Abstract
Background
Hesperidin is a citrus flavonoid with anti-inflammatory and antioxidant potential. However, its protective effects on bovine mammary epithelial cells (bMECs) exposed to oxidative stress have not been elucidated.
Results
In this study, we investigated the effects of hesperidin on H2O2-induced oxidative stress in bMECs and the underlying molecular mechanism. We found that hesperidin attenuated H2O2-induced cell damage by reducing reactive oxygen species (ROS) and malondialdehyde (MDA) levels, increasing catalase (CAT) activity, and improving cell proliferation and mitochondrial membrane potential. Moreover, hesperidin activated the Keap1/Nrf2/ARE signaling pathway by inducing the nuclear translocation of Nrf2 and the expression of its downstream genes NQO1 and HO-1, which are antioxidant enzymes involved in ROS scavenging and cellular redox balance. The protective effects of hesperidin were blocked by the Nrf2 inhibitor ML385, indicating that they were Nrf2 dependent.
Conclusions
Our results suggest that hesperidin could protect bMECs from oxidative stress injury by activating the Nrf2 signaling pathway, suggesting that hesperidin as a natural antioxidant has positive potential as a feed additive or plant drug to promote the health benefits of bovine mammary.
Graphical Abstract

Similar content being viewed by others
Introduction
Reactive oxygen species (ROS) are highly reactive and unstable molecules generated as byproducts of cellular metabolism. They can damage cellular components such as lipids, proteins, and DNA, and disrupt cellular homeostasis [1]. Bovine mammary epithelial cells (bMECs) are the main functional cells in the mammary gland that synthesize and secrete milk. ROS production is increased in bMECs during lactation due to their high metabolic rate and oxygen demand [2]. Moreover, various stress factors can also induce excessive ROS generation, such as bacterial infection, heat stress, or negative energy balance [3]. Excessive ROS can trigger cell death pathways (such as apoptosis and necrosis) and impair mammary gland function and integrity [4]. Apoptosis and oxidative stress in bMECs detrimentally affect the health and productivity of dairy cows by reducing milk yield and quality, increasing susceptibility to mastitis, and impairing reproductive performance [5].
Plant extracts, such as flavonoids, phenolic acids, terpenoids, and alkaloids, have antioxidant, anti-inflammatory, anti-apoptotic, and free radical scavenging properties [6,7,8]. Hesperidin is a flavonoid abundantly present in citrus fruits, such as oranges and lemons. It has been reported to have anti-inflammatory, anti-apoptotic, free radical scavenging, cholesterol-lowering, anti-allergic, anti-hypertensive, anti-carcinogenic, and anti-viral effects on various cells [9,10,11]. However, the protective effects of hesperidin on bMECs oxidative stress have not yet been elucidated. Previous studies have reported that hesperidin can modulate the expression of genes related to apoptosis and antioxidant response [12, 13]. For example, hesperidin alleviated oxidative stress, inflammation, and apoptosis in rat neurotoxicity or mice myocardial ischemia by regulating the B-cell lymphoma 2 (Bcl-2)/Bcl-2-associated X protein (Bax)-cysteine-aspartic acid protease 3 (caspase-3) [14] or the nuclear factor erythroid 2-related factor 2 (Nrf2) pathway [15]. We previously reported that hesperidin can inhibit hydrogen peroxide (H2O2)-induced apoptosis of bMECs by modulating the Bcl-2/Bax-caspase-3 pathway [16]. Therefore, we hypothesized that hesperidin could prevent oxidative damage in bMECs by regulating the Nrf2 pathway. The Nrf2 pathway is a major antioxidant response pathway that requires the activation of Nrf2, a transcription factor that regulates the expression of certain antioxidant enzymes, such as heme oxygenase-1 (HO-1) and NAD(P)H quinone oxidoreductase 1 (NQO1) [17].
The aim of this study was to investigate the protective effects of hesperidin on bMECs with oxidative damage caused by H2O2, a common agent for inducing oxidative stress in vitro that is widely used to establish oxidative stress models. This study could provide new insights into the pathogenesis of bovine mastitis associated with oxidative stress and the potential application of hesperidin as a natural antioxidant for bovine mammary health.
Materials and methods
Culture of bovine mammary epithelial cells
The bMECs were primary epithelial cells that were isolated from mammary tissue of lactating Holstein dairy cows by the Ruminant Research Group at the Institute of Subtropical Agriculture, Chinese Academy of Science (Changsha, Hunan, China) [18]. The bMECs were cultured in an incubator (8000DH, Thermo Fisher, USA) with a humidified atmosphere containing 5% CO2 at 37 °C in DMEM/F12 medium (Gibco, USA), supplemented with 10% (v/v) fetal bovine serum (Gibco, USA), 100 U/mL penicillin (Gibco, USA), 100 μg/mL streptomycin (Gibco, USA), 2.5 μg/mL amphotericin B (Solaibao Technology Co., Ltd., Beijing, China), 5 μg/mL hydrocortisone (Sangon Biotech Co., Ltd., Shanghai, China), 10 ng/mL epidermal growth factor (Sangon Biotech Co., Ltd., Shanghai, China), and 10 μg/mL Insulin-Transferrin-Selenium (Gibco, USA). Cells were seeded in a TC-treated multiple well plate or a culture dish (Corning, NY, USA). The medium was changed every 2 d. Cells were passaged or experiments were performed when cells reached approximately 80% confluence.
Treatment methods for cells
Hesperidin (97% purity) was provided by Jiyuan Technology Co., Ltd. (Zhangjiajie, China). Hesperidin was dissolved in dimethyl sulfoxide (DMSO, Sigma Aldrich Corporation, St. Louis, MO, USA) at a concentration of 200 mg/mL as a stock solution for storage and diluted to specific concentrations in cell culture medium for cell treatments. Specifically, a hesperidin stock solution was freshly prepared by dissolving 200 mg of hesperidin in 1 mL of DMSO (Sigma Aldrich Corporation, St. Louis, MO, USA) and filtering it using a 0.22-μm syringe filter into a clean glass screw-top vial. The hesperidin stock solution was diluted in serum-free DMEM-F12 culture medium to concentrations of 20, 40, 60, 80, 100, 120, 140, 160, 180, and 200 μg/mL, and the same volume of DMSO was added to the control groups. The final concentration of DMSO in the treatment solutions prepared above was less than 0.1% (v/v). H2O2 (3 wt%, Sigma Aldrich Corporation, St. Louis, MO, USA) was dissolved in serum-free DMEM-F12 culture medium to a specific concentration (200, 400, 600, 800, and 1,000 μmol/L) for cell treatment. In subsequent experiments, the concentration of hesperidin was fixed at 120 μg/mL for 24 h and the concentration of H2O2 was fixed at 1,000 μmol/L to treat bMECs for 8 h.
To investigate the protective effect of hesperidin on H2O2-induced bMECs injury, the bMECs were categorized into the following groups: (1) control group (CK); (2) H2O2 model group; (3) hesperidin group; and (4) H2O2 + hesperidin group. The control and hesperidin cells were pretreated with serum-free DMEM-F12 for 8 h, and the H2O2 and H2O2 + hesperidin cells were pretreated with 1,000 μmol/L H2O2 for 8 h. Subsequently, the control and H2O2 cells were treated with serum-free DMEM-F12 for 24 h, and the hesperidin and H2O2 + hesperidin cells were treated with 120 μg/mL hesperidin for 24 h.
