Abstract
COVID-19 has been a major global health concern for the past three years, and currently we are still experiencing coronavirus patients in the following years. The virus, known as SARS-CoV-2, shares a similar genomic identity with previous viruses such as SARS-CoV and MERS-CoV. To combat the pandemic, modern drugs discovery techniques such as in silico experiments for docking and virtual screening have been employed to design new drugs against COVID-19. However, the release of new drugs for human use requires two safety assessment steps consisting of preclinical and clinical trials. To bypass these steps, scientists are exploring the potential of repurposing existing drugs for COVID-19 treatment. This approach involves evaluating antiviral activity of drugs previously used for treating respiratory diseases against other enveloped viruses such as HPV, HSV, and HIV. The aim of this study is to review repurposing of existing drugs, traditional medicines, and active secondary metabolites from plant-based natural products that target specific protein enzymes related to SARS-CoV-2. The review also analyzes the chemical structure and activity relationship between selected active molecules, particularly flavonol groups, as ligands and proteins or active sites of SARS-CoV-2.
Similar content being viewed by others
Introduction
COVID-19, also known as coronavirus disease 2019, is a highly infectious illness that is caused by the novel coronavirus, which has been officially named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This disease was first discovered on December 31, 2019, in Wuhan, China, as a cluster of pneumonia cases. Later, on March 27, 2020, World Health Organization (WHO) declared the outbreak a global pandemic, as it had spread to numerous countries around the world [1, 3, 5].
An outbreak of SARS was first reported in Guangdong, China, in November 2002 [7]. This disease was later identified in Hong Kong in late February 2003, and it subsequently spread globally to North America, Europe, and other parts of Asia [9, 11]. However, phylogenetic analysis showed that SARS coronavirus (SARS-CoV) differed from previously known coronavirus [13]. In June 2012, another coronavirus-related respiratory illness, the Middle East respiratory syndrome (MERS), caused by MERS coronavirus (MERS-CoV), emerged in the Middle East, particularly in Saudi Arabia, and was spread to humans by dromedary camels [14, 16, 17]. MERS-CoV is phylogenetically related to bat coronavirus (SARS-CoV-2), the virus that causes COVID-19.
The genomic characteristics of SARS-CoV-2 indicate that it is closely related (88% identity) to two bat-derived SARS-like coronavirus, bat-SL-CoVZC45 and bat-SL-CoVZXC21, were detected in Rhinolophus pusillus bats from Zhoushan, eastern China, in 2018 [19, 21]. Additionally, Zhou et al. reported that a coronavirus strain, SARSr-Ra-BatCoV-RaTG13, isolated from Rhinolophus affinis bats in Pu'er, China, in 2013, has an overall genome identity of 96.2% to SARS-CoV-2n. This close phylogenetic relationship to RaTG13 suggests that SARS-CoV-2 originated in bats [22].
According to Lam et al. [18], receptor-binding domain (RBD) of SARS-CoV-2 spike (S) protein exhibits extremely high sequence similarity to Guangdong pangolin (97.4% amino acid similarity). The amino acids of this pangolin coronavirus, GX/P2V, are identical to the five critical residues of RBD, while RaTG13 has only one identical amino acid to SARS-CoV-2. It is worth noting that SARS-CoV-2 rapidly spread among human populations. The lack of insertion of the polybasic cleavage sites in the spike protein of pangolin coronavirus contributed to this phenomenon.
Lau et al. [23], stated that the genome backbone of SARS-CoV-2 evolved from bat coronavirus, its RBD region was likely acquired from pangolin coronavirus, causing SARS-CoV-2 to become a recombinant virus. Additionally, SARS-CoV-2 RBD has distinct evolutionary characteristics compared to other Sarbecovirus species, particularly in terms of subunit cleavage sites. While the genomic characteristics of SARS-CoV and MERS-CoV are more distant from SARS-CoV-2, with similarities of 79% and 50%, respectively, RBD of SARS-CoV-2 within lineage B was found to be closer to that of SARS-CoV [21].
However, viruses are known to enhance their infectivity by acquiring mutations that allow viruses to evade human immune responses, including those triggered by vaccines and drugs. The original strain of SARS-CoV-2 underwent mutations primarily in its spike protein (S) that resulted in the emergence of several variants, such as α, β, γ, δ, and Omicron variants (Table 1). The number and location of mutations within the spike protein influences the characteristics and potential risks of each variant in evading infection by circumventing human antibodies and immune responses [24]. Recent surge in COVID-19 cases, known as the third wave, has been attributed to the Omicron variant, specifically the B.1.1.529 strain. The high reinfection rate and greater transmissibility of this variant are believed to be due to the large number of mutations in the spike protein. Therefore, addressing the nature of the virus and developing effective treatment to overcome current and future waves of infections should be a top priority.
Structure of coronavirus
The size of the virus ranges from 20 to 300 nm and it is capable of infecting and replicating cells. It contains genes and proteins enclosed within a lipid layer envelope or a non-enveloped one. Specifically, SARS-CoV-2 has a diameter ranging from 60 to 140 nm with a spike protein size of approximately 9 to 12 nm. Its virion is spherical, sometimes pleomorphic, with a diameter of 78 nm and resembles a solar corona. Goldsmith et al. and Tshibangu et al. stated that the virus contains a helical nucleocapsid within an envelope [29, 30].
According to Wang and Liang [31], viruses associated with acute respiratory infections include influenza, parainfluenza, picornaviruses, coronavirus (CoV), adenoviruses, and respiratory syncytial viruses. The human coronavirus (HCoV) has a complex structure, with an RNA genome inside the nucleocapsid protein, coated by spike glycoproteins and an envelope on the outer side (Fig. 1). The viral envelope is composed of structural membrane containing spike (S), envelope protein (E), and membrane lipoprotein (M). The viral lipid bilayer envelope, which is sensitive to desiccation, heat, and amphiphiles such as soap and detergents, is more susceptible to sterilization outside the human cell environment than the non-enveloped virus. However, the glycoproteins in the viral envelope helps the virus bind to the receptor sites on the host membrane to avoid the human immune system. Therefore, coronavirus binds to its primary receptor, the cellular angiotensin-converting enzyme 2 (ACE2), through its spike glycoproteins. Once the spike binds to the receptor, the cell and viral membrane fuses directly, causing the virion RNA genome inside the capsid to enter the host cell or endocytosis [1, 3, 32, 33].
