Abstract
Kidney renal clear cell carcinoma (KIRC) is a main subtype of kidney cancers. Cuproptosis and ferroptosis are correlated with immune infiltration and prognosis in tumors. However, the role of Cuproptosis-related Ferroptosis genes (CRFGs) in KIRC has rarely been fully understood. Therefore, we constructed a prognostic signature based on different expression of CRFGs in KIRC. All raw data of this study were extracted from public TCGA datasets. Cuproptosis and Ferroptosis genes were collected from the previous research. Finally, a total of 36 significantly different CRFGs were identified from TCGA–KIRC cohort. Six-gene signature (TRIB3, SLC2A3, PML, CD44, CDKN2A and MIOX) was identified by LASSO Cox regression based on the significantly different CRFGs. The CRFGs signature was correlated with worse overall survival and the AUC was 0.750. Functional enrichment indicated that CRFGs were mainly enriched in metabolism, drug resistance, tumor immunity pathways. Besides, the IC50 and immune checkpoint differentially expressed between different groups. The proposed 6-CRFGs signature is a promising biomarker to predict clinical outcomes and therapeutic responses for KIRC patient.
Similar content being viewed by others
Introduction
Kidney cancer, accounting for approximately 4.2% of all malignancies, is a disease affecting human health and life [1]. Kidney renal clear cell carcinoma (KIRC) represents approximately 85% of Renal cell carcinoma (RCC) [2, 3]. Targeted treatment, chemotherapy, and immunotherapy are preferred therapy for advanced clear cell carcinoma [4]. However, the therapeutic outcomes are proved to be of unsatisfactory due to the lack of individual variance, reliable prognostic biomarkers and medication resistance [5]. Therefore, it is crucial to seek accurate predictive biomarkers to improve prognosisis of KIRC patients.
Studies demonstrated that various cell death modalities were closely involved in cancer eradication. Cuproptosis is a newly discovered mechanism of copper-induced cell death [6,7,8,9]. Evidence suggests that copper-induced cell death is vital to cancer progression, such as lung [10], thyroid [11], gallbladder [12], breast [13], and prostate [14]. Meanwhile, Ferroptosis is another type of iron-reliant cell death by reactive oxygen species (ROS) accumulation, but differs from autophagy pyroptosis, necrosis and apoptosis [15, 16]. Inhibition ferroptosis may be a promising strategy for cancer therapies, such as ovarian and lung cancer [17, 18]. Recently, it has been suggested excessive copper increased iron toxicity and the development of oxidative stress [19]. In contrary, recent investigations reported that copper affected iron metabolism in neurodegenerative diseases, immunological diseases, and cancer [20,21,22]. In addition, some studies have shown that CuO can promote apoptosis and cytotoxicity modified by reactive oxygen species (ROS) [23, 24]. The abovementioned evidence suggests that copper–iron interactions participants in various physiology and pathophysiology process including cancer progression. However, few studies have focused on copper–iron interactions in terms of cuproptosis–ferroptosis interaction in KIRC.
Here, we established a Cuproptosis-related Ferroptosis genes (CRFGs) signature. We also characterized the immune and molecular profile of CRFGs signature. The signature was effective for predicting KIRC prognosis.
Methods
Data collection and diferentially expressed gene analysis
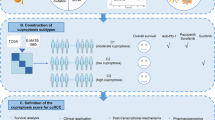
All row data of this study were extracted from public database of The Cancer Genome Atlas (TCGA) repository (72 normal samples and 541 tumor samples). All data are publicly available and open access. Therefore, the institutional review board approval was waived. The flow diagram for this study is displayed in Fig. 1A. A detailed clinical characteristics is given in Table 1. Ferroptosis and cuproptosis genes collected from the previous studies and FerrDb database [25, 26]. Finally, all genes are listed in Additional file 1: Table S1, respectively. Differentially expressed genes (DEGs) between nontumor and KIRC tissues were identified. The Pearson correlation coefficient was relationship between the cuproptosis and ferroptosis genes was calculated with p < 0.001 and correlation coefficient |r2|> 0.3.