To investigate the role of the Nrf2 pathway in the hesperidin-mediated protective effect on H2O2-induced bMECs injury, bMECs were divided into 6 groups: (1) control group; (2) H2O2 model group; (3) H2O2 + hesperidin group; (4) ML385 (the Nrf2 inhibitor) group; (5) ML385 + H2O2 group; and (6) ML385 + H2O2 + hesperidin group: the first 3 groups were cultured with serum-free DMEM-F12 for 48 h, and the last 3 groups were treated with 5 μmol/L ML385 for 48 h. Next, the control and ML385 groups were pretreated with serum-free DMEM-F12 for 8 h, and the H2O2, H2O2 + hesperidin, ML385 + H2O2 and ML385 + H2O2 + hesperidin groups were pretreated with 1,000 μmol/L H2O2 for 8 h. Subsequently, the control, H2O2, ML385 and ML385 + H2O2 groups were treated with serum-free DMEM-F12 for 24 h, and the H2O2 + hesperidin and ML385 + H2O2 + hesperidin groups were treated with 120 μg/mL hesperidin for 24 h.
Cell viability determination
Cell viability was determined using a Cell Counting Kit-8 (CCK-8; Beyotime Biotech. Inc., Shanghai, China). Briefly, cells were seeded into 96-well plates at a density of 4 × 104 cells/well, incubated for 24 h, and then starved in serum-free DMEM-F12 for 1 h. The media were then replaced with fresh DMEM/F12 containing various concentrations of H2O2 (final concentration: 0, 200, 400, 600, 800, and 1,000 μmol/L), and the cells were exposed for various time intervals (6, 8, 12 and 24 h). Then 10 μL of CCK-8 solution was added to each well and incubated for 2 h at 37 °C in a humidified atmosphere of 5% CO2. The absorbance (OD) of each well was measured by using a microplate reader (Tecan, Männedorf, Switzerland) at a wavelength of 450 nm according to the manufacturer’s instructions. To explore the optimal treatment concentration of hesperidin, we assessed the cell viability of bMECs after exposure to different concentrations (0, 20, 40, 60, 80, 100, 120, 140, 160, 180, and 200 μg/mL) of hesperidin for 24 h. The duration of the hesperidin treatment was based on previous research [19,20,21]. The results were calculated using the percentage viability according to the following formula: Viability, % = 100 × (absorbance of treatment/absorbance of control).
Detection of malondialdehyde (MDA) levels
The viability of bMECs treated with H2O2 at concentrations of 600, 800, and 1,000 μmol/L for 8 h, and at concentrations of 1,000 μmol/L for 12 h was approximately 60%–70%. These H2O2 treatment conditions were chosen to detect the MDA levels in bMECs. Cells were seeded into 6-well plates at a density of 1 × 105 cells/well for 24 h. After recovery in serum-free DMEM-F12 for 1 h, the cells were treated with H2O2 (600, 800, and 1,000 μmol/L) for 8 h or with H2O2 at a concentration of 1,000 μmol/L for 12 h. The cells were harvested, and the cellular MDA contents were determined using a spectrophotometric diagnostic kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions. The concentration of total protein in the cell sample was determined by a BCA Protein Assay kit (Beyotime, Shanghai, China). The MDA level is expressed as nmol/mg protein in relation to the cellular protein concentration.
Flow cytometer detection of cell apoptosis
The cellular apoptosis status was detected for the bMECs treated with H2O2 at a concentration of 1,000 μmol/L for 8 h that had high MDA level. Briefly, bMECs were seeded into 6-well plates at a density of 1 × 105 cells/well, incubated for 24 h, and then exposed to H2O2 to a final concentration of 1,000 μmol/L for 12 h. Subsequently, bMECs were digested with 0.25% trypsin without EDTA (Gibco, USA) and collected by centrifugation at 1,000 × g for 5 min at 4 °C in 1.5 mL microcentrifuge tubes, followed by washing twice with cold PBS. Then bMECs were stained with a combination of Annexin V-FITC and propidium iodide (PI) using a commercial kit (KeyGEN Biotech, Nanjing, China). After incubating them in the dark at room temperature for 10 min, the stained cells were analyzed using a flow cytometer (Beckman Coulter, USA).
Measurement of ROS levels
The intracellular ROS content of bMECs was measured by flow cytometry using dichlorodihydrofluorescein diacetate (DCFH-DA) as a fluorescent probe. Briefly, the bMECs were divided into four groups: control (CK), H2O2, hesperidin, and H2O2 + hesperidin. Cells were plated at a density of 1 × 105 cells/well for 24 h in 6-well plates. Then, the control and hesperidin cells were pretreated with serum-free DMEM-F12 for 8 h, and the H2O2 and H2O2 + hesperidin cells were pretreated with 1,000 μmol/L H2O2 for 8 h. Subsequently, the media of control and H2O2 cells were replaced with serum-free DMEM-F12 for 24 h, and the media of hesperidin and H2O2 + hesperidin cells were replaced with 120 μg/mL hesperidin for 24 h. Next, the cells were stained with 10 μmol/L DCFH-DA at 37 °C for 30 min in the dark. Cells were washed three times with serum-free media, and dihydrodichlorofluorescein (DCF) fluorescence was analyzed by an inverted fluorescence microscope (Leica DMI 3000B, Wetzlar, Germany). The percent fluorescence intensity, % = the mean fluorescence of each group/the mean fluorescence of the control group × 100.
Measurement of MDA, GSH-Px, CAT and SOD
To measure intracellular levels of MDA, glutathione peroxidase (GSH-Px), catalase (CAT) and superoxide dismutase (SOD) in bMECs, cells were cultured in 6-well plates (1 × 105 cells/well) and then treated as mentioned earlier in the Treatment methods for cells section. The activities of GSH-Px, CAT and SOD, and the content of MDA in cells were determined using spectrophotometric diagnostic kits from Beyotime Biotech. Inc., (Shanghai, China) according to the manufacturer’s protocols. The cells were harvested and suspended in PBS. The absorbance was detected at 340 nm for GSH-Px, 520 nm for CAT, 450 nm for SOD, and 532 nm for MDA using a microplate reader (Tecan, Männedorf, Swiss). The MDA level is expressed as nmol/mg protein relative to the cellular protein concentration. The levels of GSH-Px, CAT and SOD are expressed as U/g protein relative to the cellular protein concentration.
Determination of cell proliferation
Cell proliferation of bMECs was detected using a 5-ethynyl-2'-deoxyuridine (EdU) cell proliferation assay kit (Beyotime, Shanghai, China). The bMECs were seeded in BeyoGold 35 mm confocal dishes (Beyotime, Shanghai, China) at a density of 1 × 105 cells/dish, and then treated as described in the Treatment methods for cells section. After incubation, EdU solution was added to the medium at an equal volume and incubated for 2 h. The cells were washed twice with PBS, fixed in 4% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 for 10 min at room temperature. After washing twice with PBS, the cells were treated with 500 μL click reaction for 30 min at room temperature, incubated with 1 mL Hoechst dye for 10 min at room temperature, and then washed twice with PBS. Finally, the cells were observed using an inverted fluorescence microscope (Leica DMI 3000B, Wetzlar, Germany), and the fluorescence integrated density was analyzed with ImageJ software (1.50 version, NIH, USA).
