Abstract
Objective
It has been demonstrated that IDO1, a target of immune checkpoint inhibition, functions as an oncogene in the majority of human malignancies. IDO1’s function in human pan-cancers hasn’t been thoroughly studied, though.
Materials and methods
The Kaplan–Meier (K-M) and COX analyses were applied to the survival analysis. Furthermore, we used Spearman’s correlation analysis to examine the associations between IDO1 and microsatellite instability (MSI), DNA methyltransferases (DNMTs), tumor mutational burden (TMB), the associated genes of mismatch repair (MMR), and immune checkpoint biomarkers. Moreover, immunohistochemical analysis and qRT-PCR were used to evaluate IDO1’s expression in pan-cancer cells.
Results
The findings of this study reveal that IDO1 has abnormal expression in a number of malignancies and is related to the prognosis for UVM, LGG, KIRP, GBM, LAML, OV, READ, MESO, SARC, SKCM, and HNSC. Furthermore, the aberrant IDO1 expression was connected to the TMB, MSI, MMR, drug sensitivity, immune cells infiltrating, and tumor immune microenvironment across a variety of cancer types. The PCR results showed that in contrast to normal cells, IDO1 was found to be significantly highly expressed in breast cancer cells and hepatocellular carcinoma cells, and significantly lowly expressed in gastric cancer cells.
Conclusion
The clinical treatment of IDO1 is now better supported by a theoretical basis and guidelines provided by our study.
Similar content being viewed by others
Introduction
A growing number of people are being diagnosed with cancer and dying from it. According to the World Health Organization (WHO) study from 2022, cancer accounts for approximately 16% of fatalities worldwide and is one of the main causes of mortality, particularly for those under the age of 70 [1]. Despite advances in cancer detection and therapy, low survival rates are still caused by several issues, including cancer recurrence and medication resistance [2]. Finding new techniques for cancer diagnosis and treatment is crucial [3]. A combination immunotherapy strategy is more likely to improve survival than a single-drug therapeutic approach, according to preclinical and clinical studies [4, 5]. Therefore, it is crucial to look for new cancer markers to more accurately and early identify the clinical stage, metastasis, and prognosis of most malignancy [6, 7].
Numerous studies have demonstrated that targeting the immunometabolic enzyme indoleamine 2,3 dioxygenase 1 (IDO1) is highly expressed in multiple types of human cancer. IDO1 catalyzes the degradation of tryptophan (Trp) to kynurenine (Kyn). The IDO1 gene can be induced by interferon and is associated with mediating potent immunosuppressive effects in cancer [8, 9]. IDO1 is overexpressed in various forms of cancer and is linked to a bad prognosis. A tumor-friendly immunological microenvironment can be created by IDO1 and Kyn metabolites, and aryl hydrocarbon receptors can be activated as part of this process [10]. IDO1 stimulates CD4 + T regulatory cells (Treg) and myeloid-derived suppressor cells (MDSC) while suppressing local CD8 + T effector cells and natural killer cells [11].
Patients who get immune checkpoint blockade therapy may see some truly astonishing lengthy anti-cancer effects [12]. Recently discovered immunological checkpoint inhibitors have been approved for use in clinical settings. One of the most effective methods for finding new cancer drugs is the development of immune checkpoint inhibitors as anticancer therapeutic alternatives in recent years [13]. IDO1 inhibitors have been the focus of significant research over the past 10 years. IDO1 inhibitors have been identified and are currently investigated in clinical studies, including Indoximod, Epacadostat, BMS-986205, and PF-06840003 [14,15,16,17,18,19]. IDO1 has been linked to mediating strong immunosuppressive effects in cancer and is interferon-inducible [20]. Combining an IDO1 inhibitor with a checkpoint inhibitor is a promising way to increase the number of patients who can receive immunotherapies because it has been demonstrated that IDO1 takes part in the mechanisms of resistance to checkpoint inhibitors [21]. Strong evidence exists that a combination approach can offer synergistic effects, despite mounting evidence that a treatment targeting IDO1 alone is ineffective [8, 22,23,24,25]. IDO1 is being used as an adjuvant medication in a growing number of ongoing clinical trials in conjunction with other cancer therapy methods due to the poor efficacy of single medicines [26]. IDO1 inhibitors have so far been established, examined, and filtered in disease-related preclinical models [26, 27]. The IDO1 pathway modulator Indoximod (d-1-MT), the IDO1 vaccination, and Epacadostat appears to be well tolerated by cancer patients [28,29,30,31]. The number of trials examining IDO1 inhibition in cancer therapy is still increasing since it has been established that targeting IDO1 is secure and well-tolerated [8].
In this study, we combined The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) to investigate abnormal expression and to assess its prognostic value in multiple cancer types, providing a comprehensive picture of the prognostic value and alteration of the IDO1 gene in pan-cancer. More importantly, we investigated the relationship between IDO1 expression and various tumor immunity, microsatellite instability (MSI), tumor mutation burden (TMB), tumor immune microenvironment (TME), and tumor-infiltrating immune cells (TIICs), and further observed the correlation between IDO1and tumor immunotherapy.
Methods
Data processing and IDO1 expression analysis
We obtained data on IDO1 from the TCGA and the GTEX to examine the difference of IDO1. From the TCGA database, we collected data on somatic mutations and clinical follow-up for 33 different types of cancer. Log2 transformation was used to normalize all expression levels. We later used the HPA to display human protein expression in normal and tumor tissues, which allowed us to identify the expression of the IDO1 protein.
Analysis of mismatch repair system (MMRS) and DNA methyltransferase
It is possible that DNA MMRS defects can lead to tumorigenesis [32]. As well as altering chromatin structure and gene expression, DNMTs also affect gene expression [33]. The TCGA database was used to determine the expression levels of 5 MMR genes (EPCAM, MLH1, MSH2, MSH6, and PMS2) as well as the 4 methyltransferases (DNMT3B, DNMT2, DNMT3A, and DNMT1).
Survival and prognosis analysis
Forest plots and K-M curves were utilized to assess the relationship between IDO1 expression and patient prognosis in 33 distinct cancer types by K-M and COX regression analysis. The ROC package was then used to create ROC curves, and the area under the curve (AUC) was used to calculate the predictive power. In addition, the RMS software produced nomogram plots and calibrations for projecting OS.
Correlations between IDO1 expression and immune
The scores of these six TIICs in 33 tumors were retrieved from the TIMER database. You can access 10,897 TCGA samples in the TIMER database. Additionally, using Spearman correlation analyses, we evaluated the relationships between IDO1 expression and TMB, MSI, immune/stromal scores, TIICs, the related genes of immunological checkpoint marker, MMR, and DNMTs.
