Abstract
Background
Atresia and degeneration, a follicular developmental fate that reduces female fertility and is triggered by granulosa cell (GC) apoptosis, have been induced by dozens of miRNAs. Here, we report a miRNA, miR-423, that inhibits the initiation of follicular atresia (FA), and early apoptosis of GCs.
Results
We showed that miR-423 was down-regulated during sow FA, and its levels in follicles were negatively correlated with the GC density and the P4/E2 ratio in the follicular fluid in vivo. The in vitro gain-of-function experiments revealed that miR-423 suppresses cell apoptosis, especially early apoptosis in GCs. Mechanically speaking, the miR-423 targets and interacts with the 3'-UTR of the porcine SMAD7 gene, which encodes an apoptosis-inducing factor in GCs, and represses its expression and pro-apoptotic function. Interestingly, FA and the GC apoptosis-related lncRNA NORHA was demonstrated as a ceRNA of miR-423. Additionally, we showed that a single base deletion/insertion in the miR-423 promoter is significantly associated with the number of stillbirths (NSB) trait of sows.
Conclusion
These results demonstrate that miR-423 is a small molecule for inhibiting FA initiation and GC early apoptosis, suggesting that treating with miR-423 may be a novel approach for inhibiting FA initiation and improving female fertility.
Similar content being viewed by others
Introduction
MicroRNAs (miRNAs) are 18–23 nucleotides (nt) long, single-stranded, non-coding RNAs (ncRNAs) that are generated endogenously. miRNAs usually destabilise target mRNAs or suppress target mRNA translation by interacting with their seed sequences and the miRNA recognition elements (MREs) in the 3′-UTR of target mRNAs by entering the miRNA-induced silencing complexes (miRISCs) [1, 2]. miRNAs are distributed in almost all organs, tissues, and cells and in various organelles, such as mitochondria and exosomes, and regulate an organism’s development, tissue and organ formation, cellular status, and organelle functions [3,4,5]. Functional disorders of miRNAs often lead to developmental abnormalities, aging, and diseases [6, 7]. Although miRNAs were the first ncRNAs to be discovered, they are still the most studied because of their wide distribution, conserved sequences, simultaneous actions on multiple target mRNAs, and crucial functions [2, 4, 8]. Additionally, the main mechanism of action of other popular ncRNAs, such as circular RNAs (circRNAs) [5, 9, 10], involves acting as competing endogenous RNAs (ceRNAs) of miRNAs.
miR-423 is an intronic miRNA transcribed from the first intron of the protein-coding gene nuclear speckle splicing regulatory protein 1 (NSRP1), which encodes a splicing regulator [11]. miR-423 is ubiquitously expressed in various tissues and cells in mammals. It is a multifunctional small molecule and biomarker that contributes to various biological processes related to health and diseases [12,13,14,15]. In female mammals, miR-423 is involved in multiple reproductive system functions [16]. For instance, in the uterus, miR-423 participates in preventing trophoblast cell progression, influencing decidualization, and maintaining early pregnancy [17, 18]. In the ovary, miR-423 is associated with some ovarian functions such as 17β-estradiol (E2) synthesis by granulosa cells (GCs) [16], and its dysfunction can lead to ovarian diseases, such as polycystic ovary syndrome and ovarian hyper response [19,20,21]. We previously found that miR-423 is decreased during follicular atresia (FA) in sows using RNA-seq (unpublished). FA is a follicular developmental fate that reduces female fertility and is triggered by GC apoptosis. In the present study, we aimed to determine the role of miR-423 in GC apoptosis in sows and its mechanism of action.
Materials and methods
Animals
Ear tissues of 369 Yorkshire sows (Kangle, Changzhou, China) were obtained for extracting DNA. Data on fertility traits such as the total number of piglets born (TNB), the total number of piglets born alive (NBA), the number of stillbirths (NSB), and the litter weight (LW) were recorded for association analysis. Besides, commercial sow ovaries were obtained for collecting follicles, GCs, and follicular fluid.
Follicle collection and classification
The follicles (3–5 mm) were collected from sow ovaries and classified into healthy follicles (HF) and atretic follicles (AF) group. Translucent follicles with abundant vascularization, GC number < 4,000/mL, and the ratio of progesterone (P4) (ng/mL) and 17β-estradiol (E2) (pg/mL) levels (that is, the P4/E2 ratio) < 1.0 are classified as HF, others are classified as AF. GC number was counted using a cell counting plate (Qiujing, Shanghai, China). P4 and E2 levels were measured by radioimmunoassay using a P4[I125] and E2 Radioimmunoassay Kit (North Pharmaceutical, Beijing, China).
Quantitative real-time PCR (qPCR)
Trizol (Vazyme, Nanjing, China) was employed to prepare total RNA. cDNA was synthesized from 1 μg of total RNA using HiScript IIQ Select RT SuperMix (Vazyme) or HiScript III 1st Strand cDNA Synthesis Kit (Vazyme). A SYBR Green Master Mix Kit (Vazyme) was employed to quantify the relative transcription levels. The internal references of coding and non-coding genes are GAPDH and U6, respectively. Primers are shown in Table S1.
Cell culture and transfection
GCs were obtained from antral follicles (3–5 mm) of mature commercial sows (Zhushun, Nanjing, China). Cell culture and treatment were performed as described previously [5]. DMEM/F12 medium (Gibco, CA, USA) containing 15% fetal bovine serum (FBS) (Gibco) and RPMI-1640 medium (Hyclone, UT, USA) containing 10% FBS and 1% PS were used for culturing sow GCs, and KGN cells, a human GC line. After the cell density reached 80%–90%, Highgene reagent (ABclonal, Wuhan, China), plasmids, and mimics (10 μmol/L) were mixed in 200 μL of medium, and added into cells. Oligonucleotides were prepared by GenePharma (Shanghai, China), and shown in Table S2.
