 13 (13%) isolates were found to be resistant to tetracycline, kanamycin, streptomycin, ampicillin, chloramphenicol, trimethoprim/sulfamethoxazole, and gentamicin, respectively. Moreover, 12% of the isolates were resistant to enrofloxacin and ciprofloxacin. However, all isolates (100%) were found to be susceptible to cephalothin.
Looking at the bird species-based results, in pigeon origin isolates, tetracycline (72%) and kanamycin (48%) resistance were comparatively higher, whereas, in gull origin isolates, streptomycin (34%) and trimethoprim/sulfamethoxazole (32%) resistance was found to be relatively higher (Table 3). In terms of carrying isolates that were resistant to at least one antibiotic, a statistically significant (p < 0.001, χ2 = 18.9) difference was found between pigeons (42/50) and gull (21/50) origin isolates. However, in terms of carriage of multidrug-resistant isolates, a statistically non-significant (p = 0.50, χ2 = 0.43) higher MDR rate was found in gull isolates (16/50) than in pigeon isolates (13/50). Nine gull-origin isolates showed resistance to all antibiotics tested except cephalothin. The tetracycline, kanamycin, and streptomycin were detected in seven isolates of pigeon origin (Fig. 1).
Antibiotic resistance genes
Forty-six (46%) of E. coli isolates were found to carry one or more of the sulfonamide resistance genes investigated. sul1(16%), sul2 (26%), and sul3 (34%) were detected in both gull and pigeon origin isolates (Figs. 2 and 3). Most sulfonamide resistance genes were found in combination. For example, sul1, sul2, and sul3 (in 10 isolates), sul2 and sul3 (in 6 isolates), and sul1 and sul2 (in 4 isolates) genes were detected together. Comparing the two bird species sul1 gene’s percentage was found to be significantly higher (p = 0.006) in gull isolates (26%) than in pigeon isolates (6%). In contrast, the percentage of the sul3 gene was significantly (p < 0.001) higher in isolates of pigeon-origin (52%) (Table 4). Regarding the tetracycline resistance determinants, tet(A) and tet(B) were detected in 63% and 66% of the E. coli isolates (Figs. 4 and 5). The percentage of tet(A) gene was higher than tet(B) gene in pigeon isolates. On the other hand, the tet(B) gene percentage was higher than the tet(A) gene in gull isolates. Comparing the two bird species, a statistically significantly (p = 0.001) higher percentage of tet(A) gene was detected in the pigeon isolates. Among the aminoglycoside resistance determinants, strA/strB (24%) and aphA1 (48%) were detected in both gull and pigeon isolates (Fig. 6). The percentage of strA/strB gene harborage in gull isolates (42%) was found to be significantly higher (p < 0.001) than in pigeon isolates. On the other hand, tet(C) gene, PMQR gene determinants (qnr(A), qnr(B) and qnr(s)), and some aminoglycoside modifying enzyme determinants (aphA2, aadB and aac(3) IV) were not found in any of the isolates. Figure 7 shows the negative results obtained after subjecting PCR amplicons to agarose gel electrophoresis to determine plasmid-mediated quinolone resistance determinants.
Identification of sul1 (433 bp), sul2 (721 bp), and sul3 (244 bp) genes causing sulfonamide resistance by mPCR (M, marker (Thermo Scientific, SM0241, 100 bp DNA Ladder); P, positive control; N, negative control; 1, 4, 8, 9, 12, sul1, sul2, and sul3), 2 and 7, sul 2 and sul 3; 3 and 6, sul3; 5 and 10, sul2; 11, sul1 and sul2)
The association between phenotypic and genotypic resistance profile
Looking at the correlation between phenotypic and genotypic resistance profiles, 43.24% (16/37) of the streptomycin resistance isolates carried strA/strB gene, and the carriage of this gene was significantly (p = 0.001, χ2 = 11.92) associated with the streptomycin phenotypic resistance. Similarly, a significant association (p < 0.001, χ2 = 27.68) was found between aphA1 gene carriage and kanamycin resistance, which was found in 81.57% (31/38) of kanamycin-resistant isolates. In addition, 9 E. coli isolates that showed intermediate phenotypic resistance to kanamycin were found to carry the aphA1 gene. Thus, of the 48 aphA1 genes identified, 31 were found in kanamycin-resistant isolates, while the others were found in isolates that were intermediately resistant (9) and susceptible (8) to kanamycin.
Regarding the relationship between tetracycline resistance and its genetic resistance determinants, tet(A) and tet(B) carriage was detected in 84.61% (44/52) and 76.92% (40/52) of tetracycline-resistant isolates, respectively. The carriage of these genes was also found to be significantly (p < 0.05) associated with tetracycline phenotypic resistance. All E. coli isolates phenotypically resistant to tetracycline had tet(A), tet(B), or both. Moreover, 47.3% (9/19), 68.4% (13/19), and 36.8% (7/19) of the isolates phenotypically resistant to trimethoprim/sulfamethoxazole were found to carry sul1, sul2, and sul3 genes, respectively. Despite the phenotypic resistance expression (e.g., fluoroquinolone and gentamicin), resistance genes were not detected in some E. coli isolates. On the other hand, some phenotypically susceptible isolates were also found to carry resistance genes (Table 5).