Pathway study of IDO1 in pan-cancers
An analysis of gene sets obtained from Gene Set Enrichment Analysis (GSEA) website was used for this study. Gene Ontology (GO) and KEGG (Kyoto Encyclopedia of Genes and Genomes) co-expression analysis using the R-package ‘‘org.Hs.eg.db’’, ‘‘clusterProfiler’’, and ‘‘enrichplot’’ to visualize the significant positive and negative correlations of 5 pathways. Output categories were significance threshold at p and q < 0.05.
Cell culture
Breast cell lines MCF-7, MCF-10A, and MDA-MB-231; the liver cell line L-02, HepG2, HUH-7, and SMMC-7721 liver cell lines; and the gastric cell lines GES-1, MKN-45, AGS, and MGC-803 were all incubated in RPMI-1640 supplemented with 10% FBS (Hyclone).
RNA isolation and Q-PCR
The M5 Universal RNA Mini Kit kit's instructions were followed to extract total RNA. To confirm that the RNA concentration and purity were constant, the spectrophotometer was used to measure the absorbance values at 280 nm and 260 nm. Following the directions in the M5 Sprint qPCR RT kit with gDNA remover reverse transcription kit, RNA was reverse transcribed into cDNA. For RT-qPCR detection, we used cDNA as a template and 2 M5 HiPer SYBR Premix EsTaq (with Tli RnaseH) as a fluorescent dye. Wuhan Sevier Biotechnology Business Ltd created and created IDO1 primers.
Statistical analysis
An analysis of the expression of IDO1 in various tissues was conducted using Kruskal–Wallis tests and pair-t-tests. P < 0.05 denotes statistically significant.
Results
Differential expression analysis of IDO1
The results showed that among the thirty-three tumors associated with gene expression, IDO1 mRNA was highly expressed in twelve tumors in CESC, BRCA, ESCA, HNSC, GBM, KICH, KIRC, KIRP, PCPG, LIHC, STAD, and UCEC by TCGA database; especially overexpressed in CESC, KIRC, HNSC, and UCEC. IDO1 is lowly expressed in three tumors of LUAD, LUSC, and THCA (Fig. 1A and Additional file 2: Table S1).
Comprehensive analysis of the differential expression of IDO1. A Differential IDO1 mRNA expression in TCGA database. B Differential IDO1 mRNA expression between TCGA and GTEX databases. *P < 0.05, **P < 0.01, ***P < 0.001. C Immunohistological images comparing IDO1 gene expression between normal and tumor tissues for normal (left) as well as tumor (right) in LIHC, CESC, BRCA, KIRC, STAD, and UCEC
We also examined the IDO1 expression by integrating TCGA data with normal tissue data from the GTEx database. We discovered that 26 different tumor types over-expressed IDO1. In addition to the 12 types of tumors mentioned above, there are ACC, COAD, BLCA, DLBC, LGG, LUAD, PAAD, OV, PRAD, READ, TGCT, SKCM, THYM, and UCS, the combined data showed significant differences in IDO1 expression in CESC, DLBC, KIRC, OV, STAD and UCEC. Meanwhile, IDO1 was lowly expressed in both THCA and LAML (Fig. 1B and Additional file 2: Table S2). We use a paired t-test as shown in Annex (Additional file 1: Figure S1).
To analyze the expression of IDO1 at the protein level, we also assessed the IHC results from the HPA dataset and compared them to the TCGA data on IDO1 gene expression. The results are shown in (Fig. 1C). The six tumors analyzed with the protein level expression of IDO1 showed higher expression of IDO1 than normal tissues, which is consistent with the TCGA.
Multifaceted prognostic value of IDO1
We first used COX analysis (Fig. 2A–B and Additional file 1: Figure S2A–B)to investigate the association of IDO1 with patients’ prognosis (OS, DSS, DFI, and PFI). Next, we used K-M analysis to investigate. For OS, IDO1 exhibited an unfavorable prognostic factor for UVM, LGG, KIRP, GBM, and LAML. IDO1 was a favorable prognostic factor in OV, READ, MESO, SARC, SKCM, and HNSC. For DSS, IDO1 was an unfavorable prognostic factor in UVM, GBM, KIRP, and LGG; IDO1 was a protective prognostic factor in OV, MESO, SARC, SKCM, HNSC, and CHCL ((Fig. 2C–D and Additional file 2: Table S4). About DFI, IDO1 played an favorable prognostic factor in OV, SARC, , and BLCA (Additional file 1: Figure S2C). About PFI, IDO1 played an adverse prognostic factor in UVM, LGG, GBM, KIRP, and THYM. IDO1 played a protective prognostic factor in OV, MESO, SARC, SKCM, HNSC, BRCA, and CHOL (Additional file 1: Figure S2D); Subsequently, the prognostic value of IDO1 in IDO1-related survival (OS and RFS) was determined using Kaplan Meier plotter analysis (Fig. 3). Interestingly, we were able to verify that IDO1 had a protective prognostic role in BLCA (OS: P = 0.057; RFS: P = 0.0015), BRCA (OS: P = 0.027; RFS: P = 0.024), OV (OS: P = 0.00033; RFS: P = 0.0013), SARC (OS: P = 0.00014; RFS: P = 0.012), UCEC (RFS: P = 0.0058), CESC (OS: P = 0.017), HNSC (OS: P = 0.011), STAD (OS: P = 0.0054), LUAD (OS: P = 0.047), STSD (RFS: P = 0.025), and READ (OS: P = 0.0079). In contrast, IDO1 expression had a detrimental effect on KIRP (OS: P = 6.6e−07) and UCEC (OS: P = 0.009).
Clinical characteristics of IDO1
According to Fig. 4A–L and Additional file 2: Table S3, a high level of IDO1 expression was observed in patients under 65, especially in PCPG and OV. However, in UCS, LGG, COAD, and HNSC, IDO1 expression was higher in patients older than 65 years. In addition, stage III-IV patients who had KIRC, STAD, ACC, and KIRP were highly expressed for IDO1, while stage I–II patients showed low expression for COAD and HNSC. Then, we plotted the ROC curve of the IDO1 gene and associated cancers. The findings demonstrated that IDO1 had a moderate diagnostic accuracy (AUC between 0.6 and 0.9) for KIRP, STAD, UCEC, KICH, OSCC, LUAD, CESC, ESAD, CHOL, ESCA, HNSC, LIHC and THCA in predicting tumor or non-tumor prognosis, but a higher diagnostic accuracy (AUC > 0.9) for KIRH and GBM (Fig. 4M). This implies that IDO1 has a high degree of tumor prediction capacity.