Viruses can spread through the stages of their life cycle, which include cellular entry, translation, replication of the viral genome, and egress from the host cell to infect new cells [34]. While interferon (IFN) plays a crucial role in the host defence against viruses, [32, 35], efforts have also been focused on disrupting specific stages of the virus life cycle to inhibit and prevent viral infection. In particular, disrupting the viral envelope has been identified as a promising approach to impede viral egress [36, 37]. The lipid bilayer composition can be disrupted through lysis, exocytosis, or direct budding from the plasma membrane.
Modes of action of antiviral agents related to virus life cycle
In accordance with previous studies, various targets have been identified for developing antiviral drugs based on the virus life cycle, namely (1) inhibitors of fusion or entry, which targets the interaction between the virus and the host cell membrane, (2) uncoating inhibitors, a technique used to acidify the viral interior to weaken electrostatic interaction, (3) nucleic acid synthesis terminators, used to block viral enzymes, (4) integrase inhibitors, utilized to target the attachment of host cell DNA to the viral genome through the replication step, (5) protease inhibitors, often combined with reverse transcriptase inhibitors, and (6) release inhibitors, used to hinder or block the receptor from viral protein attachment [38,39,40,41].
Drugs targets for inhibiting viral infections are started by blocking the initial step of viral attachment to the receptor. This was achieved through receptor blockade by using a monoclonal antibody against the major cellular receptor or by employing specific inhibitor compounds [42]. For soluble receptors, blocking can be accomplished by disrupting the interaction between the glycosylated extracellular domains of the receptor and the hydrophobic transmembrane region on the virus. It usually blocks viral replication in cell culture form and prevents the attachment of the mutant virus to the receptor. This blockade effectively inhibits viral replication in cell cultures and decrease the affinity of the virus to bind to the receptor, making it less virulent and nonviable [42].
McKinlay et al. [42], suggested a mechanism for blocking viral entry into host cells by inhibiting the attachment of the virus to the host cell receptor. The first step in viral infection involves the attachment of the virus to the cellular receptor on the surface of the host cell, with the ACE2 receptor contributing to the attachment of coronavirus to the host cell, as shown in Fig. 2. The binding pocket of the virion capsid protein, containing hydrophobic amino acid side chains, interacts with the hydrophobic domains of the soluble receptor through van der Waals physical interactions. This interaction dissolves the virion capsid proteins or viral envelope in the outer space of the host cell, releasing the virion RNA genome, which becomes damaged by its surrounding conditions. The virion genome cannot replicate or maintain its genomic structure unless inside the host cell. Another mechanism for blocking the virus from releasing its viral genome during the uncoating process of the virion capsid or viral envelope is by providing inhibitor compounds that act as chelating agents within a hydrophobic pocket of the virus. This approach fills the empty pocket of the virus, preventing extensive conformational shifts that could cause the virion to disassemble. Therefore, chelating agents, such as a divalent cation, may block the release of RNA from the capsid.
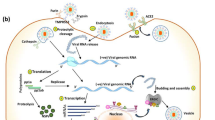
Coronavirus life cycle starts from entering a host cell (infection) until the production and release of a new virus. The words in the blue state are the steps of viral transformation inside the host cell. The activation process begins when the viral glycoprotein attaches to hACE2 and TMPRSS2 receptors. The virus fuses into the cell by endocytosis and starts to enter the cell. Following this cellular entry, the virus undergoes an uncoating process to release its viral genome. Further, structural protein synthesis is followed by RNA packaging, budding, and assembly to form a mature virion. Exocytosis releases a new infectious virion to infect a new cell. The insert box (on the bottom right) illustrates what happens to the host cell resulting in further cell death
Most inhibitors target the glycoprotein of the virion capsid or viral envelope, inhibiting viral attachment to the receptor. However, positive-stranded viral RNA is translated once the virus has attached and entered the host cell. The viral genomic RNA is released from the viral capsid through an uncoating mechanism, often facilitated by a receptor such as ACE2. Inside the host cell, viral proteins replicate and produce new ones through mRNA synthesis (Fig. 2). The virus strain, host cell type, pH, temperature, and multiplicity of infection influence this process. Typically, a virus requires five to ten hours to replicate in a single cycle [31].
SARS-CoV-2 virus consists of a large membrane glycoprotein called the structural protein (S protein), which includes several proteins such as membrane, spike, envelope, and nucleocapsid. S protein belongs to the class I viral fusion glycoproteins and is responsible for cell entry. Among the sixteen non-structural proteins (nsp1-16), three play a crucial role in the replication, transcription, and host cell recognition processes. These proteins are chymotrypsin-like protease (3CLpro), papain-like protease (PLpro), and RNA-dependent RNA polymerase (RdRp) [33, 39, 43, 44]. Cys-proteases and papain are protein degrading and processing enzymes, especially during the translation process. Chymotrypsin protease (3Cpro) contains Cys-proteases with a sulfhydryl group that cleaves the glutamine-glycine amide bond. 3CLpro is a highly conserved protease and plays a vital role in coronavirus replication by overlapping polyproteins pp 1a (486 kDa) and pp 1ab (790 kDa) in SARS-CoV [31]. Both PLpro and 3CLpro are necessary for replication, transforming polyproteins into non-structural proteins such as RdRp and helicases. 3CLpro and PLpro contain 11 and 3 cleavage sites, respectively. Therefore, 3CLpro is also known as the main protease (Mpro) and an ideal target for developing antiviral drugs [45]. 3CLpro is a highly conserved protease, and its substrate specificity is similar to the 3Cpro of the main picornavirus. Protease inhibitors can block the proteolytic process of viral polyproteins, leading to incorrect viral replication and transcription.