Genotyping of isolates by RAPD-PCR
Dendrogram analysis revealed that the pigeon-origin E. coli strains had similarities ranging from 47 to 100%, and isolates’ genotyping based on a 70% similarity threshold resulted in two single (RGA, RGB) and three multiple (RGC, RGD, RGE) genotypes (Figs. 8 and 9). Among the multiple genotypes, RGC contained 3 isolates, RGD 5 and RGE 31 isolates. In the case of gull isolate, similarities ranging from 40 to 100% were found. Genotyping of gull isolates revealed four single (RMA, RMB, RMC, RMD) and one multiple (RME) genotypes based on the 70% similarity threshold. RME contained 41 isolates (Figs. 10 and 11).
Discussion
Antibiotic resistance is a growing global challenge in both animal and human health that requires multisectoral action plans [12, 41]. A recent study by Murray et al. [14] attributed more than 1.2 million global deaths in 2019 to antimicrobial-resistant bacteria. Thus, microbial resistance to drugs of significant clinical importance is considered one of the biggest global public health threats challenging humans today [41], and it is estimated that if appropriate measures are not taken, its burden may increase and cause approximately 10 million deaths annually by 2050 [42]. Therefore, it is clear that a better understanding of antibiotic resistance situations, including resistance gene sources and vectors, is needed to reduce the potential risks associated with the increasing burden of antibiotic resistance [27, 31].
This study investigated the phenotypic and genotypic antibiotic resistance patterns of E. coli isolates isolated from gulls and pigeons. The results of the phenotypic antibiotic resistance test revealed higher antibiotic resistance to tetracycline (52%), kanamycin (38%), and streptomycin (37%). Of 100 investigated E. coli isolates, 63% were resistant to ≥ 1 antibiotic and 29% were found to be multidrug-resistant. A similar result was reported by Nowaczek et al. [52], who reported a 31.2% multidrug-resistant rate. Other researchers from Lithuania also investigated the resistance profile of E. coli obtained from various wild birds (including gulls) and reported a 33.5% MDR rate [17]. However, there are also studies reporting a high MDR rate ranging from 60 to 100% [3, 43,44,45,46] and lower MDR rates (below 20%) [47,48,49,50,51] from different parts of the world. These differences may be due to differences in bird species, other hosts with which they are in close contact, and geographical location.
Previous studies have reported a relatively high degree of resistance to tetracycline in wild bird-origin E. coli isolates from different countries [19, 25, 52,53,54]. Looking at the phenotypic resistance profiles of the isolates investigated in this study, a relatively higher degree of resistance to tetracycline (52%) was detected. This result was in agreement with recent studies reporting tetracycline resistance in E. coli isolates isolated from different wild bird species in Poland (50%), Australia (51%), Brazil (52.6%), and Italy (56%) [22, 25, 52, 55]. On the other hand, the rate of tetracycline resistance found in our study tended to be higher when compared with the results reported by Carroll et al. [50] (5.4%), Horn et al. [51] (10.91%), and Stedt et al. [28] (19%). However, other studies conducted in different parts of the world have reported high rates of tetracycline resistance, ranging from 80 to 100%, in a wide variety of wild bird species [18, 45, 46, 53]. The tetracycline group is of critical importance in the treatment of many infections in animals [12] and is also classified under “highly important antimicrobials” in the treatment of human infections [13]. The relatively high rate of tetracycline resistance detected the current study could be attributed to its widespread use in treating animal infections [56].
Resistance gene determination results revealed a significant association of tet(A) and tet(B) genes carriage with tetracycline resistance. Compared with tet(B), the percentage of tet(A) gene’s carriage was found to be relatively higher in pigeon isolates, whereas the percentage of tet(B) gene was found to be higher in gull isolates. In overall results, unlike previous studies [18, 19, 57], the percentage of the tet(B) (66%) gene was found to be relatively higher than the tet(A) (63%). However, in agreement with our findings, other researchers have reported a higher percentage of tet(B) gene than tet(A) [37, 58]. Of tetracycline-resistant isolates, 84.61% and 76.92% carried tet(A) and tet(B), respectively. In one study conducted in the Czech Republic, 55.1% (27/49) and 44.89% (22/49) of the black-headed gull (Larus michahellis) origin E. coli isolates resistant to tetracycline were reported to carry the tet(A) and tet(B) gene, respectively [48]. Another study by Merkeviciene et al. [17] reported that 85% and 18% of tetracycline-resistant E. coli isolates had tet(A) and tet(B) genes, respectively.
Looking at the aminoglycoside’s resistance profile, the streptomycin rate (37%) found in this study was comparable with a study from Australia that reported 34% resistance to this drug [59]. However, a higher resistance rate (ranging from 48 to 85%) [3, 43, 53,54,55] and a lower resistance rate (3% and 9.3%) [25, 48] have been reported in previous studies. Borges et al. (2017) reported 36.8% kanamycin resistance in Brazil, which agrees with a kanamycin resistance rate (38%) detected in this study. Moreover, the study by Barguigua et al. (2019) reported that 35% of E. coli isolates obtained from gulls were resistant to kanamycin. The kanamycin resistance rate found in this study was relatively higher when compared with the result of one study from Poland (18%) [52]. However, a higher kanamycin resistance rate (80.4%) has been reported in Turkey [30]. Among the aminoglycoside resistance genes investigated in this study, aphA1 and strA/strB were found in 48% and 24% of the E. coli isolates, respectively. Moreover, it was determined that 81.57% of the kanamycin-resistant and 43.24% of the streptomycin-resistant E. coli isolates carried aphA1 and strA/strB genes, respectively. Other researchers have also reported comparable results [17, 52].