Apoptosis analysis
An Apoptosis Detection Kit (Vazyme) was employed to measure the apoptosis rate in a flow cytometer (Becton Dickinson, NJ, USA). FlowJo v7.6 software (Stanford University, USA) was employed to analyze data of apoptosis rate.
Bioinformatics analysis
The nucleotide sequences are from the Ensembl database (https://www.ensembl.org/index.html). TargetScan (https://www.targetscan.org), miRDB (http://www.mirdb.org), and TarBase (https://dianalab.e-ce.uth.gr/html/diana/web/index.php?r=tarbasev8) were employed to search targets of miR-423. RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid) was employed to discover the MREs of miR-423. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was conducted by Kobas (http://bioinfo.org/kobasrg).
Subcellular localization
Nucleoplasmic separation of GCs was performed as described previously [5]. miR-423 levels in nuclear and cytoplasmic products were quantified by qPCR, and nuclear and cytoplasmic markers were GAPDH and U6, respectively.
Plasmids and luciferase assay
Plasmids include the pmirGLO vectors of SMAD7 3'UTR and NORHA harboring the MRE of miR-423 and its mutant version that were prepared by Tsingke (Nanjing, China), and the pcDNA3.1-SMAD7 and pcDNA3.1-NORHA vectors that were previously prepared [5, 22]. A Dual-Luciferase Reporter Assay System (Vazyme) was employed for luciferase assay.
Western blot
Total protein of GCs was obtained by RIPA lysis buffer (Biowrld, MN, USA) containing 1% PMSF (Beyotime, Nantong, China) and protease inhibitor. After detecting concentration by using a BCA Kit (Biosharp, Beijing, China), proteins were electrophoretically separated by 4%–20% SDS-PAGE and transferred to polyvinylidene fluoride membranes (Millipore, Billerica, USA). To prevent non-specific binding, the membranes were blocked with 5% BSA for 1.5 h and then incubated with primary antibody overnight at 4 °C. After incubating the second antibody for 1 h, the ECL chromogenic agent (Vazyme, Nanjing, China) was used to obtain high-resolution images in Amersham ImageQuant 800 system (Cytiva, Japan). Serve as an internal control, anti-GAPDH antibody (1:1,000 dilution) was obtained from Sangon (Shanghai, China). Other primary antibodies including anti-SMAD7 and anti-p-SMAD3 were obtained from Sangon and diluted at 1:1,000.
RNA pull-down assay
This assay was conducted as described previously [5]. C-1 magnetic beads were obtained from Life Technologies (USA). The probe labeled with biotin is carried out by Biotech (Shanghai, China), and its sequence is 5'-TTT TTT CGG GTC TGT AAT GC-3'. The RNeasy Mini Kit (QIAGEN, Germany) was employed for isolating RNA from the elution for quantifying miR-423 by qPCR.
Variant identification and association analysis
Three hundred and sixty-nine DNAs were isolated using a phenol–chloroform method and were used as the template of PCR amplification for Sanger sequencing (Sangon). Primers are listed in Table S1. The genotype of each individual is determined using Chromas v2.3 software (https://technelysium.com.au/wp/chromas) based on the sequencing peak diagram. Association analyses were performed using a general linear model as previously described [23].
Statistics
Unpaired t-test and one-way analysis of variance of Prism v8.3 software (GraphPad, USA) were employed for statistical analysis. Multiple comparisons were conducted using a Tukey method.
Results
miR-423 is involved in sow FA
Our previous RNA-seq results revealed that miR-423 expression is markedly decreased in the atretic follicles of sows (Fig. 1A). Further, it was confirmed using qPCR that atretic follicles of sows have significantly lower miR-423 levels than healthy follicles (Fig. 1B), indicating that miR-423 may be involved in the development of FA in sows. Next, we investigated the relationship between miR-423 and sow FA using in vivo experiments. Follicular miR-423 levels were negatively correlated with GC density and the P4/E2 ratio, two core biomarkers of FA in the follicular fluid (Fig. 1C–D). Additionally, follicular miR-423 levels were markedly positively correlated with E2 concentration but not with P4 concentration in the follicular fluid (Fig. 1E–F). Taken together, these results support our hypothesis that miR-423 is directly associated with sow FA.
miR-423 is strongly involved in sow follicular atresia. A and B miR-423 was markedly decreased in sow atretic follicles. A RNA-seq data. B qPCR. HF, healthy follicles; AF, atretic follicles. C–F Correlation analysis. Sow follicles and follicular fluid were collected, and the GC density in follicular fluid was detected using a cell counting plate, E2 and P4 concentration were measured by ELISA, and correlation analysis between miR-423 levels and the GC density (C), the P4/E2 ratio (D), E2 concentration (E), or P4 concentration (F) were carried out, respectively. Data represented are mean ± SEM for at last three independent experiments. **P < 0.01
miR-423 represses GC apoptosis
GC apoptosis is the fundamental cause of FA in sows [23]. To confirm the role of miR-423 in GC apoptosis, we first treated GCs with miR-423-specific mimics to increase the miR-423 levels (Fig. 2A). Flow cytometry showed that the cell apoptosis rate was considerably reduced in GCs treated with miR-423-specific mimics (Fig. 2B), indicating that miR-423 is an anti-apoptotic miRNA in GCs. Interestingly, the early and not late apoptosis rates were markedly declined in miR-423-treated GCs (Fig. 2C–E). These data suggest that miR-423 is a key modulator of early GC apoptosis and FA initiation in sows.