Drugs targeting SARS-CoV-2 spike protein can prevent membrane fusion between the spike and virus, thereby disrupting virus entry into the host cell. The spike protein also contains SARS-CoV-2 RNA-dependent RNA polymerase (RdRp), which recognizes the ACE2 receptor [33, 46]. ACE2 is a type I transmembrane metallocarboxypeptidase that plays a crucial role in the physiological renin-angiotensin system by hydrolyzing vasoconstricting angiotensin II into vasodilating angiotensin. During severe COVID-19, ACE2 expression is downregulated, leading to inflammation or cytokine storm and an increase in interleukins and other stimulating factor proteins. Therefore, modulating ACE2 expression may be a potential strategy for controlling COVID-19 symptoms.
Vaccines for COVID-19
A vaccination campaign using emergency-approved vaccines is underway in many countries. According to WHO website, there are current 13 different COVID-19 vaccines from four platforms that have been widely released and administered worldwide. These vaccines include Pfizer/BioNTech Comirnaty (BNT162b2), Moderna mRNA-1273, AstraZeneca AZD1222 and Covishield from the Serum Institute of India, Johnson & Johnson Ad26.COV2.S and Sinopharm COVID-19 vaccine from China etc. Table 2 also shows two versions of the AstraZeneca/Oxford COVID-19 vaccine produced by AstraZeneca-SKBio (Republic of Korea) and the Serum Institute of India as the ChAdOx1-S vaccine as mention on WHO website on 15 February 2021 [15]. The Pfizer/BioNTech COVID-19 (BNT162b2) vaccine is a nucleoside-modified mRNA vaccine that utilizes lipid nanoparticles to encode the prefusion SARS-CoV-2 spike protein [47]. The Ad26.COV2.S vaccine, an alternative to the ChAdOx1S recombinant vaccine produced by the Serum Institute of India, is also a recombinant vaccine that uses a vector to encode the full-length SARS-CoV-2 from incompetent adenovirus serotype 26 (Ad26) [48].
The significant advancement in vaccine development has allowed for the production of effective vaccines against SARS-CoV-2 and the development of herd immunity within the community. However, drugs and therapeutic actions are still necessary to manage and treat COVID-19 cases. Li et al. (2022), and Rehman et al. (2021), reported that majority of the vaccines recently developed use novel techniques such as messenger RNA (mRNA) to stimulate the human immune system [49, 50]. The techniques for applying messenger RNA (mRNA) as a recognizing and reactive component of the virus to create the immune system of the human body have been developed as a significant advancement in public health and vaccine development. Instead of containing a weakened or inactive form of the virus, these vaccines rely on the immune system of the body to recognize and attack the spike protein of the virus. After vaccination, vaccine particles interact with immune system cells, which deliver the mRNA message to create the spike protein in vaccinated cells. The immune system then recognizes the spike protein as foreign and produces antibodies for its destruction. This process generates an immune response that continues until all spike proteins have been eliminated, enabling the immune system to fight the virus upon infection.
The COVAX initiative, under the auspices of WHO, has facilitated the development and manufacturing of COVID-19 vaccines, aiming to ensure equitable access worldwide. In order to meet the demands of the pandemic, all recommended drugs and vaccines for SARS-CoV-2 were assessed based on the Emergency Use Listing (EUL) procedure, which ensures safety, efficacy, and quality standards. The EUL relies on a rigorous evaluation of late-phase II and phase III clinical trials, which are independently reviewed by WHO experts and teams.
WHO has emphasized that a vaccine on its own will not end the pandemic [64]. Despite the progress of the vaccination program, numerous cases associated with it have been reported, and doubts remain regarding the its long-term efficacy. The emergence of new coronavirus strains has become a significant challenge that needs to be addressed promptly. Additionally, the preparation required for administering two doses of the vaccine and booster shots is a significant task that medical and health services, pharmaceutical industries, and governments must fulfil [65].
Alerts on several medical products have been issued to the following release of COVID-19 vaccines to increase the public awareness of drugs and vaccine safety. Some rare adverse events related to vaccine use have been reported to inform individuals in making informed decisions about enhancing their immune systems. For instance, Sinovac vaccines have been linked to deafness and cerebral venous thrombosis [66, 67]. Janssen Pharmaceutical R&D team has reported that booster shots enhance immunity and maintain a safety profile of relatively 93.7% efficacy in the US [68]. However, the vaccine has been associated with rare adverse effects such as thrombocytopenia [69] and acute myocarditis [70]. The AstraZeneca vaccine has also been associated with adverse reactions such as thrombosis and blood clots [71,72,73]. Despite this, the positive benefit-risk profile of the vaccine and its tremendous potential to prevent infections and reduce deaths worldwide have outweighed the adverse effects, and it continues to be used in the public domain. Based on previous studies, Curevac, which uses an unmodified RNA-based vaccine, has low efficacy [59], with an efficacy rate of only 47% against SARS-CoV-2 [60]. Other modified mRNA-based vaccines such as Moderna and Pfizer have demonstrated significant efficacy and have been approved for emergency use during COVID-19 pandemic. In addition, mRNA vaccines are straightforward to manufacture, have a high biosafety profile, and are a safer vector than DNA, with no chance of infectious viruses [74, 75]. The development of modified mRNA-based vaccines has garnered widespread support.
As the public becomes increasingly aware of the safety and efficacy of drugs and vaccines, new natural-based alternatives are being explored. Despite the lack of specific drugs to cure COVID-19, recommendations for treating the disease have emerged from informal trials, including traditional herbal medicine. This has led to a renewed interest in repurposing or repositioning drugs, including natural products such as medicinal plants and some commercial synthetic drugs that have previously shown antiviral activity.
Repurposing antiviral agents as a potential way of drugs discovery for COVID-19
Several studies have been working to discover drugs to combat coronavirus using various methods. Some of these proposed drugs were discovered through in silico studies involving bio- and immuno-informatics, while others were discovered using conventional organic synthetic chemistry based on the retrosynthetic method [76, 77]. However, all proposed drugs must undergo preclinical and clinical testing, including a series of safety and health considerations, before they can be released commercially. These steps can take over five years to assess the safety of a drugs for consumption. Based on the emergency and pandemic nature of COVID-19, WHO has authorized health sectors and scientists to openly communicate their results and clinical trial assessments for new drugs to combat the virus.