IDO1 was correlated with TMB and MSI
The TMB is a new biomarker correlated with immune checkpoint inhibitor sensitivity, containing PD-1/PD-L1 suppression [34,35,36]. Therefore, it is interesting to investigate the relation of IDO1 expression with TMB. According to our findings, IDO1 expression in nine tumors correlated with TMB, with positive correlations including UCS, STAD, KIRC, LGG, KICH, BRCA and COAD, and negative correlations in TGCT and CHOL (Fig. 5A). MSI is a hypermutation pattern generated by errors in the mismatch repair mechanism that has been employed as a marker for PD-1 Blockade [37,38,39]. The MSI status could alter the TME of cancer patients in several ways, thus influencing the prognosis. Therefore, we investigated the relationship between IDO1 expression and MSI. The results of the study were shown by radar plots that among 10 different tumor types, only IDO1 expression in KICH and COAD was positively correlated with MSI, while others including TGCT, OV, LUSC, HNSC, GBM, ESCA, CHOL, and CESC were negatively correlated (Fig. 5B). Among the 33 different tumors, IDO1 expression correlated with at least one MMR gene expression in 27 of them. Three tumors, BRCA, LUSC, and PAAD, showed a high correlation with the expression of MMR genes (Fig. 5C). Also, we investigated the connection between IDO1 and DNMT expression, which shows a correlation between 28 tumors and DNMT expression (Fig. 5 D).
Correlation between pan-cancerous lymphocyte invasion and IDO1 gene expression
The TME plays a key role in promoting heterogeneity among tumor cells, thereby enhancing multidrug resistance and causing tumor progression and metastasis [40]. We sought to elucidate the connection between IDO1 expression and immune infiltration since TIICs have been linked to the prognosis and treatment of many forms of cancer. We used the ESTIMATE method to explore the correlation between TME and the IDO1 gene in different tumor tissues. We can see that stromal and immune cell level rises concurrently with an increase in IDO1 expression in BLCA, BRCA, CESC, CHOL, COAD, DLBC, ESCA, HNSC, LGG, KIRC, LIHC, SARC, LUSC, LUAD, PAAD, PRAD, READ, SKCM, STAD, UVM, and THCA (Fig. 6A–U). Therefore, we believe that the expression of the IDO1 gene may be related to the above tumors.
Analysis of TIICs
In cancer patients, biomarkers for checkpoint immunotherapy are key to improving prognosis [41]. Therefore, this study aims to explore the impact of IDO1 on immunotherapy and explore the correlation between the expression of IDO1 and the expression of immune checkpoints in 32 tumors. CD8+T cells are significantly positively correlated with the expression of IDO1 in all tumors except CHOL, GBM, KICH, LGG, OV, PCPG, THYM, THCA, and UCS. CD4+ T cells were significantly positively correlated with the expression of IDO1 in all tumors except ACC, DLBC, GBM, KIRP, KICH, OV, READ, PCPG, THYM, and UVM. Neutrophil cells were positively correlated with the expression of IDO1 in all tumors except CHOL, GBM, KICH, PCPG, UCS, and UVM. Myeloid dendritic cells were positively correlated with the expression of IDO1 in all but two tumors, KICH and THYM, where UVM was negatively correlated and all others were positively correlated. Macrophage was significantly positively correlated in COAD, GBM, HNSC, LGG, LUAD, LIHC, LUSC, PRAD, PAAD, SARC, TGCT, SKCM, and UCEC, but negatively correlated in DLBC, KICH, and THYM. And for B cells, except ACC, BLCA, KICH, MESO, PCPG, STAD, THYM, and UVM, IDO1 is negatively correlated in DLBC, and positively correlated in all others. (Fig. 7A). We used R software CIBERSORT to analyze the connection between IDO1 expression and the numbers of 22 TIICs. There is a significant correlation between the level of some TIICs and the expression of IDO1 in BRCA (n = 20), THCA (n = 17), LUSC (n = 15), COAD (n = 15), STAD (n = 15), TGCT (n = 14), HNSC (n = 14), CESC (n = 13), LGG (n = 13), BLCA (n = 12), UVM (n = 12), SKCM (n = 12), SARC (n = 12), LUAD (n = 11), and PRAD (n = 11) (Fig. 7B). IDO1 is positively correlated with CD274, TNFRSF9, TNFRSF4, PDCD1LG2, IDO2, and CD48 in many tumor types (Fig. 7C). Additionally, to better understand the connection between IDO1 and immunity, we used three methods to measure immune cells in the TME in pan-cancer, including EPIC, QUANTISEQ, MCP-counter (Additional file 1: Fig. S3).
Drug sensitivity correlation analysis of IDO1
Further analysis of the potential correlation between drug sensitivity and IDO1 expression was conducted using the CellMiner TM database. The results showed that for the Panobinostat, RH1, Alvocidib, Belinostat, 7 − Hydroxystaurosporine, Pralatrexate, Tamoxifen, Methotrexate, Obatoclax, and Oxaliplatin they are all correlated negatively with the expression of IDO1 (Fig. 8A–J). According to the results, chemotherapy drugs including Panobinostat and Tamoxifen, which are frequently used in clinics, may be resistant to some chemotherapeutic agents because of IDO1.
Functional analysis with GSEA
To look into the biological relevance of IDO1 expression in various tumor tissues, we analyzed the role of IDO1 in various tumors by GO functional annotation and KEGG pathway. In GO, IDO1 has multiple bioactivities in BLCA, CESC, BRCA, GBM, KIRP, HNSC, LAML and LGG with diverse modulating effects on patient prognosis; for instance: T cell tolerance induction, regulation of lymphocyte-mediated immunity, immunoglobulin complex circulating, T cell receptor complex, negative regulation of tumor necrosis factor secretion, FC receptor-mediated stimulatory signaling pathway, B cell receptor signaling pathway (Fig. 9A). Among them, most pathways are strongly related to immunity or cancer. KEGG pathway analysis revealed that IDO1 influenced signaling pathways in BLCA, MESO, CECS, OV, HNSC, SARC, SKCM, and LGG, such as cytokine receptor interaction, natural killer cell-mediated cytotoxicity, T cell receptor signaling pathway, chemokine signaling pathway (Fig. 9B). NK cell-mediated cytotoxicity, chemokine signaling pathway, T cell receptor signaling pathway, and other signaling pathways are a few of the signaling pathways connected to immunity or cancer. These routes and functions, as previously mentioned, mostly relate to tumors.