The percentage of ampicillin-resistant isolates detected in the current study (28%) was in agreement with the result of Nowaczek et al. [52], who reported 28.1% resistance to ampicillin in wild bird origin E. coli isolates. Similarly, Hasan et al. [49] reported a 29.4% ampicillin resistance in E. coli isolates isolated from gulls (Chroicocephalus brunnicephalus) in Bangladesh. On the other hand, other researchers investigating the antibiotic resistance status in wild bird-origin E. coli isolates have reported a higher ampicillin resistance ranging from 70 to 100% [3, 53, 55, 60] and lower ampicillin resistance rate (less than 20%) [19, 48].
A recent study from Malaysia reported 16.7% trimethoprim/sulfamethoxazole resistance, which is consistent with the finding of this study (19%). A relatively lower trimethoprim/sulfamethoxazole resistance rate was found in pigeon-origin isolates (6%) compared to gull isolates (32%). Comparable findings were reported from Brazil (3.9% in pigeon origin E. coli isolates) [61] and Italy (26.6% in gull origin E. coli isolates) [25]. sul1, sul2, and sul3 genes, which cause sulfonamide resistance, were detected in isolates isolated from gulls and pigeons. Other studies conducted in China [19] and Lithuania [17] have also reported the commonness of these genes in E. coli isolates isolated from wild birds. Furthermore, 96% of trimethoprim/sulfamethoxazole-resistant E. coli isolates were found to carry one or more sul genes. Comparable results were reported in a study from Portugal and Slovakia [54, 62].
Regarding the chloramphenicol resistance, 21% of the isolates were found to be resistant to this antibiotic, with 24% of the gull-origin isolates and 18% of the pigeon-origin isolates being chloramphenicol resistant. WHO classified chloramphenicol under “highly important antimicrobials” in human health [13]. The use of chloramphenicol in food animals is prohibited in many countries [56], including Turkey [63]. In this case, less exposure of wild birds to this antibiotic is expected due to lower levels of antibiotic residues in the environment [64]. Despite this, resistance to this antibiotic has been reported in E. coli isolates isolated from various animal species, including cattle [56, 65]. The possible reason for this could be the persistence of antibiotic-resistant bacteria in the environment [66]. Even though the result obtained in this study tends to be higher when compared with the result of the study from Italy (0%) [25], Czech Republic (1.9%) [48], Bangladesh (2.4%) [49], Poland (6.25%) [52], and China (11.1%) [19], a comparable finding has been reported from Singapore (19.2%) [64]. Moreover, higher resistance to this antibiotic has been reported in Bangladesh (43.64%) [3] and Portugal (41.7%) [54].
Gentamicin and ciprofloxacin are antibiotics of critical importance in treating human infections [13]. Resistance to fluoroquinolones (12%) and to gentamicin (13%) was detected at a lower rate. Enrofloxacin, gentamicin, and ciprofloxacin resistance were not detected in any of the pigeon-origin E. coli isolates. Interestingly, these results were found to be consistent with one study that analyzed the antibiotic susceptibility profile of E. coli isolates obtained from domestic pigeons (Columba livia domestica) in Turkey and reported 100% susceptibility to enrofloxacin and gentamicin [67]. Furthermore, Silva et al. [62] reported 100% gentamicin susceptibility of pigeon-origin E. coli isolates, which is consistent with the findings of this study. Ong et al. [65] reported 11.5% resistance against ciprofloxacin. Russo et al. [25] also reported 13.3% and 16.6% resistance to gentamicin and enrofloxacin, respectively. In contrast to this study’s findings, one study from Egypt reported resistance to ciprofloxacin and gentamicin at 88% and 80%, respectively [43].
Although a statistically significant association between phenotypic and genotypic antibiotic resistance was found in most cases, isolates showing phenotypic resistance without corresponding resistance genes and isolating carrying resistance genes without phenotypic resistance were also encountered. For example, all tetracycline-resistant isolates were found to carry tet(A), tet(B), or both and harboring these genes was found to be significantly (p < 0.05) associated with tetracycline phenotypic resistance. Similar results were obtained during the analysis of the association between streptomycin resistance and strA/strB gene carriage and kanamycin resistance and harborage of the aphA1 gene. On the other hand, despite the isolates’ phenotypic resistance to fluoroquinolones and gentamicin, the genes responsible for the resistance of these antibiotics could not be detected in any of the resistant isolates, which indicates as phenotypic and genotypic antibiotic resistance may not always be consistent. The same situation has been reported in previous studies [68, 69].
