Abstract
The physiological and molecular responses of five strains of Streptomyces sp. (CAI-17, CAI-68, CAI-78, KAI-26 and KAI-27), with their proven potential for charcoal rot disease control in sorghum and plant growth-promotion (PGP) in sorghum and rice, were studied to understand the mechanisms causing the beneficial effects. In this investigation, those five strains were evaluated for their PGP capabilities in chickpea in the 2012–13 and 2013–14 post-rainy seasons. All of the Streptomyces sp. strains exhibited enhanced nodule number, nodule weight, root weight and shoot weight at 30 days after sowing (DAS) and pod number, pod weight, leaf area, leaf weight and stem weight at 60 DAS in both seasons over the un-inoculated control. At crop maturity, the Streptomyces strains had enhanced stover yield, grain yield, total dry matter and seed number plant−1 in both seasons over the un-inoculated control. In the rhizosphere, the Streptomyces sp. also significantly enhanced microbial biomass carbon, dehydrogenase activity, total nitrogen, available phosphorous and organic carbon in both seasons over the un-inoculated control. Of the five strains of Streptomyces sp., CAI-17, CAI-68 and CAI-78 were superior to KAI-26 and KAI-27 in terms of their effects on root and shoot development, nodule formation and crop productivity. Scanning electron microscopy (SEM) micrographs had revealed the success in colonization of the chickpea roots by all five strains. Quantitative real-time PCR (qRT-PCR) analysis of selected PGP genes of actinomycetes revealed the selective up-regulation of indole-3-acetic acid (IAA)-related and siderophore-related genes by CAI-68 and of β-1,3-glucanase genes by KAI-26.
Similar content being viewed by others
Introduction
In recent years, the use of plant growth-promoting (PGP) bacteria has become one of the most attractive options for enhancing the sustainability of agricultural systems in many parts of the world due to their eco-friendliness, low production cost and reduced use of non-renewable resources. PGP bacteria promote plant growth through various mechanisms such as nitrogen fixation, the solubilization of phosphorus, the chelation of iron, the secretion of plant growth hormones and the inhibition of phytopathogens. Bacteria such as Bacillus sp., Pseudomonas sp. and Streptomyces sp. were found to play a major role both in mobilizing and acquiring nutrients and in controlling phytopathogens (Postma et al. 2003; Perner et al. 2006; Gopalakrishnan et al. 2011a, b, c; Jalilian et al. 2012). The presence of PGP bacteria in the rhizosphere has been shown to enhance shoot growth, root growth, root hair development, plant hormone concentrations, nitrogen fixation, the solubilization of minerals and the suppression of pathogens (Shaukat et al. 2006; Lucas et al. 2009; Richardson et al. 2009; Gopalakrishnan et al. 2012a, b). The PGP potential of Streptomyces sp. has been demonstrated on tomato, wheat, rice, bean and pea (Tokala et al. 2002; Nassar et al. 2003; El-Tarabily 2008; Sadeghi et al. 2012; Gopalakrishnan et al. 2013, 2014a). Streptomyces sp. are known to promote plant growth either by producing indole-3-acetic acid (IAA) and siderophores and/or by inhibiting soil-borne fungal pathogens (Aldesuquy et al. 1998; Trejo-Estrada et al. 1998; Tokala et al. 2002; Macagnan et al. 2008; Gopalakrishnan et al. 2011a, b, 2013; 2014a).
Previously, we have demonstrated the potential of a set of eight Streptomyces strains (CAI-21, CAI-26, MMA-32, CAI-17, CAI-68, CAI-78, KAI-26 and KAI-27) isolated from herbal vermicompost, for the bio-control of charcoal rot disease, caused by Macrophomina phaseolina (Tassi) Goid., in sorghum (Gopalakrishnan et al. 2011b) and for PGP in rice (Gopalakrishnan et al. 2012a, 2013). The objectives of this investigation were to further evaluate five of the eight Streptomyces strains (CAI-17, CAI-68, CAI-78, KAI-26 and KAI-27) for their PGP traits in chickpea under field conditions, to demonstrate gene expression profiles by qRT-PCR analysis, and to ensure colonizing ability in chickpea by scanning electron microscopy (SEM) analysis.