IDO1 expression level in immune subtypes
Based on our earlier findings, we discovered that OS in a range of malignancies is influenced by IDO1 expression levels, whether high or low. Consequently, we collected information on IDO1 expression from the TISDB website for the study of the link between molecular subtypes of the immune system and various tumors. The data demonstrated a substantial connection between IDO1 expression and immunological subtype of UVM, UCES, THCA, TGCT (uveal melanoma), STAD, SARC, SKCM, READ, OV, PAAD, LUSC, LIHC, LUAD, LGG, KIRC, KIRP, HNSC, COAD, ESCA, CESC, BRCA and BLCA (Fig. 10). Although there was no statistically significant difference in IDO1 expression between the immunological subtypes of UCS, MESO, KICH, GBM, ACC, and CHOL (data not shown).
Expression and prognosis of IDO1
We found that the factors we included were significantly associated with the OS of KIRP patients (Fig. 11A) and UVM patients (Fig. 11B). The risk factors were also included in multivariate Cox regression (Fig. 11C–D). According to the results, IDO1 is an independent prognostic factor for patients with KIRP and UVM.In the nomogram mode, the clinical characteristics of KIRP (Fig. 11E) and UVM (Fig. 11F) were incorporated. There was a good match between the predicted probability of calibration plots of KIRP (Fig. 11G) and UVM (Fig. 11H) and the results observed. Our research has developed calibration plots and time-dependent ROC curves predicting the odds of OS rates after 1 year, 3 years, and 5 years. The AUCs in terms of 1 year, 3 year, and 5 year were 0.810, 0.773, and 0.642 for KIRP patients, respectively (Fig. 11I). And for UVM patients, the statistics are 0.634, 0.739, 0.809, respectively (Fig. 11J). We also analyzed the correlation between risk score, survival time, and IDO1 expression profiles of KIRP (Fig. 11K) and UVM (Fig. 11L) patients.
Univariate Cox regression analysis in KIRP A, UVM B and multivariate Cox analysis of KIRP C, UVM D including IDO1. Nomogram for KIRP E, UVM F for OS. Time-dependent ROC curves for OS prediction of KIRP G and UVM H patients. Plots of calibration data for KIRP I and UVM J OS predictions over one, three, and 5 years. The survival time distribution, risk score, and IDO1 expression of KIRP (K) and UVM (L) patients
Single-cell sequencing analysis of IDO1 and PCR expression
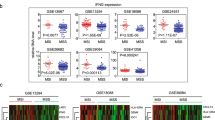
Following that, the TISCH database was used for IDO1-related single-cell analysis. We looked at IDO1 expression in tumor and stromal cells in several cancer types, including BRCA, CESC, CHOL, ESCA, LIHC, OV, SKCM, STAD, and UCEC (Fig. 12A–I). It’s interesting to note that the data showed that IDO1 is extensively co-expressed on cancer cells and stromal cells in various malignancies, particularly on dendritic cells (DC), Malignant cells, Macrophages, and Endothelial cells. The PCR results showed that in contrast to normal cells, IDO1 was poorly expressed in gastric cancer cells MKN-45, AGS, and MGC-803 by PCR (Fig. 12J). IDO1 was found to be significantly highly expressed in breast cancer cells MCF-7 and MDA-MB-231 and hepatocellular carcinoma cells HepG2, HUH-7, and SMMC-7721 in comparison to normal cells (Fig. 12K–L). The results of PCR experiments are in line with the bioinformatics analysis.
Pan-cancer single-cell sequencing analysis of IDO1 co-expression in tumor cells and stromal cells and expression levels in different cancer cell lines. The levels of IDO1 expression in tumor and stromal cells in BRCA A, CESC B, CHOL C, ESCA D, LIHC E, OV F, SKCM G, STAD H and UCEC I. J IDO1 expression in gastric cell lines; K IDO1 expression in breast cell lines; L IDO1 expression in liver cell lines
Discussion
Tumors as the number one disease threatening human health, and the goals of tumor immunology and immunotherapy are to enhance, expand, and forecast the therapeutic effectiveness of immune checkpoint inhibitor-based therapies [12]. Antitumor activity of targeted blockade of IDO1 is remarkable, and IDO1 is expected to be an emerging target for immune checkpoints [42]. In this study, we conducted bioinformatics analysis and laboratory experiments to understand the role and characteristics of IDO1 in the immunology field of tumors.
First, we examined the IDO1 expression in various tumors using the TCGA database as well as the TCGA in combination with the GTEX database. We found that IDO1 expression was upregulated in ER-HER2 and HER2 + breast, colon, and endometrial cancers [43], thyroid cancer [44], gastric cancer [45], and gynecological tumors [46]. One factor contributing to the aggressiveness of GBM is the expression of the potent IDO1 within tumors [47].
In terms of OS, IDO1 expression was unfavorable in the five cancers (GBM, KIRP, LAML, LGG, and UVM) where IDO1 expression was low, and gene expression was positively correlated with survival. Cancer cells overexpress IDO1 which suppresses T cell effector function and promotes Tregs. In severe cases of cancer, overexpression of IDO1 leads to poor survival [48]. However, it was a favorable factor in the six cancers (HNSC, MESO, OV, READ, SARC, and SKCM) where IDO1 expression was high, and gene expression was negatively correlated with survival. Related studies have shown that for HNSC patients, IDO1 was significantly higher before than after treatment and that patients with combined reduced levels of PD-L1 and IDO1 expression after treatment showed better PFS and OS [49].
Among women with HR + breast cancer (BC), high IDO1 expression was associated with poorer long-term survival. The IDO1 protein is present in most HR + BC and is an independent negative prognostic factor [50]. Trp is converted into downstream catabolites known as Kyn by IDO1 [8]. Trp and Kyn depletion and increase exert immune functions through myeloid suppressor cells, and Treg [51]. Interferon-gamma is closely associated with IDO1 expression in tumor treatment and prognosis. IDO1 was primarily expressed in endothelial cells and positively correlated with IFN and T-cell [52]. IDO1 overexpression and IFN-γ treatment increased Kyn accumulation and reduced tryptophan to induce autophagy more powerfully in cervical cancer cells [51]. IDO1 and PD-L1 expression in lung adenocarcinoma cell lines in the presence of IFN-γ and transforming growth factor-β were previously reported to be significantly linked with shorter DSS and OS [53]. Similarly, we have learned that in ESCC, prognosis and pathologic response were negatively affected by increased IDO1 expression in tumors [54]. CD40 mAb combined with epacadostat, an IDO1 inhibitor, reduced tumor growth in B16-F10 melanoma, accompanied by an increase in tumor-infiltrating T cells [52]. Therefore, focusing on the differential expression of IDO1 in different tumors and its prognosis in comparison with combination drug therapy, IDO1 could be a good immune checkpoint to start developing IDO1 inhibitor-related therapeutics to improve anti-tumor efficacy and patient survival.