Measurement of mitochondrial membrane potential
The Mitochondrial membrane potential (MMP) of bMECs was assessed by JC-1 staining using a commercial kit (KeyGEN Biotech, Nanjing, China). JC-1, 5,5',6,6'-tetrachloro-1,1',3,3'-tetraethylbenzimidazolcarbocyanine iodide, is a cationic carbocyanine dye that can permeate the cell membrane and be used as a ratiometric indicator of MMP in cells. Briefly, cells were seeded in 6-well plates at a density of 1 × 105 cells/well, incubated for 24 h, and then treated as described in the Treatment methods for cells section. Then, 1 mL of JC-1 working solution was added to the medium and incubated at 37 °C for 20 min. The cells were washed twice with 1 × incubation buffer and observed with an inverted fluorescence microscope (Leica DMI 3000B, Wetzlar, Germany) by detecting the fluorescence signals of JC-1 aggregates (red) and JC-1 monomers (green) at excitation/emission wavelengths of 525/590 nm for aggregates and 490/530 nm for monomers. The data are representative of at least three independent experiments.
RNA extraction and reverse transcription quantitative real-time PCR
Total RNA was extracted from the bMECs using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. The purity and concentration of the RNA were assessed using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA). In our study, the OD260/OD280 ratio of the total RNA was 1.9, which met the specified purity requirements. Then, for each sample, 1 μg of total RNA was used for reverse transcription using an Evo M-MLV RT Kit with gDNA Clean for qPCR (AGbio, Changsha, China). Specific primers for genes including HO-1, Nrf2, NQO1, Keap1, p38 MAPK, and GAPDH (glyceraldehyde 3-phosphate dehydrogenase) were designed using the primer 3.0 online tool (http://bioinfo.ut.ee/primer3) and GAPDH was employed as an endogenous control. The primer sequences were synthesized by Sangon Biotech. Co., Ltd. (Shanghai, China). Reverse transcription quantitative real-time PCR (RT-qPCR) was performed using the SYBR Green Premix Pro Taq HS qPCR Kit (AGbio, Changsha, China) and LightCycler 480II Real-Time PCR System (Roche Diagnostics GmbH, Mannheim, Germany). The reaction mixture contained the following components: 5.0 μL 2 × SYBR Green Pro Taq HS Premix (AGbio, Changsha, China), 1.0 μL cDNA, 0.3 μL upstream PCR primers and 0.3 μL downstream PCR primers. Nuclease-free water was added to a final volume of 10 μL. Each reaction was run in triplicate. The RT-qPCR reaction conditions consisted of an initial predegeneration step at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. Relative quantities of expression for the genes of interest among different samples were calculated using the 2−△△ct method, which compares the threshold cycle (Ct) values of the target genes and the reference gene in each sample and normalizes them to a control sample. The primers are listed in Table 1.
Protein extraction and western blotting
The total proteins of bMECs were extracted with cell lysis buffer (Beyotime, Shanghai, China). The concentration of the extracted protein was determined by the bicinchoninic acid (BCA) method (Beyotime, Shanghai, China). The protein sample was mixed with the loading buffer and then heated in a water bath at 95 °C for 10 min. Then the protein was subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The separated proteins were transferred to a polyvinylidene difluoride (PVDF) membrane using the Transfer Cell (Liuyi Biotechnology, Beijing, China). The membranes were washed three times with Tris-buffered saline with 0.1% Tween 20 (TBST) buffer for 10 min each and then blocked with Quick Block Blocking Buffer (Beyotime, Shanghai, China) for 1 h. After washing three times with TBST for 15 min each, the membranes were incubated with primary antibodies at 4 °C overnight. Anti-Nrf2 (1:4,000, Cat. No. 16396-1-AP), anti-NQO1 (1:2,000, Cat. No. 11451-1-AP), anti-HO-1 (1:1,000, Cat. No. 10701-1-AP), anti-Keap1 (1:4,000, Cat. No. 10503-2-AP), anti-p38-MAPK (1:1,000, Cat. No. 14064-1-AP), and anti-β-actin (1:5,000, Cat. No. 66009-1-Ig) antibodies were purchased from Proteintech (Wuhan, China). The following day, the membrane was incubated for an additional 2 h with horseradish peroxidase (HRP)-conjugated secondary antibody (1:5,000 dilution) at room temperature after being thoroughly washed three times with TBST for 10 min each. HRP-conjugated Affinipure Goat Anti-Rabbit IgG (H + L) (SA00001-2) and HRP-conjugated Affinipure Goat Anti-Mouse IgG (H + L) (SA00001-1) were purchased from Proteintech (Wuhan, China). Finally, immunoreactive bands were visualized using an electrochemiluminescence (ECL) kit (Beyotime, Shanghai, China) and the intensity of the protein bands was assayed by Quantity One software (Quantity One 1-D Analysis Software).
Immunofluorescence assay
The aim of the immunofluorescence assay was to detect Nrf2 protein localization in bMECs. Briefly, cells were seeded at a 1 × 105 cell density per well for 24 h in 6-well plates and then treated as mentioned earlier in the Treatment methods for cells section. After washing three times with PBS, the cells were fixed with 4% paraformaldehyde for 10 min and then permeabilized with 0.1% Triton X-100 for 10 min at room temperature. After washing three times with PBS for 10 min each, the cells were incubated with the primary antibody (anti-Nrf2; Proteintech, China) at 4 °C overnight. Then the cells were washed three times with PBS and were incubated in the secondary antibody (goat anti-rabbit IgG [H + L] Alexa Fluor-488, 33106ES60, Yeasen, Shanghai, China) for 2 h at room temperature protected from light. After washing three times with PBS, the cells were incubated with DAPI solution for nuclear staining at room temperature for 15 min in the dark. Finally, the fluorescence of the cells was observed under an inverted fluorescence microscope (Leica DMI 3000B; Germany).
Statistical analysis
Each experiment was repeated three times. The analysis of experimental results was performed using either GraphPad Prism 8.0 (GraphPad Software; San Diego, CA, USA) or SPSS 26.0 software (IBM; USA). Unless stated otherwise, all data was presented as the means ± standard error of the mean (SEM). Comparisons among groups were analyzed using one-way ANOVA followed by Tukey’s tests. Comparisons between two groups were performed using Student’s t-test. P < 0.05 was considered statistically significant. The symbol “*” was used to indicate P < 0.05; “**” was used to indicate P < 0.01, and “***” was used to indicate P < 0.001.
Results
Construction of an H2O2-induced oxidative stress model in bMECs
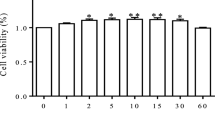
To determine the appropriate H2O2 concentration and conditions for constructing an oxidative stress model, bMECs were treated with graded concentrations of H2O2 (0, 200, 400, 600, 800, and 1,000 μmol/L) for 4 different time points (6, 8, 12, and 24 h). The CCK-8 results showed that cell proliferation decreased in a dose-dependent manner (Fig. 1A and Fig. S1). The results showed that the viability of bMECs exposed to 600, 800 and 1,000 μmol/L H2O2 for 8 h or 1,000 mol/L H2O2 for 12 h were about 60%–70%. Therefore, these treatment conditions were selected to determine the MDA content in the cells after H2O2 exposure. Treatment with 1,000 μmol/L H2O2 for 8 h significantly increased the MDA content in bMECs (P < 0.001, Fig. 1B). Meanwhile, this treatment also significantly increased the proportion of early apoptotic cells and the total proportion of apoptotic cells (P < 0.01, Fig. 1C). Hence, we selected 1,000 μmol/L H2O2 to stimulate bMECs for 8 h to establish an oxidative stress model.