Construction the cuproptosis-related ferroptosis gene signature
We developed a CRFGs signature by least absolute shrinkage and selection operator (LASSO) Cox regression. The prognostic gene signature was built by parameter λ with tenfold cross-validation. Patients were divided into KIRC-high and KIRC-low group. Prediction efficacy was evaluated with time-dependent ROC. Furthermore, a nomogram was generated to visually predict specific outcomes. Besides, the calibration and discrimination of the nomogram was assessed respectively. The GSE29609 data sets were used as external validation for the CRFGs score prediction model.
Molecular mechanisms
The protein–protein interaction (PPI) network, KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway and GO (Gene ontology) were used to predict molecular mechanisms of CRFGs.
Drug sensitivity analysis and immune cell infiltration
We assessed tumor-infiltrating immune cell by EPIC algorithms, CIBERSORT-ABS, MCP-counter, XCELL, QUANTISEQ, CIBERSORT, and TIMER. We also compared immune checkpoints and drug sensitivity from KIRC-high and KIRC-low group.
Statistical analysis
All analyses were done in R software (R 4.1.3). Continuous variables were compared by T-test. Survival analysis was performed by log-rank test. Statistical significance was set p < 0.05.
Results
Identification of CRFGs
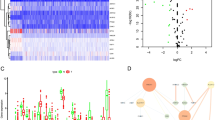
The baseline clinical of KIRC patients in TCGA dataset is listed in Table 1. In total, 8515 DEGs were identified between KIRC nontumor and tumor tissues (Fig. 1B). By analyzing the correlation coefficient of cuproptosis-related ferroptosis genes, we obtained 183 cuproptosis-related ferroptosis genes (p < 0.001 and |Cor|> 0.3). Subsequently, we screened 36 cuproptosis-related ferroptosis differential DEGs (Fig. 1C). As expected, cuproptosis-related ferroptosis genes were different from KIRC normal and tumor tissues.
Establishment of CRFGs signature
To explore whether CRFGs correlated to KIRC patient prognosis, the aforementioned 36 identified genes were analyzed by univariate Cox regression (Fig. 2A). It was demonstrated that eight cuproptosis-related ferroptosis genes with p < 0.05 were correlated to the KIRC survival. Later, a six-gene prognostic signature was generated by LASSO Cox regression using the expression value of the above mentioned 8 prognostic genes (Fig. 2B-C). The selected six genes were TRIB3, SLC2A3, PML, CD44, CDKN2A and MIOX. The risk score was calculated by relevant coefficient of 6 prognostic signature genes (Table 2). Risk score = (0.1430 × TRIB3 profile) + (0.0296 × SLC2A3 profile) + (0.3216 × PML profile) + (0.1135 × CD44 profile) + (0.2139 × CDKN2A profile) + (-0.0496 × MIOX profile). The result of PPI network showed CD44, EGFR, CA9, HIF1A, SLC2A, VEGFA, CDKN2A, CAV1 and MUC1 were hub genes(Fig. 2D). Then, patients were categorized into KIRC-low and KIRC-high groups. KIRC-high group exhibited poor survival outcome than KIRC-low group (Fig. 3A–C). The expression of signature varies across low- and high-score group (Fig. 3D). The ROC curve of 1-, 3,- and 5-year OS was 0.75, 0.675 and 0.654 (Fig. 3E). Similarly, the signature showed powerful forecasting capability for KIRC survival outcomes in validation cohorts (Additional file 2: Figure S1).
A, B Distribution of survival status based on the median risk score in TCGA set. C Kaplan–Meier survival analysis of KIRC patients between high-risk groups and low-risk groups; D Heatmap showed the differences of 6 CRFGs between high-risk and low-risk in KIRC patients; E The receiver operating characteristic (ROC) curve analyses of the prognostic CRFGs in predicting 1-, 3-, and 5-year overall survival (OS)
Analysis of independent prognostic factors
Univariate regression found that stage, age, risk score, and Grade were related to the survival of KIRC patients (p < 0.001) (Fig. 4A). Further, multivariate regression indicated that the age, stage and risk score were also correlated to KIRC survival (p < 0.05) (Fig. 4B). These results revealed that CRFGs signature could serve as prognostic marker of KIRC.