Materials and methods
Streptomyces strains
Five strains of Streptomyces spp., CAI-17 (GenBank accession number: JQ682619), CAI-68 (GenBank accession number: JQ682622), CAI-78 (GenBank accession number: JQ682623), KAI-26 (GenBank accession number: JQ682624) and KAI-27 (GenBank accession number: JQ682625), previously reported by us to have capacity for the bio-control of charcoal rot in sorghum (Gopalakrishnan et al. 2011b) and PGP in rice (Gopalakrishnan et al. 2013), were further studied in the present investigation.
Evaluation of Streptomyces sp. for PGP potential on chickpea under field conditions
This experiment was carried out during the 2012–13 and 2013–14 post-rainy cropping seasons at ICRISAT, Patancheru (17°30′ N; 78°16′ E; altitude 549 m) in peninsular India. Soils at the experimental site are classified as Vertisols (fine montmorillonitic isohyperthermic typic pallustert) having 52% clay, 21% silt and 26% sand, with an alkaline pH of 7.7 − 8.5 and an organic carbon content of 0.4 − 0.6%. The soil depth of the field used was ≥ 1.2 m, and this soil retained approximately 200 mm of plant-available water in a 120-cm (maximum rooting depth by chickpea) soil profile. The mineral content of the top 15 cm of rhizosphere soil includes 24.7 mg kg−1 soil of available nitrogen, 8.6 mg kg−1 soil of available phosphorous and 298 mg kg−1 soil of available potassium. The field was kept fallow except for this post-rainy season crop. The fields were prepared into broad beds and furrows with beds 1.2 m wide flanked by 0.3-m furrows in both seasons. Surface application and incorporation of 18 kg N ha−1 and 20 kg P ha−1 as di-ammonium phosphate (DAP) were performed in both seasons. The experiment was laid out in a randomized complete block design (RCBD) with three replicates and plot sizes of 4 m × 3 ridges (rows).
The five strains of Streptomyces spp. were cultured individually on a starch casein broth at 28°C for five days. Seeds of chickpea variety ICCV 2 (with a grain yield potential of 1.1 to 1.2 t ha−1) were treated with a Streptomyces sp. (containing 108 CFU ml−1) for 50 min and sown by hand on 11 October 2012 in the first year and 1 November 2013 in the second year in rows 30 cm apart at a depth of 4–5 cm to achieve an estimated plant stand density of at least 26 plants m−2. Streptomyces sp. (1000 ml; 108 CFU mL−1) was applied once every 15 days on the soil close to the plant until the flowering stage. Control plots were maintained without the application of Streptomyces strains. The plots were irrigated on the 21st and 49th days after sowing. The crop was kept weed-free by manual weeding. No serious insect pest or phytopathogen attacks were observed during the cropping period. The crop was harvested manually on 18 January 2013 in the first year and 6 Feb 2014 in the second year. In both the 2012–13 and 2013–14 seasons, the nodule number (plant−1), nodule weight (mg plant−1), root weight (mg plant−1) and shoot weight (g plant−1) were recorded at 30 days after sowing (DAS), and at 60 DAS, the plant height (cm), pod number (plant−1), pod weight (g plant−1), leaf area (cm−2 plant−1) and stem weight (g plant−1) were recorded. At both harvests, stover yield (t ha−1), grain yield (t ha−1), total dry matter (t ha−1), 1000-seed weight (g), pod weight (g plant−1), seed number (plant−1) and seed weight (g plant−1) were recorded. Soil samples were collected from the top 15 cm of the soil profile at the harvest stage and analysed for soil chemistry (total nitrogen [ppm], available phosphorous [ppm] and % organic carbon as per the protocols of Novozamsky et al. 1983, Olsen and Sommers 1982 and Nelson and Sommers 1982, respectively) and microbial biomass carbon (μg g−1 soil) by the fumigation method and dehydrogenase activity (μg TPF g−1 soil 24 h−1) by the triphenyl formazan production method as per the protocols of Anderson and Domsch 1989 and Casida 1977, respectively.