Effects of hesperidin and H2O2 on the cell viability, MDA levels and apoptosis of bovine mammary epithelial cells (bMECs). A Cell viability of bMECs. bMECs were treated with H2O2 (0, 200, 400, 600, 800, 1,000 μmol/L) for 8 h. B Malondialdehyde (MDA) content in bMECs. bMECs were treated with H2O2 (0, 600, 800 μmol/L) for 8 h and 1,000 μmol/L for 12 h. C The presence of apoptotic cells was detected by flow cytometry. Representative images and quantification of the early apoptosis, late apoptosis and total apoptotic rate of bMECs treated with H2O2 (1,000 μmol/L) for 8 h. D Cell viability of bMECs. bMECs were treated with hesperidin (0, 20, 40, 60, 80, 100, 120, 140, 160, 180, and 200 μg/mL) for 24 h. All data are presented as the mean ± SEM from three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001
The cytotoxicity of hesperidin against bMECs and selection of optimal hesperidin concentration for protecting bMECs from oxidative stress
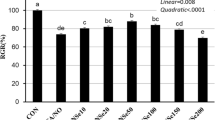
To demonstrate the toxic effect of hesperidin on bMECs, we first examined the dose effects of hesperidin (0, 20, 40, 80, 100, 120, 140, 160, 180, and 200 μg/mL) on bMECs viability via a CCK-8 assay. Hesperidin did not display cytotoxicity against the bMECs at increasing concentrations from 0 to 140 μg/mL with 20 μg/mL increments. The viability of cells treated with 120 μg/mL hesperidin was higher than that of cells treated with 140 μg/mL hesperidin. Compared with the control cells (0 μg/mL hesperidin), hesperidin at 60 and 80 μg/mL significantly increased the viability of bMECs (P < 0.05, Fig. 1D). However, hesperidin at 160 μg/mL or higher showed a toxic effect on the viability of bMECs (P < 0.05, Fig. 1D). Therefore, in this study, after taking into account the impact of hesperidin on bMECs viability and the potential concentration dependence of bioactive components of hesperidin, we finally chose a concentration of 120 μg/mL hesperidin for all subsequent experiments to investigate its protective effect against oxidative stress induced by H2O2.
Hesperidin reduces H2O2-induced oxidative stress in bMECs
The purpose of this experiment was to investigate whether hesperidin can effectively alleviate the oxidative stress caused by H2O2. The results indicated that compared with the CK group, H2O2 increased the levels of ROS and MDA, and induced oxidative stress (P < 0.001, Fig. 2A–C). Meanwhile, compared with the CK group, H2O2 decreased the levels of CAT (P < 0.001) and SOD (P < 0.05) in bMECs and impaired their antioxidant capacity (Fig. 2D and F). However, compared with the H2O2-treated group, hesperidin significantly reduced the levels of ROS and MDA induced by H2O2 (P < 0.001, Fig. 2A–C) and effectively blocked the reductions in CAT (P < 0.01, Fig. 2D). In a similar fashion, SOD and GSH-Px activities were induced by hesperidin compared to H2O2 treatment alone although the difference in SOD and GSH-Px between H2O2 group and H2O2 + hesperidin group did not reach statistical significance (P > 0.05, Fig. 2E and F).
Hesperidin ameliorates H2O2-induced oxidative stress in bMECs. bMECs were treated with H2O2 (1,000 μmol/L) for 8 h and/or hesperidin (120 μg/mL) for 24 h. A Representative images of ROS content, the scale bar represents 100 μm. B ROS content. C Malondialdehyde (MDA) content in bMECs. D Catalase (CAT) content in bMECs. E Glutathione peroxidase (GSH-Px) content in bMECs. F Superoxide dismutase (SOD) content in bMECs. All data are presented as the mean ± SEM from three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001
Hesperidin improves cell proliferation and mitochondrial function in H2O2-treated bMECs
This experiment was designed to examine the role of hesperidin in protecting the proliferation of bMECs that were damaged by H2O2. We observed that H2O2 reduced the proliferation of bMECs (P < 0.001, Fig. 3). However, hesperidin significantly rescued the cell proliferation of bMECs that were impaired by H2O2 (P < 0.001, Fig. 3). Next, we used the JC-1 fluorescent probe to measure the mitochondrial membrane potential changes of bMECs under different treatments. Figure 4A and B shows that the MMP value of the H2O2 + hesperidin group was significantly higher than that of the H2O2 group. This indicates that hesperidin can reverse the damage of cell mitochondria caused by H2O2.
Hesperidin increases cell proliferation in H2O2-treated bMECs. A Representative images presenting a cell proliferation by EdU assay. The scale bar represents 50 μm. B Quantitative analysis of an EdU assay for cell proliferation. bMECs were treated with H2O2 (1,000 μmol/L) for 8 h and/or hesperidin (120 μg/mL) for 24 h. All data are presented as the mean ± SEM from three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001
Hesperidin increases mitochondrial membrane potential in H2O2-treated bMECs. A Representative images presenting the mitochondrial membrane potential. The scale bar represents 100 μm. B Quantitative analysis of mitochondrial membrane potential. bMECs were treated with H2O2 (1,000 μmol/L) for 8 h and/or hesperidin (120 μg/mL) for 24 h. All data are presented as the mean ± SEM from three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001
Hesperidin activated the Keap1/Nrf2/ARE signaling pathway in H2O2-treated bMECs
The purpose of this experiment was to explore whether hesperidin protects bMECs from oxidative stress by activating the Keap1/Nrf2/ARE signaling pathway and promoting the mRNA and protein expression of Nrf2 downstream genes. First, we investigated whether H2O2 and hesperidin affect the cellular distribution of Nrf2. The results showed that hesperidin can induce the nuclear translocation of Nrf2, which may activate the expression of antioxidant response genes (Fig. 5A). Then, we detected the mRNA and protein expression levels of genes associated with the antioxidation ability of bMECs. We found that H2O2 inhibited the activation of the Nrf2 signaling pathway, decreased the mRNA and protein expression of NQO1 (P < 0.05, Fig. 5D and Fig. S2B), HO-1 (P < 0.05, Fig. 5E and Fig. S2C) and Nrf2 (P < 0.05, Fig. 5F and Fig. S2A), and increased the expression of Keap1 (P < 0.001, Fig. 5C and Fig. S2D), leading to a decrease in cellular antioxidant capacity. However, hesperidin counteracted the inhibitory effect of H2O2 on Nrf2 signaling, increased the mRNA and protein expression of NQO1, HO-1 and Nrf2, decreased Keap1 expression, and restored cellular antioxidant capacity (P < 0.05, Fig. 5B–F and Fig. S2A–D). Interestingly, there was no significant difference in p38 MAPK mRNA and protein expression between all the groups (Fig. 5G and Fig. S2E). This suggests that the activation of the Keap1/Nrf2/ARE signaling pathway by hesperidin is independent of the p38 MAPK pathway in bMECs under oxidative stress.