Currently, immunotherapy has shown remarkable results in the fight against malignant tumors [55]. Our research revealed that TME is crucial in promoting interstitial cancer cells, which promotes the growth and spread of cancer cells, and raises treatment resistance. IDO1 is highly expressed and promotes metastasis, drug resistance, cell proliferation, and TAM resistance through STAT3 and interleukin-6 in TAM-resistant breast cancer, [56]. In many tumors, the transcription factor AHR of growth-promoting genes is activated by KYN in parallel with IDO1 and KYN, enhancing CD28 expression and survival signaling [57]. Targeting KYN can, therefore, be used as a pathway for tumor immunotherapy. Successful immunotherapy for the treatment of malignancies is severely hampered by the immunosuppressive milieu created by Treg [58]. Treg-mediated increased glucose intake causes cellular senescence and decreases the response to T cells through cross-talk. Human Treg cells’ inhibition of glucose uptake and glycolysis as a result of TLR8 receptor signaling results in increasing anti-tumor immunity after adoptive transfer T cell therapy for melanoma [58].
Moreover, TIICs play an important anti-tumor role in TME. In the current investigation, we discovered that the levels of different TIICs, such as B cells, phagocytes, CD8T cells, CD4T cells, bone marrow dendritic cells, and NK cells, were closely linked with the levels of IDO1 expression in different cancers. The binding of CD80/CD86 on dendritic cells to CD28 on LLPC activates IDO1 [57] and promotes the production of the catabolic product Kyn, indirectly affecting the IDO1/TDO2-Kyn-AhR [21] signaling pathway. For the regulatory axis IDO1/miR-18a/NKG2D/NKG2DL in the tumor microenvironment, cytotoxicity of NK cells regulated with high expression of IDO1 was significantly reduced, regulating NK cell function [59]. According to the previous study, IDO1 expression has been shown to be positive for tumor B-cell infiltration [50]. By comparing related studies, we can confirm that IDO1 expression has a positive effect on TIICs.
Next, co-expression analysis reveals that IDO1 was highly correlated with multiple immune-related gene expressions in pan-cancer. Meanwhile, based on GO and KEGG enrichment analysis, we also found that IDO1 was involved in numerous immune mechanisms and related pathways, including T cell tolerance induction, regulation of lymphocyte-mediated immunity, immunoglobulin complex circulating, T cell receptor complex, negative regulation of tumor necrosis factor secretion, FC receptor-mediated stimulatory, B cell receptor signaling pathway signaling pathway cytokine receptor interaction, T cell receptor signaling pathway, natural killer cell-mediated cytotoxicity, chemokine signaling pathway. In summary, the differential expression of IDO1 was closely related to tumor immunity and may serve as a novel target for immunotherapy and prognostic markers.
The drawback of our current study is that further information from other open datasets is required to validate and corroborate our findings. Also, there are not enough experiments to support this, a larger sample size is needed to validate the role and mechanism of IDO1 in pan-cancer.
Conclusion
The findings of this study reveal that IDO1 has abnormal expression in a number of malignancies and is related to the prognosis for UVM, LGG, KIRP, GBM, LAML, OV, READ, MESO, SARC, SKCM, and HNSC. Furthermore, across a variety of cancer types, the aberrant IDO1 expression was connected to the TMB, MSI, MMR, medication sensitivity, and TIME. These results offer a more solid theoretical foundation for the possibility that IDO1 could be a useful prognostic biomarker and a possible indicator of immunotherapy sensitivity in a range of cancers. The clinical treatment of IDO1 is now better supported by a theoretical basis and guidelines provided by our study.
Availability of data and materials
The raw data of this study are freely available from the website TCGA Research Network (https://portal.gdc.cancer.gov/), GTEx(http://commonfund.nih.gov/GTEx/), TIMER database(https://cistrome.shinyapps.io/timer/), Kaplan–Meier plotter portal(https://kmplot.com/analysis/), cBioPortal databasehttp://cbioportal.org), and HPA (https://wwwproteinatlas.org/). All the analyzed data are included in the manuscript.
Abbreviations
- IDO1:
-
Indoleamine2,3-dioxygenase1
- PD-1:
-
Programmed death-1
- PD-L1:
-
Programmed cell death 1 ligand 1
- TCGA:
-
The Cancer Genome Atlas
- GTEx:
-
Genotypic tissue expression project
- TMB:
-
Tumor mutation burden
- MSI:
-
Microsatellite instability
- MMR:
-
Mismatch repair
- DNMTs:
-
DNA methyltransferases
- GO:
-
Gene ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- TIGIT:
-
T cell immune receptor with Ig and ITIM domains
- CTLA4:
-
Cytotoxic T Iymphocyte associate protein-4
- OS:
-
Overall survival
- DFS:
-
Disease-free survival
- ACC:
-
Adrenocortical carcinoma
- BLCA:
-
Bladder urothelial carcinoma
- BRCA:
-
Breast invasive carcinoma
- CESC:
-
Cervical squamous cell carcinoma and Endocervical adenocarcinoma
- CHOL:
-
Cholangiocarcinoma
- COAD:
-
Colon adenocarcinoma
- DLBC:
-
Lymphoid neoplasm diffuse large B-cell lymphoma
- ESCA:
-
Esophageal carcinoma
- GBM:
-
Glioblastoma multiforme
- HNSC:
-
Head and Neck squamous cell carcinoma
- KICH:
-
Kidney chromophobe
- KIRC:
-
Kidney renal clear cell carcinoma
- LAML:
-
Acute myeloid leukemia
- LGG:
-
Brain lower grade glioma
- LIHC:
-
Liver hepatocellular carcinoma
- LUAD:
-
Lung adenocarcinoma
- LUSC:
-
Lung squamous cell carcinoma
- MESO:
-
Mesothelioma
- OV:
-
Ovarian serous cystadenocarcinoma
- PAAD:
-
Pancreatic adenocarcinoma
- PCPG:
-
Pheochromocytoma and paraganglioma
- PRAD:
-
Prostate adenocarcinoma
- READ:
-
Rectum adenocarcinoma
- SARC:
-
Sarcoma
- SKCM:
-
Skin cutaneous melanoma
- STAD:
-
Stomach adenocarcinoma
- TGCT:
-
Testicular germ cell tumors
- THCA:
-
Thyroid carcinoma
- THYM:
-
Thymoma
- UCEC:
-
Uterine corpus endometrial carcinoma
- UCS:
-
Uterine carcinosarcoma
- UVM:
-
Uveal melanoma
References
Adeoye J, Akinshipo A, Thomson P, Su YX. Artificial intelligence-based prediction for cancer-related outcomes in Africa: status and potential refinements. J Glob Health. 2022;12:03017.