The possible explanation for the detection of antibiotic resistance genes in isolates without phenotypic antibiotic resistance is that the detected genes may not be expressed or non-functional [68, 70]. On the other hand, the absence of investigated resistance genes in phenotypically resistant isolates could be explained by the presence of alternative resistance mechanisms other than those investigated. For example, E. coli can develop resistance to fluoroquinolones through chromosomal mutations of gyrA and parC genes, which encode quinolone targets (DNA gyrase and topoisomerase). This resistance mechanism is vertically transmitted and is commonly known to confer high levels of resistance to quinolones, in contrast to horizontally transmitted plasmid-mediated quinolone resistance (like qnr) that confers low levels of quinolone resistance [71, 72].
RAPD-PCR is an effective method to assess the genetic diversity of many bacterial species [73] and is also being used in evaluating the genotypic relationships of wild bird origin E. coli isolates [32, 74]. Although the origin or source of the pathogen was not determined in this study, phylogenetic analysis based on RAPD-PCR results revealed genetic similarity ranging from 47 to 100% in pigeon isolates and 40 to 100% in gull isolates. The strains with high similarities (> 90%) may have been acquired from the same sources. However, this needs to be elucidated using more advanced typing techniques.
Antibiotic-resistant bacteria in wild birds are often associated with environmental contamination from animal and human wastes rather than direct exposure to antibiotics [21, 75, 76]. In particular, the level of anthropogenic impact (e.g., wastewater, waste from livestock farms, landfill) in a given area can affect the likelihood of wild birds becoming infected by antibiotic-resistant bacteria [77]. Factors such as urbanization and the loss of natural wildlife habitats, which increases the contact of birds with contaminated environments, aggravate this situation by facilitating the infection of birds with antibiotic-resistant bacteria [22]. Thus, the difference in antibiotic resistance percentage detected in this study and other studies may be attributed to the level of access of wild animals to anthropogenic sources, which may vary from country to country and even in different places in the same country, and bird’s foraging strategies that may differ according to bird species [78]. On the other hand, the low resistance percentage detected in this study (enrofloxacin, ciprofloxacin, and gentamicin resistance) and the susceptibility of all isolates to cephalothin may indicate that these bird species had less or no contact with environments contaminated with bacteria resistant to these antibiotics.
Conclusions
This study revealed that wild birds (gulls and pigeons) carry strains of E. coli that are resistant to even drugs of critical importance in human health. In addition, the isolates were found to carry resistance genes like tet (A) and tet(B), strA/strB, aphA1, sul1, sul2, and sul3, which provide resistance to drugs that have an essential role in human and veterinary medicine. These results suggest that wild birds may serve as a reservoir for multidrug-resistant bacteria and resistance genes. Given the increasing threat of antibiotic resistance in both human and animal health, the detection of MDR bacteria carrying antibiotic resistance genes from free-living wild animals deserve more attention. Specially gulls and pigeons, which are in close contact with humans, may pose a public health risk by contributing to the spread of resistant bacteria through fecal contamination of the environment. Therefore, multisectoral collaboration under the “One Health” umbrella is of critical importance in addressing antibiotic resistance challenges. Continuous monitoring of the possible antibiotic resistance reservoirs, appropriate drug prescription, awareness creation, and searching for alternative treatment options is paramount. Although this study has identified the carriage of multidrug resistance with resistance genes in E. coli isolates of gull and pigeon origin, further molecular studies are warranted to elucidate the source of infection for wild birds and possible interspecies transmission of resistant bacteria.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author on reasonable request.
References
Gargiulo A, Fioretti A, Russo TP, Varriale L, Rampa L, Paone S, et al. Occurrence of enteropathogenic bacteria in birds of prey in Italy. Lett Appl Microbiol. 2018;66:202–6. https://doi.org/10.1111/lam.12836.
Dipineto L, Russo TP, Gargiulo A, Borrelli L, De Luca Bossa LM, Santaniello A, et al. Prevalence of enteropathogenic bacteria in common quail (Coturnix coturnix). Avian Pathol. 2014;43:498–500. https://doi.org/10.1080/03079457.2014.966055.
Islam MS, Nayeem MMH, Sobur MA, Ievy S, Islam MA, Rahman S, et al. Virulence determinants and multidrug resistance of escherichia coli isolated from migratory birds. Antibiotics. 2021;10:190. https://doi.org/10.3390/antibiotics10020190.
Chung DM, Ferree E, Simon DM, Yeh PJ. Patterns of bird-bacteria associations. Ecohealth. 2018;15:627–41. https://doi.org/10.1007/s10393-018-1342-5.
Gülhan T, Boynukara B, Durmuş A, Kiziroǧlu I, Sancak YC. Enteric bacteria and some pathogenic properties of Enterococcus faecalis, Enterococcus faecium and Escherichia coli strains isolated from wild ducks and gulls. Fresenius Environ Bull. 2012;21:1961–6.
Gülhan T. Determination of biochemical, enterotoxigenic and verotoxigenic properties of Escherichia coli strains isolated from faeces of healthy animals. Yüzüncü Yıl Üniversitesi Vet Fakültesi Derg. 2003;14:102–9.