In light of the health concerns surrounding the use of drugs to treat COVID-19 and the time required for their clinical assessment, an approach to repurpose or reposition existing drugs that have previously been recognized as effective antiviral was proposed. This approach is based on certain criteria, including the similarity of the virus type or group, genomic composition, and structure. Examples of some drugs that have been repurposed in this way are shown in Fig. 3, Table 3.
The phylogenetic tree can group viruses that share similar characteristics based on their genomic composition and structure. HCoV is a member of the Coronaviridae group, an RNA virus that causes respiratory tract infections. This means that HCoV viruses have close relationships with each other, as shown in a phylogenetic tree analysis. SARS-CoV-2, as a member of this group, has an 88% similarity in identity to two bat-derived SARS-like coronavirus [20, 21]. Additionally, the viral structure can be used as a critical factor in grouping the virus. For example, SARS-CoV-2 has an enveloped viral design, similar to herpes simplex virus (HSV), human immunodeficiency virus (HIV), retrovirus, flavivirus, and hepatitis B and C virus (HBV/HCV). On the other hand, non-enveloped structures are found in human papillomavirus (HPV), poliovirus, norovirus, and rhinovirus. Respiratory tract infections can be caused by viruses such as rhinoviruses, influenza, parainfluenza, respiratory syncytial virus (RSV), enteroviruses, coronavirus, and certain strains of adenovirus.
Antiviral agents previously used to treat respiratory tract diseases are potential candidates for repurposing as drugs for COVID-19. For instance, resveratrol has been proven to reduce inflammation and levels of interferon-gamma (IFN-γ) in human RSV A2-strain virus infections. This was demonstrated in in vitro assays using 9HTEo and Hep-2 cell lines, as well as in vivo assays using BALB/c nude mice [89, 90]. Therefore, resveratrol is a promising candidate for treating COVID-19 infections.
Baicalin and baicalein from Scutellaria baicalensis Georgi have been proposed as potential treatment for COVID-19 due to their inhibitory effects on the activity of HIV-1 reverse transcriptase, which blocks HIV-1 replication [94, 97, 98]. Since HIV is also an enveloped virus like SARS-CoV-2, it is hypothesized that baicalein may also inhibit COVID-19. Su et al. [103] investigated the effects of baicalin and baicalein against SARS-CoV-2 in silico study. The results showed that baicalein interacted with the two catalytic residues of SARS-CoV-2, acting as a shield to prevent further interaction with the substrates or receptors of human cells.
The approach of repurposing drugs based on the nature similarity of the virus is considered a promising technique to identify potential treatment for COVID-19. This method is viewed as a faster alternative to developing new drugs since existing medicines that have been approved as safe for use are repurposed for COVID-19 treatment, thereby eliminating the need for additional safety assessments. Consequently, this approach saves time and expedites drugs release to the public.
The strategy of repurposing drugs is crucial in responding to the emergence of new variants of SARS-CoV-2, which result from natural mutation and evolution of the virus. The high levels of viral transmission have led to the emergence of virus variants associated with increasing viral transmissibility but not disease severity. Clinically tested drugs and vaccines should also cover variant B.1.351, which has been associated with reduced efficacy of some previously recommended ones. Therefore, the scientific response to the rising number of new SARS-CoV-2 variants must adapt quickly to develop practical antiviral activity against these emerging variants. WHO reported that efforts to suppress transmission, protect the vulnerable, and save lives in a comprehensive and coordinated manner needs to be redoubled in response to the welcoming of new variants of SARS-CoV-2 in 2021. Several new variants of SARS-CoV-2 were identified after whole-genome sequencing in samples from Brazil (SARS-CoV-2 (P1) derived from B.1.1.28 lineage, the United Kingdom (SARS-CoV-2 VUI 202012/01, some listed as SARS-CoV-2 VOC 202012/01 from cluster B.1.1.7 lineage), and South Africa (501Y.V2 variant as an N501Y mutation) at the end of 2020 and into the following year. Therefore, the acceleration of access to vaccination campaigns worldwide and the development of drugs discovery are supported.
Repurposing of existing drugs has led to individuals or groups using commercial antiviral agents, such as chloroquine or hydroxychloroquine and ivermectin, without medical prescription. Scientific studies have shown that these FDA-approved drugs, initially developed as antiviral and antiparasitic agents, have potential in inhibiting SARS-CoV-2 in in vitro and in silico assays [104,105,106,107]. While the efficacy and risks associated with the use of chloroquine and hydroxychloroquine in COVID-19 treatment have been debated [108,109,110], they were even proposed as chemoprophylaxis in some countries, with trials conducted in 2020 [111]. In June 2020, WHO announced on their website that hydroxychloroquine should be discontinued as it did not reduce mortality in hospitalized COVID-19 patients [112,113,114]. Despite this, controversy over the use of hydroxychloroquine remains, and some countries continue to use it as a COVID-19 treatment [104, 111, 115, 116]. It may be a promising candidate for further investigation as a treatment for SARS-CoV-2 [117]. Ivermectin has also demonstrated antiviral activity against SARS-CoV-2 in in vitro studies, with an IC50 of approximately 2 μM [118, 119]. Chaccour et al. conducted randomized clinical trials on the use of ivermectin as a COVID-19 treatment [120]. WHO (2021c), recommends that ivermectin should only be used in COVID-19 treatment based on clinical trials as the evidence supporting its efficacy is inconclusive [121]. Doxycycline has also been investigated for its ability to inhibit SARS-CoV-2 in in vitro studies, with an EC50 of 4.5 ± 2.9 mM when tested on the IHUMI-3 strain in Vero E6 cells [122].