Hesperidin induced the nuclear translocation of Nrf2 and activated the expression of antioxidant response genes in H2O2-treated bMECs. A Representative images of the cellular distribution of Nrf2, the scale bar represents 20 μm. B Representative western blots of Keap1, NQO1, HO-1, Nrf2, p38 MAPK, and β-actin was used as a loading control. C–G Quantitation of Keap1, NQO1, HO-1, Nrf2, and p38 MAPK protein abundance. bMECs were treated with H2O2 (1,000 μmol/L) for 8 h and/or hesperidin (120 μg/mL) for 24 h. All data are presented as the mean ± SEM from three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001
Hesperidin rescued H2O2-elicited Keap1/Nrf2/ARE signaling pathway activity through Nrf2
To verify whether the activation effect of hesperidin on the Keap1/Nrf2/ARE signaling pathway is Nrf2 dependent, ML385, a specific inhibitor of Nrf2, was used to inhibit Nrf2 expression. As shown in Fig. 6A and B, the protein level of Nrf2 was markedly decreased by treatment with ML385 compared with the control group (P < 0.01). Importantly, inhibition of Nrf2 significantly blocked the hesperidin-induced upregulation of Nrf2, HO-1 and NQO1 in H2O2-treated bMECs (P < 0.01, Fig. 6C and E–G). Meanwhile, ML385 significantly increased Keap1 expression in H2O2-treated bMECs (P < 0.001, Fig. 6D).
Nrf2 inhibitor blocked the activation effect of hesperidin on the Keap1/Nrf2/ARE signaling pathway. A Protein levels of Nrf2 and β-actin by western blot analysis. B Quantitation of Nrf2 protein abundance. C Protein levels of Keap1, NQO1, HO-1 and Nrf2 by western blot analysis. D–G Quantitation of Keap1, NQO1, HO-1, and Nrf2 protein abundance. β-actin was used as an internal control. bMECs were treated with ML385 (5 μmol/L) for 48 h and/or H2O2 (1,000 μmol/L) for 8 h and/or hesperidin (120 μg/mL) for 24 h. All data are presented as the mean ± SEM from three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001
Discussion
The mammary gland is an exocrine gland that produces milk and bMECs are the main cell type in the mammary gland of the dairy cow. In addition to their role in milk production, bMECs are also effector cells in mammary immunity [22, 23]. However, bMECs in high-yielding dairy cows are prone to oxidative stress due to their high metabolic rate [24]. Moreover, the bovine mammary gland experiences oxidative stress during the transition period. Oxidative stress can increase the level of ROS in cells, disrupt the cellular redox balance and lead to endoplasmic reticulum stress, mitochondrial dysfunction, lipid peroxidation and cell apoptosis [25, 26]. Therefore, it is important to explore effective antioxidant strategies to protect bMECs from oxidative damage and maintain bovine mammary health and lactation performance. Hesperidin is a bioflavonoid found in citrus fruits with antioxidant and anti-inflammatory properties [27]. The aim of this study was to investigate the protective effect of hesperidin on H2O2-induced oxidative stress injury in bMECs and the possible molecular mechanism. This study confirmed that hesperidin exerted antioxidant effects on bMECs.
ROS and MDA are important markers of oxidative stress, which reflect the production of free radicals and lipid peroxidation in cells [28]. Oxidative stress can cause cell dysfunction and death [29]. Therefore, reducing ROS and MDA levels is an effective way to prevent oxidative damage. Hesperidin has been shown to scavenge free radicals and inhibit lipid peroxidation [30]. In this study, we found that hesperidin could reduce the ROS and MDA levels induced by H2O2 in bMECs compared with the control group. Moreover, hesperidin increased the CAT level but had no effect on the SOD and GSH-Px levels in bMECs exposed to H2O2. This is different from previous research results. Ansar et al. found that hesperidin could reduce the damage of nano-zinc oxide to liver cells by lowering MDA levels and increasing GSH levels and antioxidant enzyme (SOD, CAT, and GSH-Px) activity [31]. This may be due to the different scavenging efficiencies of hesperidin on different types of free radicals, or the differences in the regulation of antioxidant pathways by hesperidin in various cell types. Studies have shown that hesperidin has a higher scavenging efficiency for DPPH and ABTS free radicals but a lower scavenging efficiency for superoxide anion and hydroxyl free radicals [32]. As a common reagent for inducing oxidative stress in vitro, H2O2 mainly produces hydroxyl free radicals. Hesperidin may selectively regulate different signaling pathways, such as Nrf2/ARE, NF-κB and PI3K/Akt/FOXO3, to protect cells from oxidative stress caused by different free radicals [33,34,35].
Cell proliferation and mitochondrial membrane potential are important indicators of cell viability and function [36, 37]. H2O2 is a common source of oxidative stress that can cause cell damage and death by inducing DNA damage, lipid peroxidation, protein oxidation and mitochondrial dysfunction [38, 39]. Our study demonstrated that hesperidin could improve the cell proliferation and mitochondrial membrane potential of bMECs exposed to H2O2. This is similar to the results of previous studies, which showed that hesperidin can prevent oxidative damage to cellular components and restore mitochondrial function [40]. However, several studies have found that hesperidin seems to affect the expression of cell cycle- and apoptosis-related genes by inducing the endoplasmic reticulum stress pathway, regulating the immune response-related or PI3K/Akt/mTOR signaling pathway, and inducing apoptosis and autophagy in various cancer cells, thereby inhibiting the proliferation of cancer cells [41,42,43]. This suggests that hesperidin may have different mechanisms of action on normal cells and cancer cells, and can affect different cell fates and signaling pathways, and there may be a dose–effect relationship. The specific mechanisms of action and dose–effect relationship need to be further explored.
In addition, our results indicated that hesperidin may play a protective role in bMECs through the Keap1/Nrf2/ARE signaling pathway. The Keap1/Nrf2/ARE signaling pathway is a key regulator of cellular antioxidant defense [44, 45]. Keap1 is a negative regulator of Nrf2 that binds to Nrf2 and retains it in the cytoplasm [46]. Nrf2 is a transcription factor that binds to the antioxidant response element (ARE) and activates the expression of various antioxidant genes, such as HO-1 and NQO1 [47,48,49]. HO-1 is an enzyme that catalyzes the degradation of heme, a pro-oxidant molecule known as biliverdin, carbon monoxide and iron, which have anti-inflammatory, anti-apoptotic and cytoprotective effects [50]. NQO1 can reduce quinones to hydroquinone, which can be further conjugated with glutathione and excreted from the cells [51, 52]. In our study, hesperidin induced the nuclear translocation of Nrf2 by disrupting the interaction between Nrf2 and Keap1 and increased the expression of HO-1 and NQO1 in H2O2-treated bMECs. According to previous studies, plant-derived compounds, such as sulforaphane, 6-methylsulfinylhexyl isothiocyanate and curcumin, could induce the activation of Nrf2 nuclear translocation [53,54,55]. These results are consistent with previous studies that have shown that hesperidin can protect against oxidative stress-induced cell damage by upregulating HO-1 expression via ERK/Nrf2 signaling and/or by increasing CAT and SOD activities [21, 56, 57]. However, some studies have also reported that hesperidin can modulate other signaling pathways in different cell types or under different stress conditions, including the transcription factors Foxo1 and Foxo33 or p38 MAPK, which may also be involved in the antioxidant response [58,59,60]. It has been reported that the MAPK pathways participate in Nrf2-dependent nuclear translocation through kinases such as ERK, JNK and p38 MAPK in response to various stimuli. Chen et al. found that hesperidin dose-dependently facilitated the phosphorylation of ERK1/2, but not the p38 and JNK pathways [56]. In our present study, we also found that this effect was Nrf2-dependent, but independent of the p38MAPK pathway. Therefore, further studies are needed to elucidate the precise mechanisms by which hesperidin regulates oxidative stress and antioxidant defense in bMECs.