EFSA, ECDC. The European Union One Health 2019 Zoonoses Report. EFSA J 2021;19:6406.https://doi.org/10.2903/j.efsa.2021.6406
Ahmed NA, Gulhan T. Campylobacterin wild birds: ıs ıt an animal and public health concern? Front Microbiol. 2022;12:4180. https://doi.org/10.3389/FMICB.2021.812591/BIBTEX.
Allocati N, Masulli M, Alexeyev MF, Di Ilio C. Escherichia coli in Europe: an overview. Int J Environ Res Public Health. 2013;10:6235–54. https://doi.org/10.3390/ijerph10126235.
Poirel L, Madec J-Y, Lupo A, Schink A-K, Kieffer N, Nordmann P, et al. Antimicrobial resistance in Escherichia coli. Microbiol Spectr 2018;6:ARBA-0026–2017. https://doi.org/10.1128/microbiolspec.arba-0026-2017.
Radimersky T, Frolkova P, Janoszowska D, Dolejska M, Svec P, Roubalova E, et al. Antibiotic resistance in faecal bacteria (Escherichia coli, Enterococcus spp.) in feral pigeons. J Appl Microbiol 2010;109:1687–95. https://doi.org/10.1111/j.1365-2672.2010.04797.x.
OIE. OIE List of Antimicrobial Agents of Veterinary Importance. 2019. https://www.woah.org/fileadmin/Home/eng/Our_scientific_expertise/docs/pdf/AMR/A_OIE_List_antimicrobials_July2019.pdf.
WHO. Critically ımportant antimicrobials for human medicine: ranking of medically important antimicrobials for risk management of antimicrobial resistance due to non-human use. 6th ed. Geneva: World Health Organization; 2019. https://www.who.int/publications/i/item/9789241515528.
Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399:629–55. https://doi.org/10.1016/S0140-6736(21)02724-0.
Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–81. https://doi.org/10.1111/j.1469-0691.2011.03570.x.
Pulingam T, Parumasivam T, Gazzali AM, Sulaiman AM, Chee JY, Lakshmanan M, et al. Antimicrobial resistance: prevalence, economic burden, mechanisms of resistance and strategies to overcome. Eur J Pharm Sci. 2022;170: 106103. https://doi.org/10.1016/j.ejps.2021.106103.
Merkeviciene L, Klimiene I, Siugzdiniene R, Virgailis M, Mockeliunas R, Ruzauskas M. Prevalence and molecular characteristics of multi-resistant Escherichia coli in wild birds. Acta Vet Brno. 2018;87:9–17. https://doi.org/10.2754/avb201887010009.
Hleba L, Hlebová M, Kováčik A, Šmehýl P, Hricáková N, Petrová J, et al. Escherichia coli as a carrier of tetracyclines and penicillins resistance in wild pheasant (Phasianus colchicus). J Environ Sci Heal - Part A Toxic/Hazardous Subst Environ Eng. 2020;55:1201–9. https://doi.org/10.1080/10934529.2020.1777050.
Yuan Y, Liang B, Jiang B-W, Zhu L-W, Wang T-C, Li Y-G, et al. Migratory wild birds carrying multidrug-resistant Escherichia coli as potential transmitters of antimicrobial resistance in China. PLoS ONE. 2021;16:e0261444. https://doi.org/10.1371/journal.pone.0261444.
Plaza-Rodríguez C, Alt K, Grobbel M, Hammerl JA, Irrgang A, Szabo I, et al. Wildlife as sentinels of antimicrobial resistance in Germany? Front Vet Sci. 2021;7:627821. https://doi.org/10.3389/fvets.2020.627821.
Fashae K, Engelmann I, Monecke S, Braun SD, Ehricht R. Molecular characterisation of extended-spectrum ß-lactamase producing Escherichia coli in wild birds and cattle, Ibadan. Nigeria BMC Vet Res. 2021;17:33. https://doi.org/10.1186/s12917-020-02734-4.
Borges CA, Beraldo LG, Maluta RP, Cardozo MV, Barboza KB, Guastalli EAL, et al. Multidrug-resistant pathogenic Escherichia coli isolated from wild birds in a veterinary hospital. Avian Pathol. 2017;46:76–83. https://doi.org/10.1080/03079457.2016.1209298.
Varela AR, Manageiro V, Ferreira E, Guimarães MA, Da Costa PM, Caniça M, et al. Molecular evidence of the close relatedness of clinical, gull and wastewater isolates of quinolone-resistant Escherichia coli. J Glob Antimicrob Resist. 2015;3:286–9. https://doi.org/10.1016/j.jgar.2015.07.008.
Bonnedahl J, Drobni M, Gauthier-Clerc M, Hernandez J, Granholm S, Kayser Y, et al. Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the south of France. PLoS One. 2009;4:e5958. https://doi.org/10.1371/journal.pone.0005958.
Russo TP, Pace A, Varriale L, Borrelli L, Gargiulo A, Pompameo M, et al. Prevalence and antimicrobial resistance of enteropathogenic bacteria in yellow-legged gulls (Larus michahellis) in southern Italy. Animals. 2021;11:275. https://doi.org/10.3390/ani11020275.