The use of some commercial medicines has been approved by health and medical ministries to treat COVID-19 (Fig. 4). WHO is conducting solidarity therapeutic trials in over 30 countries, enrolling nearly 12,000 patients, to find effective treatment for the disease. However, after six months of attempting these trials, WHO reported in October 2020 that remdesivir, hydroxychloroquine, lopinavir or ritonavir, and IFN regimens showed little or no effect on patients hospitalized for 28 days. Other drugs, including oseltamivir, azvudine, ribavirin, favipiravir, and auranofin, have been recommended off-label [106, 123,124,125,126]. Remdesivir, an adenosine analogue that stops RNA synthesis and acts as a false substrate for RdRp, is one of the primary drugs used to treat hospitalized patients [125, 127, 128]. It is important to note that the effectiveness of these drugs in treating COVID-19 is still being evaluated.
In December 2021, Pfizer released a new oral drugs for COVID-19 called Paxlovid, which contains nirmatrelvir (300 mg) and ritonavir (100 mg) [129,130,131,132]. Administered orally twice daily for five days, Paxlovid has demonstrated a significant reduction in COVID-19-related deaths [133]. Nirmatrelvir (PF-07321332) has shown oral activity against SARS-CoV-2 in vitro, and its potency was demonstrated in phase I clinical trials with a tolerable plasma concentration in the cell [134, 135]. The detailed computational analysis of PF-07321332 against SARS-CoV-2 Mpro has been clearly discussed in several studies [136, 137]. In October 2021, Pfizermectin, new drugs for COVID-19 treatment suspected to contain ivermectin, was also developed by Pfizer, but the company has denied repackaging ivermectin inside this new pill and selling it at a higher price than existing commercial ivermectin. It was suspected of containing ivermectin, a protease inhibitor proposed to kill parasites. Molnupiravir has been shown to successfully target the viral RdRp on the Omicron variant [138,139,140]. In early 2022, WHO recommended two new drugs for COVID-19 patients, baricitinib and sotrovimab [141], while other monoclonal antibodies, such as bebtelovimab and Evusheld (containing tixagevimab and cilgavimab), have also gained attention due to their high efficacy against the virus at mild to moderate levels [142].
Rupintrivir (AG7008) are other drugs being investigated for its potential to treat COVID-19. It contains a lactam ring that mimics Glutamine residues at the P1 position and forms a covalent bond with the active-site cysteine residue of the virus protease [143]. In vitro antiviral assays using H1-HeLa and MRC-5 cells have shown Rupintrivir to have a potent broad-spectrum antiviral activity against 48 HRV serotypes and four related picornaviruses [144]. Ramajayam et al. has proven that the fluorophenylalanine group and isoxazoyl moiety in rupintrivir may hinder its ability to bind to Arg188 in the S2 pocket and hydrophobic residues of SARS-CoV 3CLpro, respectively [143]. Therefore, its efficacy in treating COVID-19 is still being studied in detail. A similar case has also been observed with amantadine, which was previously used to treat influenza. In vitro studies have shown an inhibitory effect of amantadine on SARS-CoV-2 infected Vero E6 cells with an IC50 between 83 and 119 μM [145]. The dosage required for in vitro efficacy is not feasible in vivo due to toxicity concerns. Its therapeutic window cannot be offered, suggesting that the oral administration of amantadine appears obsolete. Several studies reported that amantadine could be administered through inhalation, as the infection of human airways by SARS-CoV-2 covers a high concentration in the nasal epithelium until distal pulmonary epithelium [146].
IFN are signaling proteins produced by host cells that have shown therapeutic potential for MERS and SARS-CoV [35, 147], making it a proposed treatment for COVID-19. The EC50 of IFN-α and IFN-β treatment on infected SARS-CoV-2 Vero cells is reported to be 1.35 IU/mL and 0.76 IU/mL, respectively [148]. In addition, glucocorticoids such as ciclesonide, dexamethasone, betamethasone, hydrocortisone, fludrocortisone, and triamcinolone are potential candidates for treating inflammation accompanying COVID-19 [149]. Other therapies such as convalescent plasma and anti-interleukin-6 (anti-IL-6) inhibitors have also been explored to combat the pandemic.
Investigations have proven that 25-hydrocholesterol, a type of oxidized cholesterol products found in various human body fluids, has the potential to inhibit COVID-19 with an IC50 of 550 nM by blocking membrane fusion [150, 151]. In addition, 25-hydrocholesterol is oxidized cholesterol products found in human peripheral blood, cerebrospinal fluid, colostrum, and milk. Several studies are considering 25-hydroxycholesterol and 27-hydroxycholesterol, which are side-chain oxysterols, as potential inhibitors of respiratory viruses against COVID-19 [152].
Clinical trials have been conducted using a drugs repurposing approach, either with a single-molecule therapy or a combination of therapies, to treat COVID-19. However, one of the studies involving 1206 randomized patients showed no improvement in the recovery of mild to moderate COVID-19 patients using a single treatment of ivermectin [153]. Combination therapies involve the simultaneous repurposing of therapeutic, antiviral, immunotherapeutic, and convalescent plasma therapies. Remdesivir is popular antiviral drugs that has received emergency approval from WHO. A combination of remdesivir and baricitinib, as immunotherapeutic agents, produces better outcomes in hospitalized patients with COVID-19 than the use of only remdesivir. The use of two antiviral drugs, remdesivir and dexamethasone, has resulted in reduced mortality for 30 days. The use of combined antiviral and antibiotic therapies has also been proven to be more effective and safer for early symptomatic patients [154].
The application of repurposed drugs has yielded some promising examples of inhibitors for SARS-CoV-2. Several studies have identified available drugs agents that can inhibit the protein and reproduction cycles of viruses [155,156,157]. In addition, clinical trials conducted on April 2020 have shown that the combination of natural products, such as honey, and Nigella sativa seeds, improved symptoms and reduced mortality without adverse effects [158]. The Ayurvedic drugs, AYUSH 64, demonstrated improved recovery and reduced hospitalization for mild-moderate symptomatic patients [159]. The potential of these natural compounds as alternative COVID-19 drugs and therapeutic agents is described in detail in the following section of this study.
In-silico study on drugs discovery for COVID-19
Modern drugs discovery relies on in-silico studies involving molecular docking and dynamics. This approach uses bioinformatics and computational modelling to design new lead compounds and enable virtual screening of bioactive metabolites [160]. However, by enabling preliminary screening activities, this technology accelerates the identification and analysis of bioactive compounds, while significantly reducing time and costs associated with laboratory work. Molecular modelling is particularly useful for repurposing existing drugs and natural products, as it predicts the affinity and binding mode of molecules to the active site of a receptor protein.