Conclusion
In conclusion, based on our results, this study found that hesperidin effectively protected against H2O2-induced oxidative damage in bMECs by inhibiting cell apoptosis, ROS overproduction and MDA formation, as well as enhancing the levels of CAT. The mechanism of action includes activating the Keap1/Nrf2/ARE pathway. Hesperidin induced the nuclear translocation of Nrf2 and the dissociation of Nrf2 from Keap1 and upregulated the expression of its downstream genes NQO1 and HO-1. Interestingly, we also found that the antioxidant effect of hesperidin was Nrf2-dependent and independent of the p38 MAPK pathway. This study provides an in vitro test basis for the application of hesperidin in the prevention and treatment of bovine oxidative stress injury in animal husbandry.
Availability of data and materials
The datasets used during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ARE:
-
Antioxidant response element
- Bax:
-
Bcl-2-associated X protein
- Bcl-2:
-
B-cell lymphoma 2
- bMECs:
-
Bovine mammary epithelial cells
- caspase-3:
-
Cysteine-aspartic acid protease 3
- CAT:
-
Catalase
- DCFH-DA:
-
2,7-Dichlorodihydrofluorescein diacetate
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- GSH-Px:
-
Glutathione peroxidase
- H2O2 :
-
Hydrogen peroxide
- HO-1:
-
Heme oxygenase-1
- Keap1:
-
Kelch-like ECH-associated protein 1
- MDA:
-
Malondialdehyde
- NQO1:
-
NAD(P)H quinone oxidoreductase 1
- Nrf2:
-
Nuclear factor erythroid 2-related factor 2
- p38MAPK:
-
P38 Mitogen-activated protein kinase
- ROS:
-
Reactive oxidative stress
- SOD:
-
Superoxide dismutase
References
Oberacker T, Fritz P, Schanz M, Alscher MD, Ketteler M, Schricker S. Enhanced oxidative DNA-damage in peritoneal dialysis patients via the TXNIP/TRX axis. Antioxidants (Basel). 2022;11(6):1124. https://doi.org/10.3390/antiox11061124.
Sun X, Jia H, Xu Q, Zhao C, Xu C. Lycopene alleviates H2O2-induced oxidative stress, inflammation and apoptosis in bovine mammary epithelial cells via the NFE2L2 signaling pathway. Food Funct. 2019;10(10):6276–85. https://doi.org/10.1039/C9FO01922G.
Yang S, Lian G. ROS and diseases: role in metabolism and energy supply. Mol Cell Biochem. 2020;467:1–12. https://doi.org/10.1007/s11010-019-03667-9.
Baratta M, Miretti S, Macchi E, Accornero P, Martignani E. Mammary stem cells in domestic animals: the role of ROS. Antioxidants (Basel). 2019;8(1):6. https://doi.org/10.3390/antiox8010006.
Sordillo LM, Aitken SL. Impact of oxidative stress on the health and immune function of dairy cattle. Vet Immunol Immunopathol. 2009;128(1–3):104–9. https://doi.org/10.1016/j.vetimm.2008.10.305.
Hernández-Cruz EY, Eugenio-Pérez D, Ramírez-Magaña KJ, Pedraza-Chaverri J. Effects of vegetal extracts and metabolites against oxidative stress and associated diseases: studies in Caenorhabditis elegans. ACS Omega. 2023;8(10):8936–59. https://doi.org/10.1021/acsomega.2c07025.
Qi Q, Chu M, Yu X, Xie Y, Li Y, Du Y, et al. Anthocyanins and proanthocyanidins: chemical structures, food sources, bioactivities, and product development. Food Rev Int. 2023;39(7):4581–609. https://doi.org/10.1080/87559129.2022.2029479.
Dias MC, Pinto D, Silva AMS. Plant flavonoids: chemical characteristics and biological activity. Molecules (Basel). 2021;26(17):5377. https://doi.org/10.3390/molecules26175377.
Agrawal PK, Agrawal C, Blunden G. Pharmacological significance of hesperidin and hesperetin, two citrus flavonoids, as promising antiviral compounds for prophylaxis against and combating COVID-19. Nat Prod Commun. 2021;16(10):1–15. https://doi.org/10.1177/1934578X211042540.
Li X, Huang W, Tan R, Xu C, Chen X, Li S, et al. The benefits of hesperidin in central nervous system disorders, based on the neuroprotective effect. Biomed Pharmacother. 2023;159:114222. https://doi.org/10.1016/j.biopha.2023.114222.
Kim J, Wie MB, Ahn M, Tanaka A, Matsuda H, Shin T. Benefits of hesperidin in central nervous system disorders: a review. Anat Cell Biol. 2019;52(4):369–77. https://doi.org/10.5115/acb.19.119.
Aggarwal V, Tuli HS, Thakral F, Singhal P, Aggarwal D, Srivastava S, et al. Molecular mechanisms of action of hesperidin in cancer: recent trends and advancements. Exp Biol Med (Maywood). 2020;245(5):486–97. https://doi.org/10.1177/15353702209036.
Varışlı B, Darendelioğlu E, Caglayan C, Kandemir FM, Ayna A, Genç A, et al. Hesperidin attenuates oxidative stress, inflammation, apoptosis, and cardiac dysfunction in sodium fluoride-induced cardiotoxicity in rats. Cardiovasc Toxicol. 2022;22(8):727–35. https://doi.org/10.1007/s12012-022-09751-9.
Yıldız MO, Çelik H, Caglayan C, Kandemir FM, Gür C, Bayav İ, et al. Neuromodulatory effects of hesperidin against sodium fluoride-induced neurotoxicity in rats: involvement of neuroinflammation, endoplasmic reticulum stress, apoptosis and autophagy. Neurotoxicology. 2022;90:197–204. https://doi.org/10.1016/j.neuro.2022.04.002.
Liu P, Li J, Liu M, Zhang M, Xue Y, Zhang Y, et al. Hesperetin modulates the Sirt1/Nrf2 signaling pathway in counteracting myocardial ischemia through suppression of oxidative stress, inflammation, and apoptosis. Biomed Pharmacother. 2021;139:111552. https://doi.org/10.1016/j.biopha.2021.111552.
Liu J, Peng C, Tang S, Han X, Tan Z. Hesperidin inhibits H2O2-induced apoptosis of bovine mammary epithelial cells by regulating Bcl-2/Bax-Caspase3 pathway. J South Agric. 2023:1–11 (in Chinese). http://kns.cnki.net/kcms/detail/45.1381.S.20230313.1018.006.html.
Zhang J, Fang Y, Tang D, Xu X, Zhu X, Wu S, et al. Activation of MT1/MT2 to protect testes and Leydig cells against cisplatin-induced oxidative stress through the SIRT1/Nrf2 signaling pathway. Cells. 2022;11(10):1690. https://doi.org/10.3390/cells11101690.
Yan Q, Tang S, Zhou C, Han X, Tan Z. Effects of free fatty acids with different chain lengths and degrees of saturability on the milk fat synthesis in primary cultured bovine mammary epithelial cells. J Agric Food Chem. 2019;67(31):8485–92. https://doi.org/10.1021/acs.jafc.9b02905.
Wang Z, Han L, ShangTing H, Chen P. Effect of hesperidin on CORT-induced apoptosis and oxidative stress of mouse hippocampal nerve cells by up-regulating miR-146a-5p. Pak J Pharm Sci. 2020;33(1):1383–8.
Li M, Lin XF, Lu J, Zhou BR, Luo D. Hesperidin ameliorates UV radiation-induced skin damage by abrogation of oxidative stress and inflammatory in HaCaT cells. J Photochem Photobiol B. 2016;165:240–5. https://doi.org/10.1016/j.jphotobiol.2016.10.037.