A The correlations between the risk score and clinicopathological factors by univariate Cox regression analysis; B The correlations between the risk score and clinicopathological factors by multivariate Cox regression analysis; C, D the nomogram and calibration curve of the CRFGs-score model, respectively
Correlation between the signature and clinical characteristics
Among the signature, five genes (TRIB3, SLC2A3, PML, CD44 and CDKN2A) unregulated in high-risk group of KIRC. The MIOX gene down-regulated in high-risk group and may be a protective genes (Additional file 3: Figure S2 A). The analysis of clinical data showed that N stage, gender, M stage, T stage and grade were different from these two groups (P < 0.05) (Additional file 3: Figure S2 B-G). However, CRFGs-based signature had poor predicted outcomes in M stage (M0, M1), T stage (T1-T4), stage (stageI-IV) and Grade (G1-G2) (P > 0.05) (Additional file 4: Figure S3. A-E). Moreover, a CRFGs risk score-based nomogram was developed as graphical calculators for predicting the prognosis in KIRC patients. The nomogram performed good calibration and accuracy (Fig. 4C-D). Therefore, the nomogram may be used in the evaluation of KIRC patients.
Functional enrichment
KEGG and GO analysis showed a significant enrichment in intrinsic apoptotic, response to oxygen levels, DNA damage, apoptotic, angiogenesis, nutrient levels and chemical stress (Fig. 5B). In summary, this signature is related to biological metabolism, drug resistance, tumor immunity and metastasis in KIRC.
Immune infiltration and drug sensitivity analysis
The immune infiltration of KIRC-high and KIRC-low is displayed in Additional file 5: Figure S4. The immune checkpoint of ICOS, BTLA, CTLA-4, CD27, CD28, CD40, LAG3, PVR, SIRPA, TIGIT, HLA-DPA1 and TNFRSF9, was upregulated in KIRC-high group (Additioanl file 6: Figure S5). Immune infiltration showed the level of CD44 and PML correlated positively to dendritic Cells, CD4-CD8 + T cells and B cells. SLC2A3, PML and CD44 were positively related to macrophages and neutrophils; The CDKN2A expression level correlated negatively to macrophages. TRIB3 was negatively associated with CD4-CD8 + T cells (Fig. 6A-F). The drug sensitivity analysis found that RDEA119, Bexarotene, Bicalutamide, Bortezomib, Cyclopamine, Embelin, Midostaurin, Dasatinib, Thapsigargin, Sorafenib, Salubrinal, and Obatoclax Mesylate were higher in low- than high-risk group (Additional file 7: Figure S6).
Discussion
In this study, we established a prognostic model that incorperate six-gene signature of CRFGs and clinical features of KIRC patients to predict survival. The developed model had moderate discrimination and good calibration abilities in predicting survival. A robust association of CRFGs signature and KIRC prognosis patients was verified by functional enrichment, immune infiltration, immune checkpoint, tumor micro-environment and drug sensitivity analyses.
The KIRC patients’ prognosis is related to clinical index, genes, proteins and so on. Cuproptosis and Ferroptosis are new types of cell death mechanisms [26, 27]. All the cuproptosis and ferroptosis-related genes are considered to be promising anti-tumor targets [28,29,30]. Emerging research suggested that cuproptosis and ferroptosis were key factors of KIRC development. To date, based on cuproptosis and ferroptosis, multiple biomarkers with fair prediction performance for predicting prognostic mRNA or lncRNA value in KIRC have been established [26, 31,32,33]. However, the association of CRFGs and prognosis of KIRC needs to be investigated, as developing a CRFGs signature for predicting KIRC prognosis and optimizing therapeutic methods is speculated to be helpful.
The prognostic signature was screened in this study (TRIB3, SLC2A3, PML, CD44, CDKN2A and MIOX). TRIB3 could inhibit mitosis in germ cell [34]. TRIB3 can promote cell proliferation in Renal Cell Carcinoma Cells by MAPK Signaling Pathway [35]. Moreover, TRIB3 induces immune evasion and reduces CD8+ T cell infiltration in colorectal cancer [36]. In breast cancer, TRIB3 supports cell stemness by regulation of SOX2 transcription [36]. SLC2A3, a membrane proteins, could inhibit tumor growth by up-regulating miR-184 [37]. In gastric cancer, SLC2A3 promotes infiltrating macrophages [38]. PML is a tumor suppressor response to environmental stimuli and crucial to antiviral defense activities [39]. CD44, known as P-glycoprotein 1, has been associated with tumor metastasis and invasion [40, 41]. Our study investigated the prognostic value of six CRFGs in KIRC. However, more intensive researches are warranted to explore potential regulatory effects for KIRC.