Colonization of Streptomyces sp.
The roots of chickpea were examined for colonization by Streptomyces sp. by SEM (Bozzola and Russell 1998). For this procedure, the seeds of chickpea variety ICCV 2 were surface-sterilized first with 2.5% sodium hypochlorite solution (for five minutes), followed by 70% ethanol (for five minutes), and then rinsed with sterilized water (eight times) before being allowed to sprout in a Petri plate overnight. The sprouted seeds were transferred into test Streptomyces strains (CAI-17, CAI-68, CAI-78, KAI-26 and KAI-27; grown in Bennett’s broth separately) for an hour before being sown in pots containing sterilized coarse sand (six seeds/8″ plastic pot). The soil was drenched with a booster dose of Streptomyces strains (5 ml per seedling; 108 CFU ml−1) one week after sowing in sand. The pots were incubated for two weeks in a greenhouse at a temperature of 24 ± 2°C. At the end of the incubation period, chickpea seedlings were removed from the sand pots and the roots were washed in phosphate buffer (0.1 M; pH 7.2). The tip of the roots were cut into pieces 5 mm long and fixed in 2.5% glutaraldehyde in phosphate buffer for 24 h at 4°C. At the end of incubation, the samples were washed with phosphate buffer, postfixed in 2% osmium tetroxide for 4 h and dehydrated using a graded series of ethanol. The dehydrated samples were dried with critical-point liquid carbon dioxide as a transition fluid. The dried materials were adhered onto aluminium specimen mounts with double-stick adhesive tape. The samples were later coated with gold-palladium in an automated sputter coater (JEOL JFC-1600) and examined with a scanning electron microscope (JOEL-JSM 5600) as per the standardized protocols at RUSKA lab, College of Veterinary Science, Rajendranagar, Hyderabad, India. Observations of the presence of Streptomyces spores on root surfaces were recorded.
Gene expression profile
RNA extraction
The five selected Streptomyces sp. (CAI-17, CAI-68, CAI-78, KAI-26 and KAI-27) were grown in Bennett’s broth at 28°C for 72 h. Total RNA was extracted from all of the Streptomyces strains using the “NucleoSpin® RNA Plant” kit (Macherey-Nagel). The quality and quantity of RNA was estimated by a Nanodrop (Thermo Scientific) and RNA integrity was determined using a 2100 Bioanalyzer (Agilent).
Quantitative real time- PCR (qRT-PCR)
qRT-PCR was performed using the Applied Biosystems 7500 Real-Time PCR System with the SYBR green chemistry (Applied Biosystems, USA) as per the manufacturer’s instructions. Well-characterized genes Spaepen et al. 2007 relating to IAA production (F: GTCACCGGGATCTTCTTCAAC; R: GATGTCGGTGTTCTTGTCCAG) and siderophore (F: ATCCTCAACACCCTGGTCTG; R: TCCTTGTACTGGTACGGGACTT) were collected from the UniprotKB database (http://www.uniprot.org/uniprot/) as described by Gopalakrishnan et al. (2014a). Similarly, genes for β-1,3-glucanase (F: CCGAACACCACCTACTCCAC; R: CCAGGTTGAGGATCAGGAAG) were also selected for the study. Gene-specific primers for real-time PCR were designed using Primer 3 software (Rosen and Skaletsky 2000). RNA polymerase principal sigma factor HrdB (SCO5820) (F: GGTCGAGGTCATCAACAAGC; R: CTCGATGAGGTCACCGAACT) was used as the endogenous control. qRT-PCR reactions were performed as described earlier (Gopalakrishnan et al. 2014a). The data obtained were analysed using the mean of the CT values of the three biological replicates that were normalized to the mean CT values of the endogenous gene. The expression ratios were calculated using the 2_∆∆Ct method. Relative transcription levels are presented graphically.