Chen MC, Ye YY, Ji G, Liu JW. Hesperidin upregulates heme oxygenase-1 to attenuate hydrogen peroxide-induced cell damage in hepatic L02 cells. J Agric Food Chem. 2010;58(6):3330–5. https://doi.org/10.1021/jf904549s.
Wu Q, Liu MC, Yang J, Wang JF, Zhu YH. Lactobacillus rhamnosus GR-1 ameliorates Escherichia coli-induced inflammation and cell damage via attenuation of ASC-independent NLRP3 inflammasome activation. Appl Environ Microbiol. 2016;82(4):1173–82. https://doi.org/10.1128/AEM.03044-15.
Li Y, Ren Q, Wang X, Luoreng Z, Wei D. Bta-miR-199a-3p inhibits LPS-induced inflammation in bovine mammary epithelial cells via the PI3K/AKT/NF-κB signaling pathway. Cells. 2022;11(21):3518. https://doi.org/10.3390/cells11213518.
Jin X, Wang K, Liu H, Hu F, Zhao F, Liu J. Protection of bovine mammary epithelial cells from hydrogen peroxide-induced oxidative cell damage by resveratrol. Oxid Med Cell Longev. 2016;2016:2572175. https://doi.org/10.1155/2016/2572175.
Sun J, Chen J, Li T, Huang P, Li J, Shen M, et al. ROS production and mitochondrial dysfunction driven by PU.1-regulated NOX4-p22phox activation in Aβ-induced retinal pigment epithelial cell injury. Theranostics. 2020;10(25):11637–55. https://doi.org/10.7150/thno.48064.
Xie JH, Lai ZQ, Zheng XH, Xian YF, Li Q, Ip SP, et al. Apoptosis induced by bruceine D in human non-small-cell lung cancer cells involves mitochondrial ROS-mediated death signaling. Int J Mol Med. 2019;44(6):2015–26. https://doi.org/10.3892/ijmm.2019.4363.
Lee D, Kim N, Jeon SH, Gee MS, Ju YJ, Jung MJ, et al. Hesperidin improves memory function by enhancing neurogenesis in a mouse model of Alzheimer’s disease. Nutrients. 2022;14(15):3125. https://doi.org/10.3390/nu14153125.
Fei W, Zhang J, Yu S, Yue N, Ye D, Zhu Y, et al. Antioxidative and energy metabolism-improving effects of maca polysaccharide on cyclophosphamide-induced hepatotoxicity mice via metabolomic analysis and Keap1-Nrf2 pathway. Nutrients. 2022;14(20):4264. https://doi.org/10.3390/nu14204264.
Ko J, Jang S, Kwon W, Kim SY, Jang S, Kim E, et al. Protective effect of GIP against monosodium glutamate-induced ferroptosis in mouse hippocampal HT-22 cells through the MAPK signaling pathway. Antioxidants (Basel). 2022;11(2):189. https://doi.org/10.3390/antiox11020189.
Nandakumar N, Jayaprakash R, Rengarajan T, Ramesh V, Balasubramanian MP. Hesperidin, a natural citrus flavonoglycoside, normalizes lipid peroxidation and membrane bound marker enzymes in 7, 12-Dimethylbenz (a) anthracene induced experimental breast cancer rats. Biomed Prev Nutr. 2011;1(4):255–62. https://doi.org/10.1016/j.bionut.2011.06.004.
Ansar S, Abudawood M, Alaraj ASA, Hamed SS. Hesperidin alleviates zinc oxide nanoparticle induced hepatotoxicity and oxidative stress. BMC Pharmacol Toxicol. 2018;19(1):65. https://doi.org/10.1186/s40360-018-0256-8.
Ekinci Akdemir FN, Gülçin İ, Karagöz B, Soslu R, Alwasel SH. A comparative study on the antioxidant effects of hesperidin and ellagic acid against skeletal muscle ischemia/reperfusion injury. J Enzyme Inhib Med Chem. 2016;31(sup4):114–8. https://doi.org/10.1080/14756366.2016.1220378.
Elhelaly AE, AlBasher G, Alfarraj S, Almeer R, Bahbah EI, Fouda MMA, et al. Protective effects of hesperidin and diosmin against acrylamide-induced liver, kidney, and brain oxidative damage in rats. Environ Sci Pollut Res Int. 2019;26(34):35151–62. https://doi.org/10.1007/s11356-019-06660-3.
Tabeshpour J, Hosseinzadeh H, Hashemzaei M, Karimi G. A review of the hepatoprotective effects of hesperidin, a flavanon glycoside in citrus fruits, against natural and chemical toxicities. Daru. 2020;28(1):305–17. https://doi.org/10.1007/s40199-020-00344-x.
Jia Y, Li J, Liu P, Si M, Jin Y, Wang H, et al. Based on activation of p62-Keap1-Nrf2 pathway, hesperidin protects arsenic-trioxide-induced cardiotoxicity in mice. Front Pharmacol. 2021;12:758670. https://doi.org/10.3389/fphar.2021.758670.
Fang X, Xia W, Li S, Qi Y, Liu M, Yu Y, et al. SIRT2 is critical for sheep oocyte maturation through regulating function of surrounding granulosa cells. Int J Mol Sci. 2022;23(9):5013. https://doi.org/10.3390/ijms23095013.
Hung CH, Lin YC, Tsai YG, Lin YC, Kuo CH, Tsai ML, et al. Acrylamide induces mitophagy and alters macrophage phenotype via reactive oxygen species generation. Int J Mol Sci. 2021;22(4):1683. https://doi.org/10.3390/ijms22041683.
Li X, Wang C, Zhu J, Lin Q, Yu M, Wen J, et al. Sodium butyrate ameliorates oxidative stress-induced intestinal epithelium barrier injury and mitochondrial damage through AMPK-mitophagy pathway. Oxid Med Cell Longev. 2022;2022:3745135. https://doi.org/10.1155/2022/3745135.
Shokolenko I, Venediktova N, Bochkareva A, Wilson GL, Alexeyev MF. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009;37(8):2539–48. https://doi.org/10.1093/nar/gkp100.
Tian M, Han YB, Zhao CC, Liu L, Zhang FL. Hesperidin alleviates insulin resistance by improving HG-induced oxidative stress and mitochondrial dysfunction by restoring miR-149. Diabetol Metab Syndr. 2021;13(1):50. https://doi.org/10.1186/s13098-021-00664-1.
Birsu Cincin Z, Unlu M, Kiran B, Sinem Bireller E, Baran Y, Cakmakoglu B. Anti-proliferative, apoptotic and signal transduction effects of hesperidin in non-small cell lung cancer cells. Cell Oncol (Dordr). 2015;38(3):195–204. https://doi.org/10.1007/s13402-015-0222-z.
Wang Y, Yu H, Zhang J, Gao J, Ge X, Lou G. Hesperidin inhibits HeLa cell proliferation through apoptosis mediated by endoplasmic reticulum stress pathways and cell cycle arrest. BMC Cancer. 2015;15:682. https://doi.org/10.1186/s12885-015-1706-y.
Jeong SA, Yang C, Song J, Song G, Jeong W, Lim W. Hesperidin suppresses the proliferation of prostate cancer cells by inducing oxidative stress and disrupting Ca2+ homeostasis. Antioxidants (Basel). 2022;11(9):1633. https://doi.org/10.3390/antiox11091633.
Zhou X, Zhao L, Luo J, Tang H, Xu M, Wang Y, et al. The toxic effects and mechanisms of nano-Cu on the spleen of rats. Int J Mol Sci. 2019;20(6):1469. https://doi.org/10.3390/ijms20061469.