The functional enrichment revealed that the CRFGs related to many immune-related biological processes and pathways. Renal cell carcinomas (RCCs) highly resistant against chemotherapies, which may be due to impaired intrinsic or extrinsic apoptotic pathways [42, 43]. p53, a key regulator response to DNA damage, associated with poor patient prognosis and aggressiveness of tumor [44, 45]. Interestingly, p53 is inactivated in KIRC and considered as inducer for DNA damage response [46].
Cuproptosis and ferroptosis are also considered a form of immunogenic cell death [47]. Tumor microenvironment (TME) is a novel factor for cancer treatment [48]. Our results showed that Treg cells and tumor-associated macrophages (TAMs) were abundant in KIRC-high group than KIRC-low group. The CD8+T cells are vital to tumor progression and have antitumor effect.[49]. In KIRC, CD8+T cells was a favorable prognostic factor [50]. Our results are consistent with this conclusion. Moreover, We witnessed the expression of immune checkpoint, including CTLA-4, BTLA, CD27, CD28, CD40, ICOS, LAG3, PVR, SIRPA, TIGIT and TNFRSF9, was higher in high risk group might be owing to the immunosuppressive microenvironment. Immune checkpoint inhibitors activate the immune cell to kill cancer cells [51]. Ipilimumab, an anti CTLA-4 drugs, was used in KIRC patients [52]. All the results revealed that the signature can be further developed to evaluate the efcacy of KIRC patients.
Predicting the drug sensitivity promoted in improving drug selectivity and increasing the success rate of therapy [53]. Surprisingly, high-risk group patients were more susceptible to RDEA119, Bexarotene, Bicalutamide, Bortezomib, Cyclopamine, Embelin, Midostaurin, Dasatinib, Thapsigargin, Sorafenib, Obatoclax. Mesylate, and Salubrinal. In early study, cyclopamine was safe and well tolerated by the mice [54]. In glioblastoma, Cyclopamine acts to suppress carcinogenesis [55]. This may provide novel therapeutic strategies in KIRC patients.
Cell death has been implicated to cancer development [56]. Cuproptosis and ferroptosisis are two distinct regulated cell death mechanisms. Such unusual mechanisms may lead to a new therapeutic opportunity for treating cancer. In our study, the predictive value of CRFGs was comparable in KIRC-high and KIRC-low group. Nonetheless, it also has some limitations. First, whether this CRFGs signature can modulate KIRC process remains unknown. Their function needs further exploration. Second, the utility of prognostic model in this study requires further validation by a large sample size based on prospective studies in future research. Last but not least, cuproptosis and ferroptosis are new fields of cancer research. All genes were used in our study may be incomplete due to more and more these genes will be discovered.
In conclusion, we established a novel CRFGs signature that can predict KIRC prognosis and further studies are needed for validation of signature.
Availability of data and materials
The data during the current study are available from TCGA and GEO database.
Abbreviations
- RCC:
-
Renal cell carcinoma
- CRFGs:
-
Cuproptosis-related Ferroptosis genes
- ccRCC:
-
Clear cell renal cell carcinoma
- LASSO:
-
least absolute shrinkage and selection operator
- KIRC:
-
Renal clear cell carcinoma
- TCGA:
-
Cancer genome atlas
- TCA:
-
Tricarboxylic acid
- ROS:
-
Reactive oxygen species
- DEGs:
-
Diferentially expressed genes
- GO:
-
Gene ontology
- FDR:
-
False discovery rate
- BP:
-
Biological processes
- CC:
-
Cellular components
- MF:
-
Molecular function
- PPI:
-
Protein–protein interaction
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- ROC:
-
Receiver operating characteristic
- C-index:
-
Concordance index
- TIMER:
-
Tumor immune estimation resource
- GDSC:
-
Genomics of Drug Sensitivity in Cancer
- CI:
-
Confidence interval
- HR:
-
Hazard ratio
- TME:
-
Tumor microenvironment
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34.