Statistical analysis
Analysis of variance) was performed by the GLM (General Linear Model) procedure of SAS software package (SAS Institute Inc 2013). Isolate were tested for significance and their means with control were compared by using Dunnett’s t-test.
Results
As all of the strains of Streptomyces spp. (CAI-17, CAI-68, CAI-78, KAI-26 and KAI-27) enhanced different plant component performances, the means of all the five strains are presented. The Streptomyces-treated plots, by a mean of the five strains, enhanced agronomic performance in terms of nodule numbers (up to 61%), nodule weight (up to 44%) and shoot weight (up to 28%) at 30 DAS and plant height (up to 5%), pod numbers (up to 79%), pod weight (up to 89%), leaf area (up to 29%), leaf weight (up to 42%) and stem weight (up to 6%) at 60 DAS in both seasons (2012–13 and 2013–2014) over the un-inoculated control plots (Tables 1 and 2). At chickpea crop maturity, the Streptomyces-treated plots also enhanced stover yield (up to 41%), grain yield (up to 16%), total dry matter (up to 26%), pod weight (up to 7%), seed number (up to 21%) and seed weight (up to 11%) in both seasons over the un-inoculated control plots (Table 3). At crop maturity, in the top 15-cm of rhizosphere soil, the Streptomyces sp. treatments enhanced the microbial biomass carbon (up to 59%) and dehydrogenase activity (up to 21%), as well as soil mineral nutrient contents, including total N (up to 8%), available P (up to 45%) and % organic carbon (up to 7%), in both seasons over the un-inoculated control plots (Tables 4 and 5).
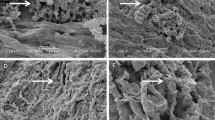
SEM analysis of chickpea roots revealed a remarkable degree of colonization by Streptomyces sp. Roots from inoculated plants exhibited significant surface colonization by Streptomyces sp., while those from un-inoculated plants did not. The sporulation of Streptomyces sp. on the surface cell layer of chickpea roots was clearly evident for all five strains. The hyphae of Streptomyces sp. were also found to penetrate the surface cell layer of chickpea roots (Figure 1).
Gene expression profiles of plant-growth-promoting genes, IAA, siderophore and β-1,3-glucanase revealed overall up-regulation in the Streptomyces strains studied. The gene IAA showed higher up-regulation in the strain CAI-68 (13.7-fold), while no significant variation in the expression pattern was observed in the other four strains. The siderophore gene was highly up-regulated in strain CAI-68 (10.8-fold), followed by KAI-27 (3.6), and β-1,3-glucanase expression was up-regulated in KAI-26 (9.3-fold) (Figure 2).
Discussion
The five strains of Streptomyces spp. (CAI-17, CAI-68, CAI-78, KAI-26 and KAI-27), which are known for biocontrol of charcoal rot in sorghum and PGP in sorghum (Gopalakrishnan et al. 2011b) and rice (Gopalakrishnan et al. 2013), were evaluated for their PGP effects in chickpea under field conditions. In the present study, nodules were observed in the roots of chickpea on both the seasons (2012–13 and 2013–2014) though rhizobia were neither inoculated on seed nor applied on soil indicating the presence of native rhizobia in the soil. Rhizobia were always noted in the Vertisols of ICRISAT (Gopalakrishnan et al. 2014b). The increase in the number of nodules per plant in 2013–14 over 2012–13 could be due to the build-up of native rhizobia specific to chickpea during the cropping season of 2012–13 and hence subsequently observed in the following season, 2013–14. Of the five Streptomyces sp. applied, CAI-78 significantly enhanced all of the agronomic characteristics and yield components the most, including the nodule numbers and nodule weight. The enhancement effects of the other strains were, in descending order, CAI-68, CAI-17, KAI-26 and KAI-27.