Computer modelling enables the efficient screening of hundreds and thousands of compounds, in experiments conducted for a brief period. Promising compounds or drugs identified during the process are further subjected to docking studies (Table 4, Fig. 5). In order to understand the mode of action of these compounds or drugs, molecular dynamics (MD) simulations were used to model their interaction with the active site of the virus [161,162,163]. However, through artificial neural network analysis, medicines are classified based on their primary role in SARS-CoV-2 infection, specifically in viral replication and immune response. This approach differentiates between antiviral agents that prevent virus replication and those modulating immunity to combat the virus without overreacting. Once a potential antiviral candidate is identified, further assays using human and non-human cell lines are necessary.
Viral entry is prevented by targeting the host receptor, ACE2 and other proteins parts of coronavirus, such as spike glycoproteins (including nsp1-16, RdRp) and proteases (Mpro and PLpro), and has been explored as a potential strategy [33, 157, 179,180,181]. In-silico investigations provide valuable parameters, including RMSD, docking scores, and binding affinities, to assess the effectiveness of antiviral agents.
A notable example of applying the in-silico approach for drugs discovery is the study conducted by Elinger et al. [182]. They successfully generated a small set of related drugs that exhibited significant activity in terms of IC50. A primary screening assay [183] of 5632 compounds was tested for their ability to inhibit SARS-CoV-2 in human epithelial colorectal adenocarcinoma cells (Caco-2) [184]. After the procedure, 271 compounds were selected based on achieving more than 75% inhibition cut-off, as determined by quantifying cell viability readouts. Subsequently, 184 compounds were further chosen based on their clinical status. Among these, 64 compounds demonstrated an IC50 value of less than 20 μM, while 19 exhibited an IC50 value of less than 1 μM.. This study highlighted six of the 64 compounds, namely camostat, nafamostat, lopinavir, mefloquine, papaverine, and cetylpyridium. However, 90% of those confirmed compounds have not been reported as SARS-CoV-2 antiviral agents in in vitro cell assays [182, 185]. The names and structures of those compounds were not disclosed in the present study.
The molecular docking-based virtual screening approach using AutoDock Vina was employed to identify potential inhibitors of 3CLpro of SARS-CoV-2 [200]. The top four compounds were selected from a pool of 2000 compounds in the ZINC database, based on their low free energy binding adherence to the Lipinski rule of five, and functional molecular interactions with the target protein. Similarly, Barage et al. utilized AutoDock Tool 1.5.6 to retrieve 3277 compounds from the ZINC database and generate 10 top compounds with the lowest binding energy against RdRp (PDB ID: 6NUR) and Nsp15 (PDB ID: 20ZK). MD simulations performed with GROMACS tools, was used to identify three compounds with the highest affinity to interact with RdRp and Nsp15 namely alectinib, naldemedine, and ergotamine [201].
Potential components of Ayurvedic medicinal plants have been assessed for their repurposing possibility as anti-COVID-19. After screening selected compounds from twelve medicinal plants, molecular docking and dynamic simulations showed that curcumin, gingerol, and quercetin were potential candidates [202]. In another study, fluoro-substituted heterocyclic ring systems were added to quercetin-based derivatives, which were then screened by in silico experiments against SARS-CoV 3CLpro (PDB ID: 6LU7) using Autodock 4.2 software. The compounds L4 (5-fluoro-2H-1,2,3-triazol-4-yl), L8 (2-fluoro-4H-1,3-oxazin-4-yl), and L14 (3-fluoropiperidin-4-yl) showed promising results, with IC values of 0.330, 0.456, and 0.50 uM, respectively. Additionally, a study on marine natural product-based drugs-like small molecules screened 14,492 compounds from the MNP library, of which 7471 compounds fulfilled Lipinski rule of five. After conducting the evaluation process through ADMET descriptor, 2033 compounds were selected for further analysis. Docking analysis and molecular dynamic simulations of 14 compounds led to the identification of six hits of phenyl propanoid compounds, including fasciospongiside A, epolactoena, constanolactone B, constanolatone F, debromo araplysillin I, and maniloside A as potential anti-COVID-19 agents [178].
The Korea Chemical Bank Drugs Repurposing (KCB-DR) database, consisting of 1,865 compounds, was used to propose potential therapeutic agents for COVID-19. GOLD virtual screening identified 149 binders based on their Goldscore and Chemscore. MD simulations were then employed to analyze the binding modes and fundamental interactions, thereby revealing seven top drugs. Based on the binding free energy approaches, ceftaroline fosamil and telaprevir emerged as potential drugs against SARS-CoV-2 with telaprevir raising safety concerns due to its side effects. In order to address this, a substructure search in the PubChem database led to the identification of 11 potential derivatives of telaprevir exhibiting desirable pharmacokinetic properties, particularly lower hepatotoxicity [203]. However, in another study, the molecular interactions and stabilities of 3,639 drugs from the SuperDRUG2 database were analyzed using PyRx and GROMACS v5.1.5. It was observed that colchicine emerged as the top binding compound against SARS-CoV-2 Mpro [204].
Sharma and Kaur investigated the potential of jensenone, a key component of eucalyptus oil, as an inhibitor for COVID-19 infection [205]. The in-silico study revealed that jensenone formed a complex structure with the main viral proteinase/chymotrypsin-like proteinase (Mpro) through hydrophobic, hydrogen bonds, and strong ionic interactions. Paul et al. explored synthetic molecules, peptidomimetic, and small molecules inhibitors targeting viral proteinases to assess its potentials as anti-SARS-CoV Mpro agents through computational approaches [206]. Another study by J. K. R. da Silva et al. [207], investigated the potential of 171 essential oil components in treating SARS-CoV-2 using molecular docking analysis. The findings showed that (E)-β-farnesene exhibited the best normalized docking score, while (E,E)-α-farnesene, (E)-β-farnesene, and (E,E)-farnesol were identified as the best docking ligands. Unfortunately, the docking energies were relatively weak, limiting their applicability to coronavirus interactions.