Velhner M, Todorović D, Novović K, Jovčić B, Lazić G, Kojić M, et al. Characterization of antibiotic resistance in Escherichia coli isolates from Black-headed gulls (Larus ridibundus) present in the city of Novi Sad. Serbia Vet Res Commun. 2021;45:199–209. https://doi.org/10.1007/s11259-021-09801-7.
Skarżyńska M, Zaja̧c M, Bomba A, Bocian Ł, Kozdruń W, Polak M, et al. Antimicrobial resistance glides in the sky—free-living birds as a reservoir of resistant Escherichia coli with zoonotic potential. Front Microbiol 2021;12:656223. https://doi.org/10.3389/fmicb.2021.656223.
Stedt J, Bonnedahl J, Hernandez J, McMahon BJ, Hasan B, Olsen B, et al. Antibiotic resistance patterns in Escherichia coli from gulls in nine European countries. Infect Ecol Epidemiol. 2014;4:21565. https://doi.org/10.3402/iee.v4.21565.
Gülhan T, Boynukara B, Ilhan Z. Memeli ve kanatlı hayvanlardan izole edilen Escherichia coli suşlarının Antibiyotiklere Duyarlılıkları. Vet Hekimleri Mikrobiyoloji Derg. 2003;3:39–44.
Yapicier OŞ, Kandir EH, Öztürk D. Antimicrobial resistance of E. coli and Salmonella isolated from wild birds in a rehabilitation center in Turkey. Arch Razi Inst 2022;77:239–49. https://doi.org/10.22092/ARI.2021.356322.1823.
Zhao H, Sun R, Yu P, Alvarez PJJ. High levels of antibiotic resistance genes and opportunistic pathogenic bacteria indicators in urban wild bird feces. Environ Pollut. 2020;266:115200. https://doi.org/10.1016/j.envpol.2020.115200.
Fadel HM, Afifi R, Al-Qabili DM. Characterization and zoonotic impact of Shiga toxin producing Escherichia coli in some wild bird species. Vet World 2017;10:1118–28. https://doi.org/10.14202/vetworld.2017.1118-1128.
Abd El-Razik KA, Abdelrahman KA, Ahmed YF, Gomaa AM, Eldebaky HA. Direct ıdentification of major pathogens of the bubaline subclinical mastitis in Egypt using PCR. J Am Sci. 2010;6:652–60.
CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. 25th ed. Wayne: Clinical and Laboratory Standards Institute; 2015.
CLSI. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. 4th ed. Wayne: Clinical and Laboratory Standards Institute; 2018.
CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI Supplement M100, Wayne: Clinical and Laboratory Standards Institute; 2020.
Kozak GK, Boerlin P, Janecko N, Reid-Smith RJ, Jardine C. Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario. Canada Appl Environ Microbiol. 2009;75:559. https://doi.org/10.1128/AEM.01821-08.
Cattoir V, Poirel L, Rotimi V, Soussy C-J, Nordmann P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother. 2007;60:394–7. https://doi.org/10.1093/JAC/DKM204.
Pławińska-Czarnak J, Wódz K, Piechowicz L, Tokarska-Pietrzak E, Bełkot Z, Bogdan J, et al. Wild duck (Anas platyrhynchos) as a source of antibiotic-resistant Salmonella enterica subsp. diarizonae O58—the first report in Poland. Antibiotics 2022;11:530. https://doi.org/10.3390/antibiotics11040530.
Versalovic J, Koeuth T, Lupski R. Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial enomes. Nucleic Acids Res. 1991;19:6823–31. https://doi.org/10.1093/NAR/19.24.6823.
WHO. Antimicrobial resistance 2021. https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance. Accessed 31 Dec 2021.
O’Neill J. Tackling drug-resistant infections globally: final report and recommendations: the review on antimicrobial resistance. 2016. https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf.
Tawakol M, Younes A. Role of migratory birds in transmission of E. coli ınfection to commercial poultry. Alexandria J Vet Sci 2019;62:72–81. https://doi.org/10.5455/ajvs.57309.
Atterby C, Börjesson S, Ny S, Järhult JD, Byfors S, Bonnedahl J. ESBL-producing Escherichia coli in Swedish gulls—a case of environmental pollution from humans? PLoS One. 2017;12:e0190380. https://doi.org/10.1371/journal.pone.0190380.
Mohamed-Yousif IM, Abu J, Abdul-Aziz S, Zakaria Z, Rashid A, Awad EA. Occurrence of antibiotic resistant C. Jejuni and E. Coli in wild birds, chickens, environment and humans from orang asli villages in sungai siput, Perak, Malaysia. Am J Anim Vet Sci 2019;14:158–69. https://doi.org/10.3844/ajavsp.2019.158.169.
Barguigua A, Rguibi Idrissi H, nayme K, Timinouni M. Virulence and antibiotic resistance patterns in E. coli, Morocco. Ecohealth 2019;16:570–5. https://doi.org/10.1007/S10393-019-01404-8.
Elsohaby I, Samy A, Elmoslemany A, Alorabi M, Alkafafy M, Aldoweriej A, et al. Migratory wild birds as a potential disseminator of antimicrobial-resistant bacteria around al-asfar lake, eastern saudi arabia. Antibiotics. 2021;10:260. https://doi.org/10.3390/antibiotics10030260.