The highest magnitude of enhanced soil biological activities and mineral nutrient properties was found with CAI-78 treatment, followed by CAI-68, CAI-17, KAI-26 and KAI-27. The enzyme dehydrogenase is regarded as an indicator of total life in the soil and a strong indicator of biological activity. Similarly, increased microbial biomass C and dehydrogenase activity have been reported in fields upon the use of biological options for soil fertilization and insect-pests control (Carpenter-Boggs et al. 2000; Sharma and Singh 2004). Superior soil nutrient status (N, P, K and % organic carbon) has been reported on organic farms compared to soils treated with chemical pesticides and fertilizers (Sharma and Singh 2004; Singh et al. 2004). In the present study, the enhancement of soil biological and mineral nutrient properties with Streptomyces treated plots over un-inoculated control could be due to the presence of more microbial activity in the rhizosphere which leads to beneficial functions for crops such as plant growth-promotion, nitrogen fixation, phosphate solubilization and protection against pathogens.
The PGP influence of Streptomyces sp. had previously been reported in various agriculturally important crops (Tokala et al. 2002; Nassar et al. 2003; El-Tarabily 2008; Sadeghi et al. 2012; Gopalakrishnan et al. 2014a). PGP agents having a broad spectrum of activities offer effective and novel strategies not only for the improvement of crop growth and yield but also for the control of insect pests and pathogens that affect crop plants. In addition to suppressing plant pathogens and insect pests by secreting secondary metabolites, such as antibiotics, some PGP agents also elicit induced systemic resistance against a broad range of insects and pathogens (Jetiyanon and Kloepper 2002; Ryu et al. 2007). Although the Streptomyces spp. used in this study excrete antimicrobial compounds, antagonism tests by the poison food technique indicate that none of the strains inhibit the growth of nodulating bacteria Rhizobium leguminosarum on yeast extract mannitol agar media (data not shown). Thus, the Streptomyces spp., appears to be compatible with Rhizobium.
Colonization of chickpea roots by Streptomyces spp., at the right time and place is essential for enhanced PGP activity. Successful host-microbe interaction is essential, which depends on the presence of a sufficient population of bacteria, as well as the rhizosphere competence, root-colonizing ability and PGP capability of the bacteria (Lugtenberg and Dekkers 1999). The data for grain and stover yield, root mass and other agronomical traits, the chemical and biological activities of the rhizosphere soil along with the SEM micrograph clearly establish that the PGP effects of the Streptomyces spp., had been caused by successful colonization of the inoculated chickpea roots.
The mechanism by which the five Streptomyces sp. could consistently enhance agronomical and yield traits on sorghum, rice (from our previous study) and chickpea can be attributed to their enzymatic activities, such as the ability to produce siderophores and indole acetic acid and β-1,3-glucanase (Gopalakrishnan et al. 2011b; 2013). Siderophores are known to form stable organic complexes with heavy metals such as Cu, Cd, In, U, Np, Al, Pb, Zn and Ga and increase the soluble metal concentration (Rajkumar et al. 2010) in the soil system, thus alleviating the various heavy metal stresses imposed on plants. Siderophores also act as solubilizing agents of iron from minerals when iron nutrition becomes a limitation (Indiragandhi et al. 2008). IAA-producing microorganisms are known to stimulate seed germination, initiate adventitious and lateral root formation and increase root length and surface area, thereby providing the host plant greater access to soil nutrients and water (Ahemad and Kibret 2014). The cell wall of plant pathogens is composed of layers of β-1,3-glucan (as in the case of Fusarium oxysporum, the causal agent of wilt disease in many crops) and lysis of these layers by β-1,3-glucanase-producing microorganisms leads to the leakage of cell contents and the collapse of the pathogenic fungi (Singh et al. 1999).