The Searching off-lAbel drugs aNd NEtwoRk (SAveRUNNER) is an interesting approach for repurposing existing drugs to treat COVID-19. This method evaluates the interaction between drugs and target protein based on their location and position in the same network neighbourhoods. Recent study utilized 14 COVID-19-related diseases to generate 282 repurposing drugs of 1875 FDA-approved drugs from DrugBank v5.1.6. Ruxolitinib has the potential to inhibite JAK and H1-antihistamines that play a vital role in controlling immune responses [208]. Asides from SAveRUNNER, other in-silico approaches for identifying potential repurposing drugs include network module separation and the RWR algorithm. Both approaches highlight the disease module of H1N1 flu and SARS-CoV-2 infection [209]. The development of network-based mechanisms involves multimodal technology using artificial intelligence, network diffusion, and proximity algorithms. [210], stated that 76 of the 77 drugs achieved viral effects through indirect viral protein binding targets by perturbing the host subcellular network. Molecular docking through computational approaches was used to observe the interaction patterns of binding viral proteins to host targets. Indeed, network-based perturbations is induced by altering the virus ability to enter the cell or replicate within the cell. This advanced approach of in-silico method for drugs repurposing is beneficial in developing a faster and cheaper strategy for drugs discovery schemes.
However, computer modelling is not the only approach for determining drugs as reliable antiviral agents, even when it shows a strong binding mode to the active sites of the virus. Vatansever et al. [211] stated that calculated binding energy does not necessarily correlate strongly with the actual IC50 values. Computer modelling is the only approach used to obtain detailed information in relation to predicting the mode of antiviral action. The next crucial step is to conduct in vitro and in vivo assays in preclinical trials. These assessments help to identify a small number of drugs or compounds for further evaluation in clinical trials. As public awareness of health and safety increases and the challenges posed by viral infections persist, there is a growing need for alternative, nature-based medications. This alternative treatment can complement existing approaches and offer potential solutions for viral infections that are difficult to cure or present challenges during treatment.
Natural products for treatment of viral infection
Medicinal plants encompass all plants or herbs whose components exhibit biological activities. These bioactive compounds, when extracted from medicinal plants, can be considered as lead compounds. In recent times, there has been a growing interest in novel natural approaches to treating viral infections, driven by increased public awareness and concern for safety and health issues in comparison to synthetic drugs. The utilization of natural products as remedies for various infectious diseases often stems from the fields of ethnobotany, phytochemistry, and local wisdom, giving rise to ethnopharmacology. However, it is important to note that in many cases, there are insufficient or even lack of scientific evidences to substantiate the health-related information or knowledge associated with these natural remedies.
Discovery of alternative drugs for treating viral respiratory diseases such as COVID-19 has led to repurposing of natural products with new pharmacological properties. Medicinal plants from the Lamiaceae, Cupressaceae, and Zingiberaceae families, as well as isolated natural products such as ritonavir, chloroquine phosphate, arbidol, and ribavirin, have shown potential antiviral activities against some viruses [212]. These natural products have also been found to be beneficial in preventing and relieving the symptoms of COVID-19. Other natural products compounds, such as vitamin D (calcitriol), vitamin C (ascorbic acid), lactoferrin, quercetin, resveratrol, hanfangchin A (tetrandrine), glycyrrhizin, artemisinin, colchicine, and berberine, are current under clinical trials for treatment of COVID-19 [213]. In addition, Panyod et al. [5] stated that the use of immunomodulator foods and herbs containing large amounts of vitamins C and D, flavonoids, and essential oils, helps strengthen the immune system and acts as air disinfectants or sanitizers to prevent aerosol transmission of the virus. The use of rich and bulky spices found in tropical lands, such as cinnamon, cloves, mint, lemon, and balm, also offers possibilities for discovering bioactive natural molecules suitable against viruses [214,215,216,217,218,219].
Several food sources have been found to play a role in the immunomodulatory system by reducing inflammation. For example, the fruit extract of Embelia schimperi (Myrsinaceae), which contains benzoquinones, has been proven to exhibit potent HCV-PR (hepatitis C virus protease) activity [220]. Pomegranate peel extract (PPE), which contains polyphenols, has also been found to have immunomodulatory effects [221,222,223]. In addition, Ali et al. [224], and Wen et al. [95], reported that among 221 phytocompounds, some diterpenoids, sesquiterpenoids, triterpenoids, and lignoids were potent inhibitors against SARS-CoV on Vero E6 cells. Other possible sources of natural products active compounds in treating human diseases include endophytes and medicinal plants. [105], stated some molecules obtained from medicinal plants that have been claimed to be effective against SARS-CoV-2 in virtual assays or clinically applied, although there is no scientific proof.
Traditional herbal medicines
Traditional herbal remedies are widely used as complementary or alternative medicines in many countries, particularly in the context of eastern medicine. These remedies consist of traditional medicinal preparations derived from single or combined medicinal plants. Despite the lack of comprehensive studies, these therapies have been employed for centuries in treatment of various ailments. Traditional Chinese Medicine (TCM), which boasts a history of over 2000 years, and Ayurveda are two prominent herbal remedies enjoying trust and popularity not only within their countries of origin, China and India respectively, but also in other parts of the world.
In a study conducted by Yang et al. [227], various TCM herb formulae and extracts were identified for their potential in treating SARS-CoV infections, along with TCM-derived compounds exhibiting anti-HCoV activities. Notably, the Yin Qiao San formula demonstrated positive therapeutic effects against upper respiratory tract infections, while Ma Xin Gan Shi Tang exhibited anti-SARS-CoV activity. Several TCM compounds were found to possess antiviral properties, particularly against SARS-CoV, MERS, and SARS-CoV-2. These include plant-derived phenolic compounds from Isatis indigotica root extract, litchi seed extract, herbacetin, rhoifolin, pectolinarin, quercetin, epigallocatechin gallate, and gallocatechin gallate. Glycyrrhizin from Glycyrrhizae radix, water extract of Houttuynia cordata, and emodin derived from Rheum and Polygonum genera also exhibited antiviral activity (Fig. 6). Yi et al. [228], conducted study on 121 Chinese herbal medicines and reported that tetra-O-galloyl-β-D-glucose and luteolin were two active constituents effective against the wild type of SARS-CoV. Another review reported that TCM is obtained from a single preparation rather than the combination of medicinal plants or Chinese medical formulas. Xi et al. [229], specifically identified components of TCM herbs as potential agents against antiviral pneumonia, while An et al. [225] listed various TCM treatment along with their initial symptoms, outcomes, and effects on antiviral diseases. It is important to note that the evidence supporting TCM treatment relies on clinical evidence obtained from their practical use.