Dolejska M, Cizek A, Literak I. High prevalence of antimicrobial-resistant genes and integrons in Escherichia coli isolates from Black-headed gulls in the Czech Republic. J Appl Microbiol. 2007;103:11–9. https://doi.org/10.1111/J.1365-2672.2006.03241.X.
Hasan B, Melhus Å, Sandegren L, Alam M, Olsen B. The gull (Chroicocephalus brunnicephalus) as an environmental bioindicator and reservoir for antibiotic resistance on the coastlines of the bay of Bengal. Microb Drug Resist. 2014;20:466–71. https://doi.org/10.1089/mdr.2013.0233.
Carroll D, Wang J, Fanning S, Mcmahon BJ. Antimicrobial resistance in wildlife: ımplications for public health. Zoonoses Public Health. 2015;62:534–42. https://doi.org/10.1111/zph.12182.
Horn RV, Bezerra WGA, Lopes ES, Teixeira RSC, Silva ING, Bona MD, et al. Antimicrobial susceptibility and diarrheagenic diagnosis of Escherichia coli and Salmonella enterica isolated from feral pigeons (Columba livia) captured in Fortaleza. Brazil Pesqui Vet Bras. 2018;38:2155–8. https://doi.org/10.1590/1678-5150-PVB-5633.
Nowaczek A, Dec M, Stępień-Pyśniak D, Urban-Chmiel R, Marek A, Różański P. Antibiotic resistance and virulence profiles of Escherichia coli strains ısolated from wild birds in Poland. Pathogens. 2021;10:1059. https://doi.org/10.3390/PATHOGENS10081059.
Nabil NM, Erfan AM, Tawakol MM, Haggag NM, Naguib MM, Samy A. Wild birds in live birds markets: potential reservoirs of enzootic avian ınfluenza viruses and antimicrobial resistant Enterobacteriaceae in Northern Egypt. Pathogens. 2020;9:196. https://doi.org/10.3390/PATHOGENS9030196.
Radhouani H, Poeta P, Gonçalves A, Pacheco R, Sargo R, Igrejas G. Wild birds as biological indicators of environmental pollution: antimicrobial resistance patterns of Escherichia coli and Enterococci isolated from common buzzards (Buteo buteo). J Med Microbiol. 2012;61:837–43. https://doi.org/10.1099/jmm.0.038364-0.
Mukerji S, Stegger M, Truswell AV, Laird T, Jordan D, Abraham RJ, et al. Resistance to critically important antimicrobials in Australian silver gulls (Chroicocephalus novaehollandiae) and evidence of anthropogenic origins. J Antimicrob Chemother. 2019;74:2566–74. https://doi.org/10.1093/jac/dkz242.
Marinho CM, Santos T, Gonçalves A, Poeta P, Igrejas G. A decade-long commitment to antimicrobial resistance surveillance in Portugal. Front Microbiol. 2016;7:1650. https://doi.org/10.3389/FMICB.2016.01650/BIBTEX.
Islam MS, Sobur MA, Rahman S, Ballah FM, Ievy S, Siddique MP, et al. Detection of blaTEM, blaCTX-M, blaCMY, and blaSHV genes among extended-spectrum beta-lactamase-producing Escherichia coli ısolated from migratory birds travelling to Bangladesh. Microb Ecol. 2021;83:942–50. https://doi.org/10.1007/s00248-021-01803-x.
Bryan A, Shapir N, Sadowsky MJ. Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains ısolated from diverse human and animal sources. Appl Environ Microbiol. 2004;70:2503. https://doi.org/10.1128/AEM.70.4.2503-2507.2004.
Smith HG, Clarke RH, Larkins JA, Bean DC, Greenhill AR. Wild Australian birds and drug-resistant bacteria: characterisation of antibiotic-resistant Escherichia coli and Enterococcus spp. Emu - Austral Ornithol. 2019;119:384–90. https://doi.org/10.1080/01584197.2019.1591162.
Holko I, Doležalová M, Pavlíčková S, Gal R, Valenta T, Holkova T. Antimicrobial-resistance in Escherichia coli isolated from wild pheasants (Phasianus colchicus). Vet Ital 2019;55:169–72. https://doi.org/10.12834/VetIt.887.4400.3.
Silva VL, Nicoli JR, Nascimento TC, Diniz CG. Diarrheagenic Escherichia coli strains recovered from urban pigeons (columba livia) in Brazil and their antimicrobial susceptibility patterns. Curr Microbiol. 2009;59:302–8. https://doi.org/10.1007/s00284-009-9434-7.
Handrova L, Kmet V. Antibiotic resistance and virulence factors of Escherichia coli from eagles and goshawks. J Environ Sci Heal - Part B. 2019;54:605–14. https://doi.org/10.1080/03601234.2019.1608103.
Aslantaş O, Gövce N. Investigation of antimicrobial resistance in pigeons (Columba livia domestica) using indicator bacteria. J Hell Vet Med Soc 2020;71:2095–106. https://doi.org/10.12681/jhvms.23632.
Ong KH, Khor WC, Quek JY, Low ZX, Arivalan S, Humaidi M, et al. Occurrence and antimicrobial resistance traits of Escherichia coli from wild birds and rodents in Singapore. Int J Environ Res Public Health. 2020;17:5606. https://doi.org/10.3390/ijerph17155606.