Our previous results documented the production of IAA (0.22 – 0.95%), β-1,3 glucanase (0.2 – 2.92 units) and siderophore (qualitative; 1–3 rating scale) by all of these Streptomyces strains (Gopalakrishnan et al. 2011b; Gopalakrishnan et al. 2013). Further characterization on gene expression profiles of these PGP genes revealed overall up-regulation in all the Streptomyces strains. However, no significant change in expression profiles of all three genes, IAA, siderophore and β-1,3-glucanase, was observed in isolates CAI-17 and CAI-78. The relative expression profiles of these genes in CAI-17 and CAI-78 do not correlate with the PGP capabilities observed under field conditions. This may be due to analysis of the gene expression profiles in only one biological replication of the strains. The exact role of these genes is to be precisely elucidated with more biological replications. However, validation of the IAA, siderophore and β-1,3-glucanase genes confirmed the insights gained from in vitro PGP attributes of the studied Streptomyces strains. Hence, it is concluded that the Streptomyces strains studied in this investigation contains multiple PGP traits and can therefore be exploited not only for PGP but also for biological control of plant pathogens.
The five Streptomyces spp. used in this study are apparently well adapted not only to the sorghum and rice rhizospheres, as reported earlier, but also to the chickpea rhizosphere, as demonstrated in the current investigation. Additionally, the five Streptomyces spp. contain a broad range of PGP abilities and demonstrate multiple beneficial actions. Therefore, the five Streptomyces spp. used in this study are potential candidates for the discovery of novel secondary metabolites and their usefulness in host plant resistance against a range of pathogens and insect pests. These Streptomyces sp. strains can be useful in furthering the use of eco-friendly bio-pesticides and bio-fertilizers in integrated pest, disease and nutrition management programs. However, there is a need to determine the effectiveness of these Streptomyces strains under multi-location trials and to understand the nature of their interaction with other native soil micro-flora and micro-fauna with host plants and the environment.
References
Ahemad M, Kibret M (2014) Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J King Saud University 26:1–20
Anderson TH, Domsch KH (1989) Ratios of microbial biomass carbon to total organic carbon in arable soils. Soil Biol Biochem 21:471–479
Aldesuquy HS, Mansour FA, Abo-Hamed SA (1998) Effect of the culture filtrates of Streptomyces on growth and productivity of wheat plants. Folia Microbiol 43:465–470
Bozzola JJ, Russell LD (1998) “Specimen Preparation for Scanning Electron Microscopy”. 3rd Chapter. In: Electron Microscopy Principals and Techniques for Biologists, 2nd edn. Jones and Barlett publishers, Sudbury, Massachusetts, pp 19–24, 54–55, 63–67
Carpenter-Boggs L, Kennedy AC, Reganold JP (2000) Organic and biodynamic management: Effects on soil biology. Soil Sci Soc Am J 64:1651–1659
Casida LE (1977) Microbial metabolic activity in soil as measured by dehydrogenase determinations. Appl Environ Microbiol 34:630–636
El-Tarabily KA (2008) Promotion of tomato (Lycopersicon esculentum Mill.) plant growth by rhizosphere competent 1-aminocyclopropane-1-carboxylic acid deaminase–producing Streptomycete actinomycetes. Plant Soil 308:161–174
Gopalakrishnan S, Humayun P, Kiran BK, Kannan IGK, Vidya MS, Deepthi K, Rupela O (2011a) Evaluation of bacteria isolated from rice rhizosphere for biological control of charcoal rot of sorghum caused by Macrophomina phaseolina (Tassi) Goid. World J Microbiol Biotechnol 27:1313–1321