miR-423 represses GC apoptosis. A Overexpression efficiency assay. GCs were transfected with miR-423-specific mimics, miR-423 level was detected by qPCR. B–E Detection of GC apoptosis rate. Q1, injured or dead cells. Q2, early apoptosis cells. Q3, live cells. Q4, late apoptosis cells. GCs were transfected with miR-423-specific mimics, apoptosis was detected by flow cytometry (B), and total apoptosis rate (C), early apoptosis rate (D) and late apoptosis rate (E) was calculated, respectively. NC, negative control. Data represented are mean ± SEM for three independent experiments. *P < 0.05. ns, not significant
SMAD7 is a potential target of miR-423
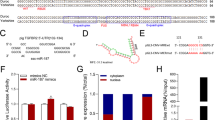
Next, we characterised the porcine miR-423, ssc-miR-423, transcribed from intron 1 of the protein-encoding gene NSRP1 on pig chromosome 12 (Fig. S1). The precursor sequence of ssc-miR-423 is 80 bp long and is highly homologous with the precursor sequence of other vertebrate miR-423, particularly the mature and seed sequences (Fig. 3A). Subcellular localization analysis showed that miR-423 mainly exists in the cytoplasm of sow GCs (Fig. 3B), indicating that miR-423 suppresses GC apoptosis, possibly through an RNA interference mechanism. Based on this assumption, we employed three algorithms to predict the potential targets of miR-423 with putative MREs in the 3′-UTR. A total of 943 potential targets were identified (Fig. 3C), which were significantly enriched in FA- and GC apoptosis-related signaling pathways, such as the oestrogen, insulin, and Ras signaling pathways (Fig. 3D–E). Interestingly, among the 13 common potential targets observed, SMAD7 is a well-known inductor of sow GC apoptosis [22, 24]. RNAhybrid prediction showed that miR-423 has a strong binding ability to the porcine SMAD7 3′-UTR (Fig. 3F), indicating that SMAD7 is a potential target of miR-423 in swine.
SMAD7 is a potential target of miR-423. A Alignments of pre-miR-423 sequences in mammals. B Subcellular distribution of miR-423 in GCs. The levels of internal reference genes (U6 in the nucleus and GAPDH in the cytoplasm) and miR-423 were detected by qPCR, respectively. C–E Potential function prediction of miR-423. The potential targets of miR-423 were predicted by three algorithms (TarBase, miRDB, TargetScan) (C), GO (D) and KEGG (E) analysis with miR-423 targets were performed, respectively. F The minimal free energy (mfe) between miR-423 and the 3'-UTR of the porcine SMAD7 gene was predicted by RNAhybrid. The heatmap shows the −log10 (P value) of the terms. Data represented are mean for three independent experiments
SMAD7 directly targets miR-423 in GCs
An MRE of miR-423 was discovered at 2,062–2,084 nt in the porcine SMAD7 3'-UTR (NM_001244175), which is fully complementary to the seed sequence of miR-423 (Fig. 4A). To determine whether miR-423 targets SMAD7, two reporter vectors of SMAD7 3′-UTR were prepared (Fig. 4B). Co-transfection assay with miR-423 mimics revealed that miR-423 induced downregulation of the reporter vector of SMAD7 3′-UTR, whereas it had no effect on the reporter with mutated MRE of miR-423 (Fig. 4C), suggesting that SMAD7 is a direct target of miR-423. Compared with the control group, miR-423-treated sow GCs had significantly diminished SMAD7 levels (Fig. 4D and E, and Fig. S2), suggesting that miR-423 can directly interact with SMAD7 in sow GCs.
SMAD7 is a direct target of miR-423 in GCs. A Schematic showing the MRE of miR-423 in the 3'UTR of the porcine SMAD7 gene. B Schematic annotation of the reporter vectors of SMAD7 3'UTR with wild-type and mutated MRE of miR-423. C Luciferase assay. GCs were co-transfected with the reporter vectors and miR-423-specific mimics, luciferase activity was determined. D–E miR-423 regulates SMAD7 levels in GCs. GCs were transfected with miR-423-specific mimics, qPCR and western blotting were performed to detect the levels of SMAD7 mRNA (D) and protein (E). wt, wild type; mut, mutation; NC, negative control. Data represented are mean ± SEM for three independent experiments. **P < 0.01
miR-423 suppresses GC apoptosis through SMAD7
The above results indicated that miR-423 represses GC apoptosis and targets SMAD7 in GCs. To further determine whether miR-423 supresses GC apoptosis via SMAD7, miR-423-specific mimics and SMAD7 overexpression vector were co-transfected into GCs. Flow cytometry showed that SMAD7 impeded the function of miR-423 (Fig. 5A), which is inhibition of cell apoptosis, and total apoptosis, early apoptosis, and late apoptosis exhibited the same trend (Fig. 5B–D). This suggested that miR-423 regulates apoptosis by suppressing SMAD7 expression in sow GCs.
miR-423 regulates GC apoptosis through SMAD7. GCs were co-transfected with miR-423-specific mimics and SMAD7 overexpression vector, and apoptosis was detected by flow cytometry (A), and total apoptosis rate (B), early apoptosis rate (C) and late apoptosis rate (D) was calculated, respectively. oe, overexpression; NC, negative control. Data represented are mean ± SEM for three independent experiments. ***P < 0.001. **P < 0.01. *P < 0.05
lncRNA NORHA sponges miR-423 in GCs
Interestingly, an MRE of miR-423 was discovered at 693–728 nt in the porcine NORHA transcript (Fig. 6A), a long non-coding RNA (lncRNA) related to sow FA [23]. RNAhybrid demonstrated that miR-423 has a strong binding ability to the porcine NORHA transcript (Fig. 6B). To test whether NORHA interacts with miR-423 in sow GCs, RNA pull-down experiments were performed using a biotin-labelled NORHA probe, and physical interactions between NORHA and miR-423 were observed (Fig. 6C). Furthermore, luciferase assay revealed that miR-423 downregulated the reporter of NORHA with the wild-type but not the mutated MRE of miR-423 (Fig. 6E), indicating that NORHA interacts with miR-423 via its MRE motif. Additionally, NORHA suppressed the expression of SMAD7 targets by miR-423 in GCs (Fig. 6F–G). Taken together, these results suggest that the lncRNA NORHA is a ceRNA of miR-423 in sow GCs.