Makarov DA, Ivanova OE, Karabanov SY, Gergel MA, Pomazkova A V. Antimicrobial resistance of commensal Escherichia coli from food-producing animals in Russia. Vet World 2020;13:2053–61. https://doi.org/10.14202/vetworld.2020.2053-2061.
Andersson DI, Hughes D. Persistence of antibiotic resistance in bacterial populations. FEMS Microbiol Rev. 2011;35:901–11. https://doi.org/10.1111/J.1574-6976.2011.00289.X.
Aşkar Ş, Sakarya F, Yildirim M. The potential risk in epizootiology of bacterial zoonozis: pigeon (Columba livia domestica) feces. Kafkas Univ Vet Fak Derg. 2011;17:13–6.
Zhang S, Chen S, Rehman MU, Yang H, Yang Z, Wang M, et al. Distribution and association of antimicrobial resistance and virulence traits in Escherichia coli isolates from healthy waterfowls in Hainan. China Ecotoxicol Environ Saf. 2021;220:112317. https://doi.org/10.1016/J.ECOENV.2021.112317.
Rahman MM, Husna A, Elshabrawy HA, Alam J, Runa NY, Badruzzaman ATM, et al. Isolation and molecular characterization of multidrug-resistant Escherichia coli from chicken meat. Sci Rep. 2020;10:21999. https://doi.org/10.1038/s41598-020-78367-2.
Diren Sigirci B, Celik B, Başaran Kahraman B, Bagcigil AF, Ak S. Tetracycline resistance of Enterobacteriaceae ısolated from feces of synanthropic birds. J Exot Pet Med. 2019;28:13–8. https://doi.org/10.1053/J.JEPM.2017.12.003.
Esmaeel NE, Gerges MA, Hosny TA, Ali AR, Gebriel MG. Detection of chromosomal and plasmid-mediated quinolone resistance among Escherichia coli isolated from urinary tract infection cases; Zagazig University Hospitals. Egypt Infect Drug Resist. 2020;13:413–21. https://doi.org/10.2147/IDR.S240013.
Ehwarieme DA, Whiliki OO, Ejukonemu FE. Occurrence of plasmid mediated fluoroquinolone resistance genes amongst enteric bacteria isolated from human and animal sources in Delta state. Nigeria AIMS Microbiol. 2021;7:75–95. https://doi.org/10.3934/microbiol.2021006.
Li W, Raoult D, Fournier PE. Bacterial strain typing in the genomic era. FEMS Microbiol Rev. 2009;33:892–916. https://doi.org/10.1111/j.1574-6976.2009.00182.x.
Awadallah MAI, Merwad AM, Mohamed RE. Prevalence of zoonotic Escherichia coli and Salmonellae in wild birds and humans in Egypt with emphasis on RAPD-PCR fingerprinting of E. coli. Glob Vet. 2013;11:781–8.
Ahlstrom CA, van Toor ML, Woksepp H, Chandler JC, Reed JA, Reeves AB, et al. Evidence for continental-scale dispersal of antimicrobial resistant bacteria by landfill-foraging gulls. Sci Total Environ. 2021;764:144551. https://doi.org/10.1016/j.scitotenv.2020.144551.
Ahlstrom CA, Bonnedahl J, Woksepp H, Hernandez J, Reed JA, Tibbitts L, et al. Satellite tracking of gulls and genomic characterization of faecal bacteria reveals environmentally mediated acquisition and dispersal of antimicrobial-resistant Escherichia coli on the Kenai Peninsula. Alaska Mol Ecol. 2019;28:2531–45. https://doi.org/10.1111/MEC.15101.
Dolejska M, Literak I. Wildlife is overlooked in the epidemiology of medically important antibiotic-resistant bacteria. Antimicrob Agents Chemother 2019;63. https://doi.org/10.1128/AAC.01167-19.
Ramey AM, Ahlstrom CA. Antibiotic resistant bacteria in wildlife: perspectives on trends, acquisition and dissemination, data gaps, and future directions. J Wildl Dis. 2020;56:1–15. https://doi.org/10.7589/2019-04-099.
Acknowledgements
The authors thank Prof. Dr. Alper Çiftci and Merve Gizem Sezener (research assistant) for their invaluable technical support. Moreover, we are grateful to Associate Prof. Dr. Yeliz Tanriverdi Çayci for providing positive controls for qnr genes. NAA is a recipient of the Turkiye Scholarship and thus thanks the Presidency of Turks Abroad and Related Communities (YTB).
Funding
This study was financially supported by Ondokuz Mayis University (project number: PYO VET. 1904.21.024).
Author information
Authors and Affiliations
Contributions
NAA: conceptualization and designing of the study protocols, laboratory works, writing, and analysis. TG: conceptualization and designing of the study protocols, funding acquisition, supervision, laboratory works, and reviewing and editing.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ahmed, N.A., Gulhan, T. Determination of antibiotic resistance patterns and genotypes of Escherichia coli isolated from wild birds. Microbiome 12, 8 (2024). https://doi.org/10.1186/s40168-023-01729-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40168-023-01729-1