Gopalakrishnan S, Humayun P, Vadlamudi S, Vijayabharathi R, Bhimineni RK, Rupela O (2012a) Plant growth-promoting traits of Streptomyces with biocontrol potential isolated from herbal vermicompost. Biocontrol Sci Technol 22:1199–1210
Gopalakrishnan S, Kiran BK, Humayun P, Vidya MS, Deepthi K, Rupela O (2011b) Biocontrol of charcoal-rot of sorghum by actinomycetes isolated from herbal vermicompost. Afr J Biotechnol 10:18142–18152
Gopalakrishnan S, Pande S, Sharma M, Humayun P, Kiran BK, Sandeep D, Vidya MS, Deepthi K, Rupela O (2011c) Evaluation of actinomycete isolates obtained from herbal vermicompost for biological control of Fusarium wilt of chickpea. Crop Prot 30:1070–1078
Gopalakrishnan S, Vadlamudi S, Apparla S, Bandikinda P, Vijayabharathi R, Bhimineni RK, Rupela O (2013) Evaluation of Streptomyces spp. for their plant growth-promotion traits in rice. Can J Microbiol 59:534–539
Gopalakrishnan S, Vadlamudi S, Bandikinda P, Sathya A, Vijayabharathi R (2014b) Plant growth-promoting traits of Pseudomonas geniculata isolated from chickpea nodules. 3Biotech. DOI 10.1007/s13205-014-0263-4
Gopalakrishnan S, Vadlamudi S, Bandikinda P, Sathya A, Vijayabharathi R, Rupela O, Kudapa H, Katta K, Varshney RK (2014b) Evaluation of Streptomyces strains isolated from herbal vermicompost for their plant growth-promotion traits in rice. Microbiol Res 169:40–48
Gopalakrishnan S, Upadhyaya HD, Vadlamudi S, Humayun P, Vidya MS, Alekhya G, Singh A, Vijayabharathi R, Bhimineni RK, Seema M, Rathore A, Rupela O (2012b) Plant growth-promoting traits of biocontrol potential bacteria isolated from rice rhizosphere. SpringerPlus 1:71
Indiragandhi P, Anandham R, Madhaiyan M, Sa TM (2008) Characterization of plant growth-promoting traits of bacteria isolated from larval guts of diamondback moth Plutella xylostella (Lepidoptera; Plutellidae). Curr Microbiol 56:327–333
Jalilian J, Modarres-Sanavy SAM, Saberali SF, Sadat-Asilan K (2012) Effects of the combination of beneficial microbes and nitrogen on sunflower seed yields and seed quality traits under different irrigation regimes. Field Crop Res 127:26–34
Jetiyanon K, Kloepper JW (2002) Mixtures of plant growth promoting rhizobacteria for induction of systemic resistance against multiple plant diseases. Biol Control 24:285–291
Lucas JA, Solano BR, Montes F, Ojeda J, Megias M, Manero FJG (2009) Use of two PGPR strains in the integrated management of blast disease in rice (Oryza sativa) in Southern Spain. Field Crop Res 114:404–410
Lugtenberg BJJ, Dekkers LC (1999) What makes Pseudomonas bacteria rhizosphere competent? Environ Microbiol 1:9–13
Macagnan D, Romeiro RDA, Pomella AMV, deSouza JT (2008) Production of lytic enzymes and siderophores, and inhibition of germination of basidiospores of Moniliophthora (ex Crinipellis) perniciosa by phylloplane actinomycetes. Biol Control 47:309–314
Nassar AH, El-Tarabily KA, Sivasithamparam K (2003) Growth promotion of bean (Phaseolus vulgaris L.) by a polyamine producing isolate of Streptomyces griseoluteus. Plant Growth Reg 40:97–106
Nelson DW, Sommers LE (1982) Total Organic Carbon and Organic matter. In: Page AL, Miller RH, Keeney DR (eds) Methods of Soil Analysis, Part 3, Chemical and Microbiological Properties. SSSA, Madison, WI, pp 39–579
Novozamsky I, Houba VJG, Van ECKR, vanVark W (1983) A novel digestion technique for multiple element analysis. Comm Soil Sci Plant Anal 14:239–249
Olsen SR, Sommers LE (1982) Phosphorus. In: Page AL (ed) Methods of Soil Analysis, Agron No 9, Part 2, ‘Chemical and Microbial properties’, 2nd edn. American Society of Agronomy, Madison WI, USA, pp 403–430