Li DM, Wu YX, Hu ZQ, Wang TC, Zhang LL, Zhou Y, et al. Lactoferrin prevents chronic alcoholic injury by regulating redox balance and lipid metabolism in female C57BL/6J mice. Antioxidants (Basel). 2022;11(8):1508. https://doi.org/10.3390/antiox11081508.
Luo J, Yan D, Li S, Liu S, Zeng F, Cheung CW, et al. Allopurinol reduces oxidative stress and activates Nrf2/p62 to attenuate diabetic cardiomyopathy in rats. J Cell Mol Med. 2020;24(2):1760–73. https://doi.org/10.1111/jcmm.14870.
Wang Y, Ma G, Wang XF, Na L, Guo X, Zhang J, et al. Keap1 recognizes EIAV early accessory protein Rev to promote antiviral defense. PLoS Pathog. 2022;18(2):e1009986. https://doi.org/10.1371/journal.ppat.1009986.
Ma X, Luo Q, Zhu H, Liu X, Dong Z, Zhang K, et al. Aldehyde dehydrogenase 2 activation ameliorates CCl4 -induced chronic liver fibrosis in mice by up-regulating Nrf2/HO-1 antioxidant pathway. J Cell Mol Med. 2018;22(8):3965–78. https://doi.org/10.1111/jcmm.13677.
Colares JR, Hartmann RM, Schemitt EG, Fonseca SRB, Brasil MS, Picada JN, et al. Melatonin prevents oxidative stress, inflammatory activity, and DNA damage in cirrhotic rats. World J Gastroenterol. 2022;28(3):348–64. https://doi.org/10.3748/wjg.v28.i3.348.
Detsika MG, Nikitopoulou I, Veroutis D, Vassiliou AG, Jahaj E, Tsipilis S, et al. Increase of HO-1 expression in critically ill COVID-19 patients is associated with poor prognosis and outcome. Antioxidants (Basel). 2022;11(7):1300. https://doi.org/10.3390/antiox11071300.
Siegel D, Gustafson DL, Dehn DL, Han JY, Boonchoong P, Berliner LJ, et al. NAD(P)H:quinone oxidoreductase 1: role as a superoxide scavenger. Mol Pharmacol. 2004;65(5):1238–47. https://doi.org/10.1124/mol.65.5.1238.
Choi YH. The cytoprotective effects of ethanol extract of Ecklonia cava against oxidative stress are associated with upregulation of Nrf2-mediated HO-1 and NQO-1 expression through activation of the MAPK pathway. Gen Physiol Biophys. 2016;35(1):45–53. https://doi.org/10.4149/gpb_2015029.
Morimitsu Y, Nakagawa Y, Hayashi K, Fujii H, Kumagai T, Nakamura Y, et al. A sulforaphane analogue that potently activates the Nrf2-dependent detoxification pathway. J Biol Chem. 2002;277(5):3456–63. https://doi.org/10.1074/jbc.M110244200.
Balogun E, Hoque M, Gong P, Killeen E, Green CJ, Foresti R, et al. Curcumin activates the haem oxygenase-1 gene via regulation of Nrf2 and the antioxidant-responsive element. Biochem J. 2003;371(3):887–95. https://doi.org/10.1042/bj20021619.
Fahey JW, Haristoy X, Dolan PM, Kensler TW, Scholtus I, Stephenson KK, et al. Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[a]pyrene-induced stomach tumors. Proc Natl Acad Sci U S A. 2002;99(11):7610–5. https://doi.org/10.1073/pnas.112203099.
Chen M, Gu H, Ye Y, Lin B, Sun L, Deng W, et al. Protective effects of hesperidin against oxidative stress of tert-butyl hydroperoxide in human hepatocytes. Food Chem Toxicol. 2010;48(10):2980–7. https://doi.org/10.1016/j.fct.2010.07.037.
Zhu C, Dong Y, Liu H, Ren H, Cui Z. Hesperetin protects against H2O2-triggered oxidative damage via upregulation of the Keap1-Nrf2/HO-1 signal pathway in ARPE-19 cells. Biomed Pharmacother. 2017;88:124–33. https://doi.org/10.1016/j.biopha.2016.11.089.
Kwatra M, Ahmed S, Gawali B, Panda SR, Naidu VGM. Hesperidin alleviates chronic restraint stress and lipopolysaccharide-induced hippocampus and frontal cortex damage in mice: role of TLR4/NF-κB, p38 MAPK/JNK, Nrf2/ARE signaling. Neurochem Int. 2020;140:104835. https://doi.org/10.1016/j.neuint.2020.104835.
Selim NM, Elgazar AA, Abdel-Hamid NM, El-Magd MRA, Yasri A, Hefnawy HME, et al. Chrysophanol, physcion, hesperidin and curcumin modulate the gene expression of pro-inflammatory mediators induced by LPS in HepG2: in silico and molecular studies. Antioxidants (Basel). 2019;8(9):371. https://doi.org/10.3390/antiox8090371.
Yun H, Park S, Kim MJ, Yang WK, Im DU, Yang KR, et al. AMP-activated protein kinase mediates the antioxidant effects of resveratrol through regulation of the transcription factor FoxO1. FEBS J. 2014;281(19):4421–38. https://doi.org/10.1111/febs.12949.
Acknowledgements
The authors thank Smart Medical Art (SMART): (https://smart.servier.com) for providing the drawing materials, where the orange tree, sun and grass symbols in the abstract figure are taken from SMART, and the others are homemade.
Funding
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDA26040304).
Author information
Authors and Affiliations
Contributions
XH conceived the project and designed the experiment. QH and JL conducted the experiment, collected and analysed samples, analysed data and drafted the manuscript. CP analysed samples. XH and ZT performed funding acquisition, project administration, data curation, writing-review & editing and conceptualization, and supervision of the study.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors read and agree to the content of this paper and its publication.
Competing interests
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company could be construed as influencing the content of this paper.
Supplementary Information
Additional file 1:
Fig. S1. Effects of H2O2 on the viability of bMECs. A bMECs were treated with H2O2 (0, 200, 400, 600, 800, 1,000 μmol/L) for 6 h. B bMECs were treated with H2O2 (0, 200, 400, 600, 800, 1,000 μmol/L) for 12 h. C bMECs were treated with H2O2 (0, 200, 400, 600, 800, 1,000 μmol/L) for 24 h. All data are presented as the mean ± SEM from three independent experiments. *P< 0.05, **P< 0.01, and ***P< 0.001.
Additional file 2:
Fig. S2. Hesperidin induces the nuclear translocation of Nrf2 and activates the mRNA expression of antioxidant response genes in H2O2-treated bMECs. A Nrf2 mRNA expression. B NQO1 mRNA expression. C HO-1 mRNA expression. D Keap1 mRNA expression. E p38 MAPK mRNA expression. bMECs were treated with H2O2 (1,000 μmol/L) for 8 h and/or hesperidin (120 μg/mL) for 24 h. All data are presented as the mean ± SEM from three independent experiments. *P< 0.05, **P< 0.01, and ***P< 0.001.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Huang, Q., Liu, J., Peng, C. et al. Hesperidin ameliorates H2O2-induced bovine mammary epithelial cell oxidative stress via the Nrf2 signaling pathway. J Animal Sci Biotechnol 15, 57 (2024). https://doi.org/10.1186/s40104-024-01012-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40104-024-01012-9