NORHA is an ceRNA of miR-423 in GCs. A Schematic showing the MRE of miR-423 in the porcine NORHA transcript. B The minimal free energy (mfe) between miR-423 and NORHA tanscript was predicted by RNAhybrid. C RNA pull-down analysis of the physical interaction miR-423 and NORHA transcript in GCs. D Schematic annotation of the reporter vectors of NORHA with the wild-type and mutated MRE of miR-423. E Luciferase assay. KGN cells were co-transfected with the reporter vectors and miR-423-specific mimics, luciferase activity was determined. F–G NORHA regulates SMAD7 expression in GCs. GCs were transfected with the pcDNA3.1-NORHA, the levels of NORHA (F), and SMAD7 mRNA (G) were detected by qPCR. wt, wild type; mut, mutation; NC, negative control. Data represented are mean ± SEM for three independent experiments. *P < 0.05. ns, not significant
miR-423 mediated the regulation of SMAD7 levels and GC apoptosis by NORHA
Next, we investigated whether miR-423 mediates the promotion of SMAD7 expression by NORHA. Co-transfection with miR-423-specific mimics and the NORHA overexpression vector revealed that miR-423 mimics reversed the upregulation of SMAD7 caused by NORHA overexpression (Fig. 7A–B, and Fig. S3), indicating that NORHA enhanced SAMD7 expression through miR-423 sponging. SMAD7 is an antagonist of the TGF-β pathway; therefore, we investigated whether the activity of the TGF-β pathway is regulated by the NORHA/miR-423 axis. NORHA significantly diminished the levels of phosphorylated SMAD3 (p-SMAD3), the marker for the activity of the TGF-β pathway; however, this process was reversed by the miR-423 mimics (Fig. 7C and Fig. S4), indicating that NORHA inactivates the TGF-β pathway through miR-423 sponging. Both NORHA [23] and miR-423 are pro-apoptotic factors in GCs; therefore, we speculated that NORHA induces GC apoptosis through miR-423 sponging. As expected, the co-transfection experiment using miR-423-specific mimics and the NORHA overexpression vector confirmed that miR-423 repressed NORHA-induced GC apoptosis (Fig. 7D–G). These data suggest that NORHA induces SMAD7 expression and cell apoptosis through miR-423 sponging in sow GCs.
miR-423 mediated NORHA regulation of SMAD7 levels and GC apoptosis. A–C GCs were co-transfected with the pcDNA3.1-NORHA and miR-423 mimics, and the levels of SMAD7 mRNA (A) were detected by qPCR, and the levels of SMAD7 protein (B) and p-SMAD3 protein (C) were detected by western blotting. D–G GCs were co-transfected with the pcDNA3.1-NORHA and miR-423 mimics, and apoptosis was detected by flow cytometry (D), and total apoptosis rate (E), early apoptosis rate (F) and late apoptosis rate (G) was calculated, respectively. oe, overexpression; NC, negative control. Data represented are mean ± SEM for three independent experiments. *P < 0.05
miR-423 is a candidate gene for sow NSB trait
miR-423 was located in three QTLs (ID: 4,577, 18,349, and 21,839) for sow fertility traits, which were identified by searching a pig QTL database (Fig. 8A). To ascertain whether miR-423 is a candidate gene for sow fertility traits, we first scanned miR-423 variants using pooled-DNA sequencing. A T ins/del was observed at –703 nt in the promoter, and three genotypes, ins/ins (II), ins/del (ID), and del/del (DD), and two alleles, I and D, were identified in Yorkshire sows (Fig. 8B–D). Association analysis found that the NSB of DD sows was 44.4% (0.32/0.72) lower than that of II sows for primiparity (P < 0.05) (Fig. 8E–H). Together, these results suggest that miR-423 is a candidate gene for the NSB trait in Yorkshire sows.
miR-423 is a candidate gene for sow fertility traits. A Schematic diagram of the location of miR-423 in QTLs for sow fertility traits. B A T ins/del was identified at −703 nt in the promoter of miR-423. C Peak plot of three genotypes for the variant g.−703indelT. D–G Association analysis between polymorphism of the variant g.−703indelT with the TNB trait (D), the NBA trait (E), the NSB trait (F) and the LW trait (G) of Yorkshire sows for primiparity. TNB, the total number of piglets born; NBA, the total number of piglets born alive; NSB, the number of stillbirths; LW, the litter weight. n = 369. Data are represented as the least squares means ± SEM. *P < 0.05
Discussion
Apoptosis of GCs in follicles is the basic physiological mechanism underlying FA, a condition that affects female fertility. Thus, the mechanism behind GC apoptosis and its regulation have gained increasing attention in the field of female reproduction. GC apoptosis has been reported to be controlled by several factors, including hormones, cytokines, components of the cell death pathway, and ncRNAs like miRNAs, lncRNAs, and circRNAs [25,26,27,28]. Among these ncRNAs, miRNAs are widely involved in the regulation of GC apoptosis in various mammals, such as miR-23a and miR-27a in humans [29], miR-126-3p in rats [30], miR-21 in cattle [31], miR-450-5p and miR-202-5p in goats [32], miR-346 in sheep [33], and miR-143 and miR-29c in sows [34, 35]. However, the vast majority of these miRNAs induce GC apoptosis and FA, and only few miRNAs have inhibitory effects and can be used as potential small-molecule regulators to promote follicular development and ovulation and improve female fertility [25, 27]. In this study, we identified a new inhibitor of sow GC apoptosis and FA initiation: miR-423. In other normal cell types such as bone marrow mesenchymal stem cells [36], cardiomyocyte [37], and human umbilical vein endothelial cells [38], miR-423 has been shown to function as an anti-apoptotic modulator. Thus, our study reports a potential small molecule for alleviating FA and improving female fertility. It may even be developed as a potential non-hormonal small-nucleotide drug for treating human ovarian diseases, as it has a highly conserved sequence among mammals.