Perner H, Schwarz D, George E (2006) Effect of mycorrhizal inoculation and compost supply on growth and nutrient uptake of young leek plants grown on peat-based substrates. HORTSCI 41:628–632
Postma J, Montanari M, Van den Boogert PHJF (2003) Microbial enrichment to enhance disease suppressive activity of compost. Eur J Soil Biol 39:157–163
Rajkumar M, Ae N, Prasad MNV, Freitas H (2010) Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol 28:142–149
Richardson AE, Barea JM, Mcneill AM, Combaret CP (2009) Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 321:305–339
Rosen S, Skaletsky HJ (2000) Primer 3 on the WWW for General Users and for Biologist Programmers. In: Krawetz S, Misener S (eds) Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 365–386
Ryu CM, Murphy CF, Reddy MS, Kloepper JW (2007) A two strain mixture of rhizobacteria elicits induction of systemic resistance against Pseudomonas syringae and Cucumber mosaic virus coupled to promotion of plant growth on Arabidopsis thaliana. J Microbiol Biotechnol 17:280–286
Sadeghi A, Karimi E, Dahazi PA, Javid MG, Dalvand Y, Askari H (2012) Plant growth promoting activity of an auxin and siderophore producing isolate of Streptomyces under saline soil condition. World J Microbiol Biotechnol 28:1503–1509
SAS Institute Inc. 2013. SAS/STAT® 12.3 User’s Guide. Cary, NC
Sharma PD, Singh M (2004) Problems and prospects of Organic farming. Bull Indian Soc Soil Sc 22:14–41
Singh Y, Singh B, Ladha JK, Khind CS, Khera TS, Bueno CS (2004) Effects of residue decomposition on productivity and soil fertility in rice-wheat rotation. Soil Sci Soc Am J 68:854–864
Shaukat K, Affrasayab S, Hasnain S (2006) Growth responses of Triticum aestivum to plant growth promoting Rhizobacteria used as a biofertilizer. Res J Microbiol 1:330–338
Singh PP, Shin YC, Park CS, Chung YR (1999) Biological control of Fusarium wilt of cucumber by chitinolytic bacteria. Phytopathology 89:92–99
Spaepen S, Vanderleyden J, Remans R (2007) Indole‐3‐acetic acid in microbial and microorganism‐plant signaling. FEMS Microbiol Rev 31:425–448
Tokala RK, Strap JL, Jung CM, Crawford DL, Salove MH, Deobald LA, Bailey JF, Morra MJ (2002) Novel plant-microbe rhizosphere interaction involving Streptomyces lydicus WYEC108 and the pea plant (Pisum sativum). Appl Environ Microbiol 68:2161–2171
Trejo-Estrada SR, Paszczynski A, Crawford DL (1998) Antibiotics and enzymes produced by the biocontrol agent Streptomyces violaceusniger YCED-9. J Ind Microbiol Biotechnol 21:81–90
Acknowledgements
This work has been undertaken as part of the CGIAR Research Program on Grain Legumes. We thank Dr. M Lakshman, Associate Professor, Ruska Lab, College of Veterinary Science, Rajendranagar, Hyderabad, for SEM analysis and all of the staff of the biocontrol unit of ICRISAT including M/s PVS Prasad, P Manohar, B Nagappa, D Barath, A Jabbar and S Rohini for their significant contributions in the laboratory and field studies. We also thank Dr L Krishnamurthy, Consultant, Grain Legumes, ICRISAT for his critical review and useful suggestions for improving this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SG, VS, GA, BH, HK and RKV carried out the lab and field studies. AR done the statistical analysis and SG drafted the manuscript. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits use, duplication, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Gopalakrishnan, S., Srinivas, V., Alekhya, G. et al. The extent of grain yield and plant growth enhancement by plant growth-promoting broad-spectrum Streptomyces sp. in chickpea. SpringerPlus 4, 31 (2015). https://doi.org/10.1186/s40064-015-0811-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40064-015-0811-3