Znaor A, Lortet-Tieulent J, Laversanne M, Jemal A, Bray F. International variations and trends in renal cell carcinoma incidence and mortality. Eur Urol. 2015;67(3):519–30.
Yin L, Li W, Xu A, et al. SH3BGRL2 inhibits growth and metastasis in clear cell renal cell carcinoma via activating hippo/TEAD1-Twist1 pathway. EBioMedicine. 2020;51: 102596.
Lalani AA, McGregor BA, Albiges L, et al. Systemic treatment of metastatic clear cell renal cell carcinoma in 2018: current paradigms, use of immunotherapy, and future directions. Eur Urol. 2019;75(1):100–10.
Fendler A, Bauer D, Busch J, et al. Inhibiting WNT and NOTCH in renal cancer stem cells and the implications for human patients. Nat Commun. 2020;11(1):929.
Tsvetkov P, Coy S, Petrova B, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins [published correction appears in Science. Science. 2022;375(6586):1254–61.
Kahlson MA, Dixon SJ. Copper-induced cell death. Science. 2022;375(6586):1231–2.
Brewer GJ. Copper control as an antiangiogenic anticancer therapy: lessons from treating Wilson’s disease. Exp Biol Med (Maywood). 2001;226(7):665–73.
Larson CA, Blair BG, Safaei R, Howell SB. The role of the mammalian copper transporter 1 in the cellular accumulation of platinum-based drugs. Mol Pharmacol. 2009;75(2):324–30.
Zhang X, Yang Q. Association between serum copper levels and lung cancer risk: a meta-analysis. J Int Med Res. 2018;46(12):4863–73.
Baltaci AK, Dundar TK, Aksoy F, Mogulkoc R. Changes in the serum levels of trace elements before and after the operation in thyroid cancer patients. Biol Trace Elem Res. 2017;175(1):57–64.
Basu S, Singh MK, Singh TB, Bhartiya SK, Singh SP, Shukla VK. Heavy and trace metals in carcinoma of the gallbladder. World J Surg. 2013;37(11):2641–6.
Ding X, Jiang M, Jing H, et al. Analysis of serum levels of 15 trace elements in breast cancer patients in Shandong China. Environ Sci Pollut Res Int. 2015;22(10):7930–5.
Saleh SAK, Adly HM, Abdelkhaliq AA, Nassir AM. Serum levels of selenium, zinc, copper, manganese, and iron in prostate cancer patients. Curr Urol. 2020;14(1):44–9.
Xie Y, Hou W, Song X, et al. Ferroptosis: process and function. Cell Death Differ. 2016;23(3):369–79.
Manz DH, Blanchette NL, Paul BT, Torti FM, Torti SV. Iron and cancer: recent insights. Ann N Y Acad Sci. 2016;1368(1):149–61.
Chen P, Li X, Zhang R, et al. Combinative treatment of β-elemene and cetuximab is sensitive to KRAS mutant colorectal cancer cells by inducing ferroptosis and inhibiting epithelial-mesenchymal transformation. Theranostics. 2020;10(11):5107–19.
Wang Y, Zhao G, Condello S, et al. Frizzled-7 identifies platinum-tolerant ovarian cancer cells susceptible to ferroptosis. Cancer Res. 2021;81(2):384–99.
Klevay LM. IHD from copper deficiency: a unified theory. Nutr Res Rev. 2016;29(2):172–9.
LaVaute T, Smith S, Cooperman S, et al. Targeted deletion of the gene encoding iron regulatory protein-2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nat Genet. 2001;27(2):209–14.
Gupta A, Lutsenko S. Human copper transporters: mechanism, role in human diseases and therapeutic potential. Future Med Chem. 2009;1(6):1125–42.
Wee NK, Weinstein DC, Fraser ST, Assinder SJ. The mammalian copper transporters CTR1 and CTR2 and their roles in development and disease. Int J Biochem Cell Biol. 2013;45(5):960–3.
Gao J, Zhao N, Knutson MD, Enns CA. The hereditary hemochromatosis protein, HFE, inhibits iron uptake via down-regulation of Zip14 in HepG2 cells. J Biol Chem. 2008;283(31):21462–8.