Generally, miRNAs induce target mRNA decay or prevent target mRNA translation by recognising and interacting with its 3′-UTR through the assemblage of miRISCs with argonaut proteins (AGO1 to AGO4) and other silencing factors [39, 40]. Here, we identified that SMAD7 is a target of miR-423 in sow GCs and mediates the regulatory effect of miR-423 on the activity of the TGF-β pathway and cell apoptosis. SMAD7 is an inhibitory SMAD protein in the TGF-β pathway, which is strongly related to mammalian ovarian functions including GC states (proliferation or apoptosis) and cycle, secretion of steroid hormones (e.g., E2, P4, and androgen), oocyte-GC communication, and folliculogenesis [22, 41,42,43,44]. In Tibetan sheep, miR-21 and let-7b, two miRNAs involved in FA and GC apoptosis, have been found to suppress SMAD7 expression in GCs [45]. In addition to miR-423, SMAD7 is a functional target of other miRNAs related to FA and GC apoptosis, such as miR-92a [22], miR-181b [24], and miR-21–5p [44]. miR-424 and miR-503 directly target SMAD7 to control the proliferation and cycle progression of GCs in bovine [43]. These findings suggest that SMAD7 is a vital mediator and hub gene of the miRNA network regulating GC functions, FA, and female fertility. Furthermore, our findings report a potential small-molecule activator of the TGF-β pathway for scientific research and clinical practice.
LncRNAs are ncRNAs that are more than 200 nt in length. They often block the inhibition of miRNAs on target mRNAs by acting as ceRNAs of miRNAs in the cytoplasm [10, 46]. NORHA, a cytoplasmic lncRNA in sow GCs, is a potential ceRNA of multiple miRNAs that are differentially expressed during sow FA [23]. In this study, we demonstrated that cytoplasmic NORHA maintains the levels of SMAD7 (a target of miR-423) and its function in inducing apoptosis in sow GCs by miR-423 sponging. miR-96, miR-182, miR-183, and miR-187 have been previously confirmed to be sponged by NORHA in sow GCs [5, 23]. miR-423 is also sponged by other lncRNAs in other cell types, such as LINC00680 in oesophageal squamous cell carcinoma cells [46] and AFAP1-AS1 in nasopharyngeal carcinoma cells [47]. A recent report showed that circRNA-0000081 maintains the levels of PDPK1 (a target of miR-423) in gastric cancer cells via miR-423 sponging, thereby affecting cell proliferation, migration, and invasion potential [48]. In the present study, we discovered a new signaling pathway, the NORHA/miR-423/SMAD7 pathway, that contributes to FA and GC apoptosis and enriches the ceRNA network of the NORHA-miRNAs in GCs.
Increasing evidence indicates that miRNAs also serve as candidate genes for many important traits, including fertility and disease traits [25, 49, 50]; however, they have shorter sequences and relatively fewer variants than protein-encoding genes and lncRNAs. In domestic animals, some miRNA variants are associated with economically important traits. For example, g.17104G > A of miR-208b is associated with the proportions of type I and IIb fibre numbers and drip loss in Berkshire, Landrace, and Yorkshire pigs [51], a G > A variant of miR-22 is related to meat colour parameters in Suhuai pigs [52], g.–113C > A of miR-9 is related to litter size in Markhoz goats [53], and g.–13G > A and g.–24T > G of miR-27a-3p are associated with litter size in Hu sheep [54]. In the present study, we identified a novel variant (g.–703indelT) of miR-423, an inhibitor of sow GC apoptosis, that is markedly associated with the NSB trait. miR-423 is the second miRNA candidate identified to be associated with fertility traits in swine. Recently, miR-23a has also been reported as a candidate miRNA, because its variants (g.–398C > T, g.–283G > C, and g.–271C > T) are associated with sow fertility traits [25, 55]. Additionally, because miR-423 is the first candidate gene identified among the three QTLs (ID: 4,577, 18,349, and 21,839) for sow fertility traits, these QTLs are miRNA-related QTLs (mirQTLs).
Conclusions
To conclude, we demonstrate that miR-423 is involved in the sow FA initiation by repressing the early apoptosis of GCs. SMAD7 and NORHA was identified as a new target and a new ceRNA of miR-423, and the three form a signaling pathway that controls FA and GC apoptosis (Fig. S5). Importantly, we demonstrate that miR-423 is a new candidate miRNA for sow fertility traits.
These results demonstrate that miR-423 is a potential small molecule for inhibiting FA initiation and GC apoptosis, suggesting that treating with miR-423 may be a novel approach for inhibiting FA initiation and improving female fertility.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AF:
-
Atretic follicle
- ceRNA:
-
Competing endogenous RNA
- FA:
-
Follicular atresia
- GC:
-
Granulosa cell
- HF:
-
Healthy follicle
- LW:
-
Litter weight
- miRNA:
-
MicroRNA
- miRISC:
-
miRNA-induced silencing complexe
- MRE:
-
miRNA recognition element
- NBA:
-
Total number of piglets born alive
- ncRNA:
-
Noncoding RNA
- NSB:
-
Number of stillbirths
- TNB:
-
Total number of piglets born
References
Lee YY, Kim H, Kim VN. Sequence determinant of small RNA production by DICER. Nature. 2023;615(7951):323–30. https://doi.org/10.1038/s41586-023-05722-4.