Tang J, Pearce L, O’Donnell-Tormey J, Hubbard-Lucey VM. Trends in the global immuno-oncology landscape nature reviews. Drug Disc. 2018;17(12):922.
Qiu W, Ding K, Liao L, Ling Y, Luo X, Wang J. Analysis of the expression and prognostic value of MSH2 in pan-cancer based on bioinformatics. Biomed Res Int. 2021;2021:9485273.
Khalil DN, Smith EL, Brentjens RJ, Wolchok JD. The future of cancer treatment: immunomodulation, CARs, and combination immunotherapy. Nat Rev Clin Oncol. 2016;13(5):273–90.
Smyth MJ, Ngiow SF, Ribas A, Teng MW. Combination cancer immunotherapies tailored to the tumour microenvironment. Nat Rev Clin Oncol. 2016;13(3):143–58.
Wang Y, Lin K, Xu T, Wang L, Fu L, Zhang G, Ai J, Jiao Y, Zhu R, Han X, Cai H. Development and validation of prognostic model based on the analysis of autophagy-related genes in colon cancer. Aging. 2021;13(14):19028–47.
Wang Y, Fu L, Lu T, Zhang G, Zhang J, Zhao Y, Jin H, Yang K, Cai H. Clinicopathological and prognostic significance of long non-coding RNA MIAT in human cancers: a review and meta-analysis. Front Genet. 2021;12: 729768.
Zhai L, Ladomersky E, Lenzen A, Nguyen B, Patel R, Lauing KL, Wu M, Wainwright DA. IDO1 in cancer: a Gemini of immune checkpoints. Cell Mol Immunol. 2018;15(5):447–57.
Lou Q, Liu R, Yang X, Li W, Huang L, Wei L, Tan H, Xiang N, Chan K, Chen J, Liu H. miR-448 targets IDO1 and regulates CD8(+) T cell response in human colon cancer. J Immunother Cancer. 2019;7(1):210.
Chen B, Alvarado DM, Iticovici M, Kau NS, Park H, Parikh PJ, Thotala D, Ciorba MA. Interferon-induced IDO1 mediates radiation resistance and is a therapeutic target in colorectal cancer. Cancer Immunol Res. 2020;8(4):451–64.
Prendergast GC, Malachowski WJ, Mondal A, Scherle P, Muller AJ. Indoleamine 2,3-dioxygenase and its therapeutic inhibition in cancer. Int Rev Cell Mol Biol. 2018;336:175–203.
Miao Y, Wang J, Li Q, Quan W, Wang Y, Li C, Wu J, Mi D. Prognostic value and immunological role of PDCD1 gene in pan-cancer. Int Immunopharmacol. 2020;89(Pt B): 107080.
Alsaab HO, Sau S, Alzhrani R, Tatiparti K, Bhise K, Kashaw SK, Iyer AK. PD-1 and PD-L1 checkpoint signaling inhibition for cancer immunotherapy: mechanism, combinations, and clinical outcome. Front Pharmacol. 2017;8:561.
Tang K, Wu YH, Song Y, Yu B. Indoleamine 2,3-dioxygenase 1 (IDO1) inhibitors in clinical trials for cancer immunotherapy. J Hematol Oncol. 2021;14(1):68.
Mitchell TC, Hamid O, Smith DC, Bauer TM, Wasser JS, Olszanski AJ, Luke JJ, Balmanoukian AS, Schmidt EV, Zhao Y, Gong X, Maleski J, Leopold L, Gajewski TF. Epacadostat plus pembrolizumab in patients with advanced solid tumors: phase I results from a multicenter, open-label phase I/II trial (ECHO-202/KEYNOTE-037). J Clin Oncol. 2018;36(32):3223–30.
Luke JJ, Tabernero J, Joshua A, Desai J, Varga AI, Moreno V, Gomez-Roca CA, Markman B, De Braud FG, Patel SP, Carlino MS, Siu LL, Curigliano G, Liu Z, Ishii Y, Wind-Rotolo M, Basciano PA, Azrilevich A, Gelmon KA. BMS-986205, an indoleamine 2, 3-dioxygenase 1 inhibitor (IDO1i), in combination with nivolumab (nivo): updated safety across all tumor cohorts and efficacy in advanced bladder cancer (advBC). J Clin Oncol. 2019;37:358–358.
Nayak A, Hao Z, Sadek R, Vahanian N, Ramsey WJ, Kennedy E, Mautino M, Link C, Bourbo P, Dobbins R, Adams K, Diamond A, Marshall L, Munn DH, Janik J, Khleif SN. A Phase I study of NLG919 for adult patients with recurrent advanced solid tumors. J Immunother Cancer. 2014;2(Suppl 3):P250.
Qian F, Villella J, Wallace PK, Mhawech-Fauceglia P, Tario JD Jr, Andrews C, Matsuzaki J, Valmori D, Ayyoub M, Frederick PJ, Beck A, Liao J, Cheney R, Moysich K, Lele S, Shrikant P, Old LJ, Odunsi K. Efficacy of levo-1-methyl tryptophan and dextro-1-methyl tryptophan in reversing indoleamine-2,3-dioxygenase-mediated arrest of T-cell proliferation in human epithelial ovarian cancer. Cancer Res. 2009;69(13):5498–504.
Tumang J, Gomes B, Wythes M, Crosignani S, Bingham P, Bottemanne P, Cannelle H, Cauwenberghs S, Chaplin J, Dalvie D. PF-06840003: a highly selective IDO-1 inhibitor that shows good in vivo efficacy in combination with immune checkpoint inhibitors. Cancer Res. 2016;76:4863–4863.
Uyttenhove C, Pilotte L, Théate I, Stroobant V, Colau D, Parmentier N, Boon T, Van den Eynde BJ. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9(10):1269–74.
Cheong JE, Sun L. Targeting the IDO1/TDO2-KYN-AhR pathway for cancer immunotherapy—challenges and opportunities. Trends Pharmacol Sci. 2018;39(3):307–25.
Hou DY, Muller AJ, Sharma MD, DuHadaway J, Banerjee T, Johnson M, Mellor AL, Prendergast GC, Munn DH. Inhibition of indoleamine 2,3-dioxygenase in dendritic cells by stereoisomers of 1-methyl-tryptophan correlates with antitumor responses. Cancer Res. 2007;67(2):792–801.