Doguer C, Ha JH, Collins JF. Intersection of Iron and Copper Metabolism in the Mammalian Intestine and Liver. Compr Physiol. 2018;8(4):1433–61.
Zhang G, Sun J, Zhang X. A novel Cuproptosis-related LncRNA signature to predict prognosis in hepatocellular carcinoma. Sci Rep. 2022;12(1):11325.
Bian Z, Fan R, Xie L. A novel cuproptosis-related prognostic gene signature and validation of differential expression in clear cell renal cell carcinoma. Genes (Basel). 2022;13(5):851.
Zhang Y, Shi J, Liu X, et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat Cell Biol. 2018;20(10):1181–92.
Wang W, Green M, Choi JE, et al. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569(7755):270–4.
Liu Y, Liu Y, Ye S, Feng H, Ma L. Development and validation of cuproptosis-related gene signature in the prognostic prediction of liver cancer. Front Oncol. 2022;12: 985484.
Bao JH, Lu WC, Duan H, et al. Identification of a novel cuproptosis-related gene signature and integrative analyses in patients with lower-grade gliomas. Front Immunol. 2022;13: 933973.
Sun Z, Li T, Xiao C, et al. Prediction of overall survival based upon a new ferroptosis-related gene signature in patients with clear cell renal cell carcinoma. World J Surg Oncol. 2022;20(1):120.
Yu J, Mao W, Xu B, Chen M. Construction and validation of an autophagy-related long noncoding RNA signature for prognosis prediction in kidney renal clear cell carcinoma patients. Cancer Med. 2021;10(7):2359–69.
Xing Q, Ji C, Zhu B, Cong R, Wang Y. Identification of small molecule drugs and development of a novel autophagy-related prognostic signature for kidney renal clear cell carcinoma. Cancer Med. 2020;9(19):7034–51.
Hong B, Zhou J, Ma K, et al. TRIB3 promotes the proliferation and invasion of renal cell carcinoma cells via activating MAPK signaling pathway. Int J Biol Sci. 2019;15(3):587–97.
Liu J, Zhang W, Chuang GC, et al. Role of TRIB3 in regulation of insulin sensitivity and nutrient metabolism during short-term fasting and nutrient excess. Am J Physiol Endocrinol Metab. 2012;303(7):E908–16.
Eyers PA, Keeshan K, Kannan N. tribbles in the 21st century: the evolving roles of tribbles Pseudokinases in biology and disease. Trends Cell Biol. 2017;27(4):284–98.
Xu H, Wang X, Wu J, et al. Long non-coding RNA LINC01094 promotes the development of clear cell renal cell carcinoma by upregulating SLC2A3 via MicroRNA-184. Front Genet. 2020;11: 562967.
Yao X, He Z, Qin C, et al. SLC2A3 promotes macrophage infiltration by glycolysis reprogramming in gastric cancer. Cancer Cell Int. 2020;20:503.
Lin YC, Lu LT, Chen HY, et al. SCP phosphatases suppress renal cell carcinoma by stabilizing PML and inhibiting mTOR/HIF signaling. Cancer Res. 2014;74(23):6935–46.
Zhou Q, Li J, Ge C, Chen J, Tian W, Tian H. SNX5 suppresses clear cell renal cell carcinoma progression by inducing CD44 internalization and epithelial-to-mesenchymal transition. Mol Ther Oncolytics. 2021;24:87–100.
Yaghobi Z, Movassaghpour A, Talebi M, et al. The role of CD44 in cancer chemoresistance: a concise review. Eur J Pharmacol. 2021;903: 174147.
Heikaus S, van den Berg L, Kempf T, Mahotka C, Gabbert HE, Ramp U. HA14-1 is able to reconstitute the impaired mitochondrial pathway of apoptosis in renal cell carcinoma cell lines. Cell Oncol. 2008;30(5):419–33.
Toth C, Funke S, Nitsche V, Liverts A, Zlachevska V, Gasis M, Wiek C, Hanenberg H, Mahotka C, Schirmacher P, Heikaus S. The role of apoptosis repressor with a CARD domain (ARC) in the therapeutic resistance of renal cell carcinoma (RCC): the crucial role of ARC in the inhibition of extrinsic and intrinsic apoptotic signalling. Cell Commun Signal. 2017;15(1):16.