Bartel DP. Metazoan microRNAs. Cell. 2018;173(1):20–51. https://doi.org/10.1016/j.cell.2018.03.006.
Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478):eaau6977. https://doi.org/10.1126/science.aau6977.
Garcia-Martin R, Wang G, Brandao BB, Zanotto TM, Shah S, Kumar Patel S, et al. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature. 2022;601(7893):446–51. https://doi.org/10.1038/s41586-021-04234-3.
Yang L, Du X, Wang S, Lin C, Li Q, Li Q. A regulatory network controlling ovarian granulosa cell death. Cell Death Discov. 2023;9:70. https://doi.org/10.1038/s41420-023-01346-9.
Zhou T, He C, Lai P, Yang Z, Liu Y, Xu H, et al. miR-204-containing exosomes ameliorate GVHD-associated dry eye disease. Sci Adv. 2022;8(2):eabj9617. https://doi.org/10.1126/sciadv.abj9617.
Rivera J, Gangwani L, Kumar S. Mitochondria localized microRNAs: an unexplored miRNA niche in Alzheimer’s disease and aging. Cells. 2023;12(5):742. https://doi.org/10.3390/cells12050742.
Lee YY, Lee H, Kim H, Kim VN, Roh SH. Structure of the human DICER-pre-miRNA complex in a dicing state. Nature. 2023;615(7951):331–8. https://doi.org/10.1038/s41586-023-05723-3.
Liu Y, Yang Y, Xu C, Liu J, Chen J, Li G, et al. Circular RNA circGlis3 protects against islet beta-cell dysfunction and apoptosis in obesity. Nat Commun. 2023;14:351. https://doi.org/10.1038/s41467-023-35998-z.
Zhang C, Yi X, Hou M, Li Q, Li X, Lu L, et al. The landscape of m1A modification and its posttranscriptional regulatory functions in primary neurons. Elife. 2023;12(10):e85324. https://doi.org/10.7554/eLife.85324.
D’Souza A, Pearman CM, Wang Y, Nakao S, Logantha S, Cox C, et al. Targeting miR-423-5p reverses exercise training-induced HCN4 channel remodeling and sinus bradycardia. Circ Res. 2017;121(9):1058–68. https://doi.org/10.1161/CIRCRESAHA.117.311607.
Xiao B, Wang Y, Li W, Baker M, Guo J, Corbet K, et al. Plasma microRNA signature as a noninvasive biomarker for acute graft-versus-host disease. Blood. 2013;122(19):3365–75. https://doi.org/10.1182/blood-2013-06-510586.
Farr RJ, Rootes CL, Rowntree LC, Nguyen THO, Hensen L, Kedzierski L, et al. Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection. PLoS Pathog. 2021;17(7):e1009759. https://doi.org/10.1371/journal.ppat.1009759.
Xue P, Huang S, Han X, Zhang C, Yang L, Xiao W, et al. Exosomal miR-101-3p and miR-423-5p inhibit medulloblastoma tumorigenesis through targeting FOXP4 and EZH2. Cell Death Differ. 2022;29:82–95. https://doi.org/10.1038/s41418-021-00838-4.
Wang Q, Wang H, Yan H, Tian H, Wang Y, Yu W, et al. Suppression of osteoclast multinucleation via a posttranscriptional regulation-based spatiotemporally selective delivery system. Sci Adv. 2022;8(26):eabn3333. https://doi.org/10.1126/sciadv.abn3333.
Xie S, Zhang Q, Zhao J, Hao J, Fu J, Li Y. MiR-423-5p may regulate ovarian response to ovulation induction via CSF1. Reprod Biol Endocrinol. 2020;18(1):26. https://doi.org/10.1186/s12958-020-00585-0.
Lv Y, Gao S, Zhang Y, Wang L, Chen X, Wang Y. miRNA and target gene expression in menstrual endometria and early pregnancy decidua. Eur J Obstet Gynecol Reprod Biol. 2016;197:27–30. https://doi.org/10.1016/j.ejogrb.2015.11.003.
Guo L, Liu Y, Guo Y, Yang Y, Chen B. MicroRNA-423-5p inhibits the progression of trophoblast cells via targeting IGF2BP1. Placenta. 2018;74:1–8. https://doi.org/10.1016/j.placenta.2018.12.003.
Guo Y, Sun J, Lai D. Role of microRNAs in premature ovarian insufficiency. Reprod Biol Endocrinol. 2017;15(1):38. https://doi.org/10.1186/s12958-017-0256-3.
Xu X, Guan R, Gong K, Xie H, Shi L. Circ_FURIN knockdown assuages Testosterone-induced human ovarian granulosa-like tumor cell disorders by sponging miR-423-5p to reduce MTM1 expression in polycystic ovary syndrome. Reprod Biol Endocrinol. 2022;20(1):32. https://doi.org/10.1186/s12958-022-00891-9.
Xie S, Batnasan E, Zhang Q, Li Y. MicroRNA expression is altered in granulosa cells of ovarian hyperresponders. Reprod Sci. 2016;23(8):1001–10. https://doi.org/10.1177/1933719115625849.