Koblish HK, Hansbury MJ, Bowman KJ, Yang G, Neilan CL, Haley PJ, Burn TC, Waeltz P, Sparks RB, Yue EW, Combs AP, Scherle PA, Vaddi K, Fridman JS. Hydroxyamidine inhibitors of indoleamine-2,3-dioxygenase potently suppress systemic tryptophan catabolism and the growth of IDO-expressing tumors. Mol Cancer Ther. 2010;9(2):489–98.
Wainwright DA, Chang AL, Dey M, Balyasnikova IV, Kim CK, Alex Tobias Y, Cheng JW, Kim JQ, Lingjiao Zhang Y, Han MS, Lesniak. Durable therapeutic efficacy utilizing combinatorial blockade against IDO, CTLA-4, and PD-L1 in mice with brain tumors. Clin Cancer Res Off J Am Assoc Cancer Res. 2014;20(20):5290–301.
Holmgaard RB, Zamarin D, Munn DH, Wolchok JD, Allison JP. Indoleamine 2,3-dioxygenase is a critical resistance mechanism in antitumor T cell immunotherapy targeting CTLA-4. J Exp Med. 2013;210(7):1389–402.
Vacchelli E, Aranda F, Eggermont A, Sautès-Fridman C, Tartour E, Kennedy EP, Platten M, Zitvogel L, Kroemer G, Galluzzi L. Trial watch: IDO inhibitors in cancer therapy. Oncoimmunology. 2014;3(10): e957994.
Röhrig UF, Majjigapu SR, Vogel P, Zoete V, Michielin O. Challenges in the discovery of indoleamine 2,3-Dioxygenase 1 (IDO1) inhibitors. J Med Chem. 2015;58(24):9421–37.
Iversen TZ, Engell-Noerregaard L, Ellebaek E, Andersen R, Larsen SK, Bjoern J, Zeyher C, Gouttefangeas C, Thomsen BM, Holm B, Thor Straten P, Mellemgaard A, Andersen MH, Svane IM. Long-lasting disease stabilization in the absence of toxicity in metastatic lung cancer patients vaccinated with an epitope derived from indoleamine 2,3 dioxygenase. Clin Cancer Res. 2014;20(1):221–32.
Soliman HH, Jackson E, Neuger T, Dees EC, Harvey RD, Han H, Ismail-Khan R, Minton S, Vahanian NN, Link C, Sullivan DM, Antonia S. A first in man phase I trial of the oral immunomodulator, indoximod, combined with docetaxel in patients with metastatic solid tumors. Oncotarget. 2014;5(18):8136–46.
Perez RP, Riese MJ, Lewis KD, Saleh MN, Daud A, Berlin J, Lee JJ, Mukhopadhyay S, Zhou L, Serbest G. Epacadostat plus nivolumab in patients with advanced solid tumors: preliminary phase I/II results of ECHO-204. Am Soc Clin Oncol. 2017. https://doi.org/10.1200/JCO.2017.35.15_suppl.3003.
Siu LL, Gelmon K, Chu Q, Pachynski R, Alese O, Basciano P, Walker J, Mitra P, Zhu L, Phillips P. Abstract CT116: BMS-986205, an optimized indoleamine 2, 3-dioxygenase 1 (IDO1) inhibitor, is well tolerated with potent pharmacodynamic (PD) activity, alone and in combination with nivolumab (nivo) in advanced cancers in a phase 1/2a trial. Cancer Res. 2017;77:116–116.
Krzyzewska IM, Maas SM, Henneman P, Lip KVD, Venema A, Baranano K, Chassevent A, Aref-Eshghi E, van Essen AJ, Fukuda T, Ikeda H, Jacquemont M, Kim HG, Labalme A, Lewis SME, Lesca G, Madrigal I, Mahida S, Matsumoto N, Rabionet R, Rajcan-Separovic E, Qiao Y, Sadikovic B, Saitsu H, Sweetser DA, Alders M, Mannens MMAM. A genome-wide DNA methylation signature for SETD1B-related syndrome. Clin Epigenet. 2019;11(1):156.
Zheng M. Dose-dependent effect of tumor mutation burden on cancer prognosis following immune checkpoint blockade: causal implications. Front Immunol. 2022;13: 853300.
Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, Schrock A, Campbell B, Shlien A, Chmielecki J, Huang F, He Y, Sun J, Tabori U, Kennedy M, Lieber DS, Roels S, White J, Otto GA, Ross JS, Garraway L, Miller VA, Stephens PJ, Frampton GM. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34.
Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377(25):2500–1.
Zhu J, Zhang T, Li J, Lin J, Liang W, Huang W, Wan N, Jiang J. Association between tumor mutation burden (TMB) and outcomes of cancer patients treated with pd-1/pd-l1 inhibitions: a meta-analysis. Front Pharmacol. 2019;10:673.
Dudley JC, Lin MT, Le DT, Eshleman JR. Microsatellite instability as a biomarker for PD-1 blockade. Clin Cancer Res. 2016;22(4):813–20.
Puliga E, Corso S, Pietrantonio F, Giordano S. Microsatellite instability in gastric cancer: between lights and shadows. Cancer Treat Rev. 2021;95: 102175.
Silveira AB, Bidard FC, Kasperek A, Melaabi S, Tanguy ML, Rodrigues M, Bataillon G, Cabel L, Buecher B, Pierga JY, Proudhon C, Stern MH. High-accuracy determination of microsatellite instability compatible with liquid biopsies. Clin Chem. 2020;66(4):606–13.
Baghban R, Roshangar L, Jahanban-Esfahlan R, Seidi K, Ebrahimi-Kalan A, Jaymand M, Kolahian S, Javaheri T, Zare P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun Signal. 2020;18(1):59.
Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016;17(12):e542–51.
He X, He G, Chu Z, Wu H, Wang J, Ge Y, Shen H, Zhang S, Shan J, Peng K, Wei Z, Zou Y, Xu Y, Zhu Q. Discovery of the first potent IDO1/IDO2 dual inhibitors: a promising strategy for cancer immunotherapy. J Med Chem. 2021;64(24):17950–68.
Panda A, Ganesan S. Genomic and immunologic correlates of indoleamine 2,3-dioxygenase pathway expression in cancer. Front Genet. 2021;12: 706435.
Moretti S, Menicali E, Nucci N, Voce P, Colella R, Melillo RM, Liotti F, Morelli S, Fallarino F, Macchiarulo A, Santoro M, Avenia N, Puxeddu E. Signal transducer and activator of transcription 1 plays a pivotal role in RET/PTC3 oncogene-induced expression of indoleamine 2,3-dioxygenase 1. J Biol Chem. 2017;292(5):1785–97.
Xiang Z, Li J, Song S, Wang J, Cai W, Hu W, Ji J, Zhu Z, Zang L, Yan R, Yu Y. A positive feedback between IDO1 metabolite and COL12A1 via MAPK pathway to promote gastric cancer metastasis. J Exp Clin Cancer Res. 2019;38(1):314.