Nientiedt M, Müller K, Nitschke K, et al. B-MYB-p53-related relevant regulator for the progression of clear cell renal cell carcinoma. J Cancer Res Clin Oncol. 2021;147(1):129–38.
Vieler M, Sanyal S. p53 Isoforms and Their Implications in Cancer. Cancers (Basel). 2018;10(9):288.
Mitchell TJ, Rossi SH, Klatte T, Stewart GD. Genomics and clinical correlates of renal cell carcinoma. World J Urol. 2018;36(12):1899–911.
Catanzaro E, Feron O, Skirtach AG, Krysko DV. Immunogenic cell death and role of nanomaterials serving as therapeutic vaccine for personalized cancer immunotherapy. Front Immunol. 2022;13:925290.
Bi K, He MX, Bakouny Z, et al. Tumor and immune reprogramming during immunotherapy in advanced renal cell carcinoma. Cancer Cell. 2021;39(5):649-661.e5.
Ma J, Zheng B, Goswami S, et al. PD1Hi CD8+ T cells correlate with exhausted signature and poor clinical outcome in hepatocellular carcinoma. J Immunother Cancer. 2019;7(1):331.
Matsuki M, Hirohashi Y, Nakatsugawa M, et al. Tumor-infiltrating CD8+ T cells recognize a heterogeneously expressed functional neoantigen in clear cell renal cell carcinoma. Cancer Immunol Immunother. 2022;71(4):905–18.
Dong MB, Wang G, Chow RD, et al. Systematic immunotherapy target discovery using genome-scale in vivo CRISPR screens in CD8 T cells. Cell. 2019;178(5):1189-1204.e23.
Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016;17(12):e542–51.
Mucaki EJ, Zhao JZL, Lizotte DJ, Rogan PK. Predicting responses to platin chemotherapy agents with biochemically-inspired machine learning. Signal Transduct Target Ther. 2019;4:1.
Risso-Ballester J, Galloux M, Cao J, et al. A condensate-hardening drug blocks RSV replication in vivo. Nature. 2021;595(7868):596–9.
Eimer S, Dugay F, Airiau K, et al. Cyclopamine cooperates with EGFR inhibition to deplete stem-like cancer cells in glioblastoma-derived spheroid cultures. Neuro Oncol. 2012;14(12):1441–51.
Strasser A, Vaux DL. Cell death in the origin and treatment of cancer. Mol Cell. 2020;78(6):1045–54.
Funding
This work was supported by Grants from National Natural Science Foundation of China (Foundation number: 81970448) and National Key R&D Program of China (Foundation number: 2018YFC1106504).
Author information
Authors and Affiliations
Contributions
GL and CL designed the research; LW and GL collected and analysis the data. GL and CL wrote and revised the manuscript. ZZ and BG participated in manuscript revision. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
Not applicable.
Competing interests
The authors declare that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
The gene list of 19 cuproptosis-related genes and 254 ferroptosis-related genes.
Additional file 2: Figure S1.
Distribution of survival status based on the median risk score in validation set;The receiver operating characteristiccurve analyses of the prognostic CRFGs in predicting 1-, 3-, and 5-year overall survival;Kaplan–Meier survival analysis of KIRC patients between high-risk groups and low-risk groups.
Additional file 3: Figure S2.
Correlation between signature and clinical characteristics.
Additional file 4: Figure S3.
Kaplan–Meier curves of OS diferences stratifed by gender, age, grade, N stage, T stage, or M stage between the high-risk groups and low-risk groups.
Additional file 5: Figure S4.
Immune cells infiltration between high-risk groups and low-risk groups.
Additional file 6: Figure S5.
The relationship between prognostic signature and immune checkpoints.
Additional file 7: Figure S6.
Drug sensitivity analysis.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Luo, G., Wang, L., Zheng, Z. et al. Cuproptosis-Related Ferroptosis genes for Predicting Prognosis in kidney renal clear cell carcinoma. Eur J Med Res 28, 176 (2023). https://doi.org/10.1186/s40001-023-01137-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40001-023-01137-z