Liu J, Yao W, Yao Y, Du X, Zhou J, Ma B, et al. MiR-92a inhibits porcine ovarian granulosa cell apoptosis by targeting Smad7 gene. FEBS Lett. 2014;588(23):4497–503. https://doi.org/10.1016/j.febslet.2014.10.021.
Yao W, Pan Z, Du X, Zhang J, Liu H, Li Q. NORHA, a novel follicular atresia-related lncRNA, promotes porcine granulosa cell apoptosis via the miR-183-96-182 cluster and FoxO1 axis. J Anim Sci Biotechnol. 2021;12:103. https://doi.org/10.1186/s40104-021-00626-7.
Yao W, Pan Z, Du X, Zhang J, Li Q. miR-181b-induced SMAD7 downregulation controls granulosa cell apoptosis through TGF-beta signaling by interacting with the TGFBR1 promoter. J Cell Physiol. 2018;233(9):6807–21. https://doi.org/10.1002/jcp.26431.
Wang S, Li Y, Zeng Q, Yang L, Du X, Li Q. A mutation in endogenous saRNA miR-23a influences granulosa cells response to oxidative stress. Antioxidants (Basel). 2022;11(6):1174. https://doi.org/10.3390/antiox11061174.
Fan HY, Liu Z, Shimada M, Sterneck E, Johnson PF, Hedrick SM, et al. MAPK3/1 (ERK1/2) in ovarian granulosa cells are essential for female fertility. Science. 2009;324(5929):938–41. https://doi.org/10.1126/science.1171396.
Zhou X, He Y, Pan X, Quan H, He B, Li Y, et al. DNMT1-mediated lncRNA IFFD controls the follicular development via targeting GLI1 by sponging miR-370. Cell Death Differ. 2023;30(2):576–88. https://doi.org/10.1038/s41418-022-01103-y.
Stringer JM, Alesi LR, Winship AL, Hutt KJ. Beyond apoptosis: evidence of other regulated cell death pathways in the ovary throughout development and life. Hum Reprod Update. 2023;29(4):434–56. https://doi.org/10.1093/humupd/dmad005.
Nie M, Yu S, Peng S, Fang Y, Wang H, Yang X. miR-23a and miR-27a promote human granulosa cell apoptosis by targeting SMAD5. Biol Reprod. 2015;93(4):98. https://doi.org/10.1095/biolreprod.115.130690.
Qu Q, Liu L, Cui Y, Liu H, Yi J, Bing W, et al. miR-126-3p containing exosomes derived from human umbilical cord mesenchymal stem cells promote angiogenesis and attenuate ovarian granulosa cell apoptosis in a preclinical rat model of premature ovarian failure. Stem Cell Res Ther. 2022;13(1):352. https://doi.org/10.1186/s13287-022-03056-y.
Sabry R, Williams M, Werry N, LaMarre J, Favetta LA. BPA decreases PDCD4 in bovine granulosa cells independently of miR-21 inhibition. Int J Mol Sci. 2022;23(15):8276. https://doi.org/10.3390/ijms23158276.
Feng G, Liu J, Lu Z, Li Y, Deng M, Liu G, et al. miR-450-5p and miR-202-5p synergistically regulate follicle development in black goat. Int J Mol Sci. 2022;24(1):401. https://doi.org/10.3390/ijms24010401.
Yao X, El-Samahy MA, Li X, Bao Y, Guo J, Yang F, et al. LncRNA-412.25 activates the LIF/STAT3 signaling pathway in ovarian granulosa cells of Hu sheep by sponging miR-346. FASEB J. 2022;36(9):e22467.
Du X, Liu L, Wu W, Li P, Pan Z, Zhang L, et al. SMARCA2 is regulated by NORFA-miR-29c, a novel pathway that controls granulosa cell apoptosis and is related to female fertility. J Cell Sci. 2020;133(23):jcs249961.
Du X, Zhang L, Li X, Pan Z, Liu H, Li Q. TGF-beta signaling controls FSHR signaling-reduced ovarian granulosa cell apoptosis through the SMAD4/miR-143 axis. Cell Death Dis. 2016;7(11):e2476. https://doi.org/10.1038/cddis.2016.379.
Yang S, Guo J, Zhou L, Xing H, Wang X, Dong C. miR-148b-3p, miR-337-5p and miR-423-5p expression in alveolar ridge atrophy and their roles in the proliferation and apoptosis of OMMSCs. Exp Ther Med. 2018;16(6):5334–42. https://doi.org/10.3892/etm.2018.6850.
Luo H, Li X, Li T, Zhao L, He J, Zha L, et al. microRNA-423-3p exosomes derived from cardiac fibroblasts mediates the cardioprotective effects of ischaemic post-conditioning. Cardiovasc Res. 2019;115(7):1189–204. https://doi.org/10.1093/cvr/cvy231.
Zhao X, Wang C, Liu M, Meng F, Liu K. LncRNA FENDRR servers as a possible marker of essential hypertension and regulates human umbilical vein endothelial cells dysfunction via miR-423-5p/Nox4 axis. Int J Gen Med. 2022;15:2529–40. https://doi.org/10.2147/IJGM.S338147.
Srinivasan B, Samaddar S, Mylavarapu SVS, Clement JP, Banerjee S. Homeostatic scaling is driven by a translation-dependent degradation axis that recruits miRISC remodeling. PLoS Biol. 2021;19(11):e3001432. https://doi.org/10.1371/journal.pbio.3001432.
Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436(7051):740–4. https://doi.org/10.1038/nature03868.
Young JM, McNeilly AS. Inhibin removes the inhibitory effects of activin on steroid enzyme expression and androgen production by normal ovarian thecal cells. J Mol Endocrinol. 2012;48(1):49–60. https://doi.org/10.1530/JME-11-0134.