Zhou Q, Cao FH, Liu H, Zuo MZ. Comprehensive analysis of the prognostic value and immune function of the IDO1 gene in gynecological cancers. Am J Transl Res. 2021;13(4):2041–59.
Zhai L, Ladomersky E, Dostal CR, Lauing KL, Swoap K, Billingham LK, Gritsina G, Wu M, McCusker RH, Binder DC, Wainwright DA. Non-tumor cell IDO1 predominantly contributes to enzyme activity and response to CTLA-4/PD-L1 inhibition in mouse glioblastoma. Brain Behav Immun. 2017;62:24–9.
Komiya T, Huang CH. Updates in the clinical development of epacadostat and other indoleamine 2,3-dioxygenase 1 inhibitors (IDO1) for human cancers. Front Oncol. 2018;8:423.
Economopoulou P, Kladi-Skandali A, Strati A, Koytsodontis G, Kirodimos E, Giotakis E, Maragoudakis P, Gagari E, Maratou E, Dimitriadis G, Kotsantis I, Vagia E, Anastasiou M, Gkotzamanidou M, Kavourakis G, Lianidou E, Psyrri A. Prognostic impact of indoleamine 2,3-dioxygenase 1 (IDO1) mRNA expression on circulating tumour cells of patients with head and neck squamous cell carcinoma. ESMO Open. 2020;5(3): e000646.
Carvajal-Hausdorf DE, Mani N, Velcheti V, Schalper KA, Rimm DL. Objective measurement and clinical significance of IDO1 protein in hormone receptor-positive breast cancer. J Immunother Cancer. 2017;5(1):81.
Liu M, Wang X, Wang L, Ma X, Gong Z, Zhang S, Li Y. Targeting the IDO1 pathway in cancer: from bench to bedside. J Hematol Oncol. 2018;11(1):100.
Georganaki M, Ramachandran M, Tuit S, Núñez NG, Karampatzakis A, Fotaki G, van Hooren L, Huang H, Lugano R, Ulas T, Kaunisto A, Holland EC, Ellmark P, Mangsbo SM, Schultze J, Essand M, Tugues S, Dimberg A. Tumor endothelial cell up-regulation of IDO1 is an immunosuppressive feed-back mechanism that reduces the response to CD40-stimulating immunotherapy. Oncoimmunology. 2020;9(1):1730538.
Kozuma Y, Takada K, Toyokawa G, Kohashi K, Shimokawa M, Hirai F, Tagawa T, Okamoto T, Oda Y, Maehara Y. Indoleamine 2,3-dioxygenase 1 and programmed cell death-ligand 1 co-expression correlates with aggressive features in lung adenocarcinoma. Eur J Cancer. 2018;101:20–9.
Jiao R, Zheng X, Sun Y, Feng Z, Song S, Ge H. IDO1 expression increased after neoadjuvant therapy predicts poor pathologic response and prognosis in esophageal squamous cell carcinoma. Front Oncol. 2020;10:1099.
Anderson AC, Kuchroo VK. 2021. Introduction to the Special Issue Immuno oncology. Semin Immunol. 52 101483.
Zhao X, Jiang Y, Xu M, Hu J, Feng N, Deng H, Lu C, Huang T. Indoleamine 2,3-dioxygenase 1 regulates breast cancer tamoxifen resistance through interleukin-6/signal transducer and activator of transcription. Toxicol Appl Pharmacol. 2022;440: 115921.
Lightman SM, Peresie JL, Carlson LM, Holling GA, Honikel MM, Chavel CA, Nemeth MJ, Olejniczak SH, Lee KP. Indoleamine 2,3-dioxygenase 1 is essential for sustaining durable antibody responses. Immunity. 2021;54(12):2772-2783.e2775.
Li L, Liu X, Sanders KL, Edwards JL, Ye J, Si F, Gao A, Huang L, Hsueh EC, Ford DA, Hoft DF, Peng G. TLR8-mediated metabolic control of human treg function: a mechanistic target for cancer immunotherapy. Cell Metab. 2019;29(1):103-123.e105.
Zhang J, Han X, Hu X, Jin F, Gao Z, Yin L, Qin J, Yin F, Li C, Wang Y. IDO1 impairs NK cell cytotoxicity by decreasing NKG2D/NKG2DLs via promoting miR-18a. Mol Immunol. 2018;103:144–55.
Acknowledgements
The authors thank the Key Laboratory of Molecular Diagnostics and Precision Medicine for Surgical Oncology in Gansu Province and the DaVinci Surgery System Database (DSSD, www.davincisurgerydatabase.com) for their help and support in the methodology and pan-cancer analysis process.
Funding
This work was funded by the 2021 Central-Guided Local Science and Technology Development Fund (ZYYDDFFZZJ-1), Gan Su Joint Scientific Research Fund Major Project under Grant (No.23JRRA1537), National Natural Science Foundation of China (No. 82360498), Gansu Da Vinci robot high-end diagnosis and treatment team construction project, Natural Science Foundation of Gansu Province (No. 22JR11RA257, 22JR5RA692, 21JR7RA633, 21JR1RA038), Research project of Traditional Chinese Medicine of Gansu province (GZKZ-2022-6), Key Research and Development Plan of Gansu Province (No. 21YF5FA169), and Gansu Province Excellent Doctor Fund Project (23JRRA1320).
Author information
Authors and Affiliations
Contributions
KL and HC conceived and designed the study, and revised the manuscript. YW, FL, TY, XL, XL, RL, ZL, CZ, and YZ conducted all data collection and analysis and compiled charts. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethical approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Competing interests
The authors declare no competing conflicts of interest at the time of publication of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Figure S1. IDO1 expression in contrast between paired normal and non-tumor specimens. Figure S2. The forest maps of IDO1 expression level with survival in different cancers. Figure S3. Research on TIICs and IDO1 expression was done using the A EPIC, B , C MCP-counter methods
Additional file 2: Table S1.
Expression levels of IDO1 comparing tumor and normal tissues from TCGA database. Table S2. Expression levels of IDO1 comparing tumor and normal tissues from TCGA and GTEx database. Table S3. Clinical annotation and Pathological features of the individual patient in TCGA database. Table S4. Expression levels of IDO1 comparing tumor and normal tissues in K-M survival curves(OS).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Lin, K., Wang, Y., Liu, F. et al. Comprehensive analysis of the prognostic value and immunological role of IDO1 gene in pan-cancer. Eur J Med Res 29, 206 (2024). https://doi.org/10.1186/s40001-024-01766-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40001-024-01766-y