Gao Y, Wen H, Wang C, Li Q. SMAD7 antagonizes key TGFbeta superfamily signaling in mouse granulosa cells in vitro. Reproduction. 2013;146(1):1–11. https://doi.org/10.1530/REP-13-0093.
Pande HO, Tesfaye D, Hoelker M, Gebremedhn S, Held E, Neuhoff C, et al. MicroRNA-424/503 cluster members regulate bovine granulosa cell proliferation and cell cycle progression by targeting SMAD7 gene through activin signalling pathway. J Ovarian Res. 2018;11(1):34. https://doi.org/10.1186/s13048-018-0410-3.
Zhang X, Chen Y, Yang M, Shang J, Xu Y, Zhang L, et al. MiR-21-5p actions at the Smad7 gene during pig ovarian granulosa cell apoptosis. Anim Reprod Sci. 2020;223:106645. https://doi.org/10.1016/j.anireprosci.2020.106645.
Zhang T, Huo S, Wei S, Cui S. miR-21, miR-125b, and let-7b contribute to the involution of atretic follicles and corpus lutea in Tibetan sheep ovaries. Anim Sci J. 2022;93(1):e13756. https://doi.org/10.1111/asj.13756.
Xue ST, Zheng B, Cao SQ, Ding JC, Hu GS, Liu W, et al. Long non-coding RNA LINC00680 functions as a ceRNA to promote esophageal squamous cell carcinoma progression through the miR-423-5p/PAK6 axis. Mol Cancer. 2022;21(1):69. https://doi.org/10.1186/s12943-022-01539-3.
Lian Y, Xiong F, Yang L, Bo H, Gong Z, Wang Y, et al. Long noncoding RNA AFAP1-AS1 acts as a competing endogenous RNA of miR-423-5p to facilitate nasopharyngeal carcinoma metastasis through regulating the Rho/Rac pathway. J Exp Clin Cancer Res. 2018;37(1):253. https://doi.org/10.1186/s13046-018-0918-9.
Jiang F, Hu X, Cao H, Shen X. Hsa_circ_0000081 promotes the function of gastric cancer through sponging hsa-miR-423-5p to influence 3-phosphoinositide-dependent kinase 1 expression. Bioengineered. 2022;13(4):8277–90. https://doi.org/10.1080/21655979.2022.2053796.
Zhao D, Wu K, Sharma S, Xing F, Wu SY, Tyagi A, et al. Exosomal miR-1304-3p promotes breast cancer progression in African Americans by activating cancer-associated adipocytes. Nat Commun. 2022;13:7734. https://doi.org/10.1038/s41467-022-35305-2.
Bustos BI, Billingsley K, Blauwendraat C, Gibbs JR, Gan-Or Z, Krainc D, et al. Genome-wide contribution of common short-tandem repeats to Parkinson’s disease genetic risk. Brain. 2023;146(1):65–74. https://doi.org/10.1093/brain/awac301.
Kim JM, Lim KS, Hong JS, Kang JH, Lee YS, Hong KC. A polymorphism in the porcine miR-208b is associated with microRNA biogenesis and expressions of SOX-6 and MYH7 with effects on muscle fibre characteristics and meat quality. Anim Genet. 2015;46(1):73–7. https://doi.org/10.1111/age.12255.
Wang H, Shen Z, Huang R, Zhao A, Jiang J, Li P, et al. A polymorphism in porcine miR-22 is associated with pork color. Front Vet Sci. 2022;9:939440. https://doi.org/10.3389/fvets.2022.939440.
Mahmoudi P, Rashidi A, Rostamzadeh J, Razmkabir M. A novel variant in the promoter region of miR-9 gene strongly affects litter size in Markhoz goats. Theriogenology. 2020;158:50–7. https://doi.org/10.1016/j.theriogenology.2020.09.008.
Gui H, Li F, Chen C, Zhu Q, Zhang C, Zhang J, et al. miR-27a-3p targets NR5A2 to regulate CYP19A1 expression and 17-beta estradiol synthesis in ovine granulosa cells. Anim Reprod Sci. 2023;248:107160. https://doi.org/10.1016/j.anireprosci.2022.107160.
Wang S, Li Y, Du X, Li Q. Two single nucleotide variants in the miR-23a promoter affect granulosa cell apoptosis. Anim Genet. 2023;54:207–10. https://doi.org/10.1111/age.13284.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Key R&D Program of China (2022YFD1600903), the National Natural Science Foundation of China (32072693), and the College Students’ Innovative Entrepreneurial Training Plan Program (202110307028).
Author information
Authors and Affiliations
Contributions
Conception and design: QL. Collection and assembly of data: YL, ZZ, SW and LY. Data analysis and interpretation: YL, SW and XD. Manuscript writing: YL and QL. Final approval of manuscript: All authors.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All sow experiments were approved by the Animal Ethics Committee at Nanjing Agriculture University, China.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
Supplementary Information
Additional file 1.
Table S1. Primers designed for reverse-transcription and qPCR. Table S2. Oligonucleotides used in this study.
Additional file 2. Fig. S1.
Chromosome and genome localization of miR-423 in mammals. Fig. S2. Western blot images of miR-423 regulates SMAD7 levels in GCs. Fig. S3. Western blot images of miR-423 mediated NORHA regulation of SMAD7 levels in GCs. Fig. S4. Western blot images of miR-423 mediated the regulation of p-SMAD3 levels by NORHA in GCs. Fig. S5. Working model of miR-423 in sow GCs.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, Y., Zhang, Z., Wang, S. et al. miR-423 sponged by lncRNA NORHA inhibits granulosa cell apoptosis. J Animal Sci Biotechnol 14, 154 (2023). https://doi.org/10.1186/s40104-023-00960-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40104-023-00960-y