Some parts of medicinal plants are used as traditional herbal therapies to alleviate the symptoms of respiratory diseases, and the effectiveness of some of these remedies has been scientifically proven. However, there are other herbal remedies that have not been listed in international monographs [225, 226]. All pictures were obtained from the Google search engine and were not recognized as the author’s ownership or copyright
TCM is believed to treat COVID-19 by inhibiting the replication and transcription of SARS-CoV-2 through various mechanisms such as blocking the viral functions of RdRp, 3CLpro, spike protein, and PLpron. Additionally, it can hinder the binding of the virus to host cells by acting on ACE2 and TMPRSS2. TCM also has the potential to reduce cytokine production, prevent immune system impairment, and abnormal blood clotting following SARS-CoV-2 post-infection.
Shuanghuanglian is a popular traditional Chinese patent medicine that is formulated from the extraction of three Chinese herbal medicines, namely Lonicera japonica Thunb, Scutellaria baicalensis Georgi, and Forsythia suspense (Thunb.) Vahlv. The key constituents of this medicine are chlorogenic acid, phillyrin, and baicalin. Despite being a traditional medicine, Shuanghuanglian has undergone scientific investigations in China to assess its antiviral activity. Su et al. [45] reported that the inhibitory effect of Shuanghuanglian on SARS-CoV-2 3CLpro is primarily attributed to its significant components, baicalin and baicalein (an aglycone of baicalin). The IC50 value for this inhibition was found to be only 0.94 μM. However, it should be noted that these two compounds exhibited less than 50% inhibition activity against SARS-CoV-2 PLpro at a concentration of 50 μM.
The potent inhibitory activity of baicalein against SARS-CoV-2 3CLpro can be attributed to its structural features, including three phenolic hydroxyl groups, a carbonyl group, and a free phenyl ring. These features allow baicalein to form multiple hydrogen bonds and hydrophobic interactions with amino acid residues, both in the main and side chains of the viral active sites. While both baicalein and baicalin demonstrated significant inhibition, antiviral activity of Shuanghuanglian was found to be limited in Vero E6 cells. Su et al. [45], reported that this could be attributed to the low permeability of the cell membrane to the components of the preparation.
Ni et al. [230], reported a case in which a combination of Shuanghuanglian and Western medicine was used to treat three family members suffering from COVID-19, resulting in a positive therapeutic effect. However, the use of this method needs to be approached with caution due to the need for early treatment and potential errors, such as combining antibiotics and antiviral drugs. In addition to health concerns, some TCM herbs contain nephrotoxins and mutagens, such as aristolochic acids found in Aristolochia and related plants [231]. The regulation of these herbs varies among nations, China, Taiwan, and the US are some countries that have unregulated their usage.
Pudilan Xiaoyan Oral Liquid (PDL), another TCM preparation, has been the subject of study for disease enrichment analyses. It has shown promising potential in treating asthma and chronic obstructive pulmonary disease, with similar significance levels to COVID-19 (p = 2.4E−03 and p = 2.45E−03, respectively). PDL contains four herbs, including Indigowoad Root (Isatis indigotica), Bunge Corydalis (Corydalis Bungeana), Mongolian Dandelion (Taraxacum Mongolicum), and Scutellaria Amoena (Scutellaria Baicalensis) [232]. Interestingly, there is also a commercially available oral liquid in the United States known as Respiratory Detox Shot (RDS), which is a food supplement containing ingredients commonly used in TCM. These ingredients include Panax ginseng, Lonicera japonica, Forsythia suspensa, Glycyrrhiza uralensis, Scrophularia ningpoensis, etc. The effect of RDS on SARS-CoV-2 was investigated, resulting in an IC50 value at a 1:40 dilution [233].
Ayurveda, an ancient traditional medicine system dating back to the Vedic period (1500 to 500 BCE), has emerged as a potential remedy for mitigating the severity of COVID-19. Care [234], reported that the Ayurvedic approach, focuses on both preventive and curative aspects, tailored to different stages of the disease. Maurya et al. [235], have virtually screened natural products from Ayurveda to identify compounds capable of modulating the immune system and blocking the entry of SARS-CoV-2. Three plants of critical significance in Ayurvedic medicine, especially in Rasayana therapy, are Whitania somnifera, Tinospora cordifolia, and Asparagus racemosus. These plants contain active steroid compounds such as ashwagandhonolides, whitacoagin, withaferin, and withanone, which have shown potential against various proteins associated with SARS-CoV-2, including spike glycoprotein (RBD), RdRp, and Mpro [236]. Ayush-64, an Ayurvedic formulation used clinically for its anti-malarial, anti-inflammatory, and antipyretic properties, has demonstrated favourable binding energy to Mpro, with values of approximately − 8.4, − 7.5, and − 7.4 kcal/mol, corresponding to molecules akuammicine N-Oxide (from Alstonia scholaris), akuammiginone, and echitaminic acid, respectively [237]. Nimbin and curcumin, active compounds found in Ayurvedic formulations, have exhibited higher binding affinity than nafamostat, a synthetic protease inhibitor [235]. Figure 7 shows some of the active compounds found in traditional Ayurvedic medicines that have demonstrated efficacy against SARS-CoV-2 in in vitro assays.
In Romania, the native flora, including medicinal plants such as dandelion, daisy, and fat grass [238], was utilized as part of traditional medicine during current pandemic [225, 239]. These plants are rich in flavonoids, saponins, tannins, sterols, fatty acids, coumarin, and vitamins. Moreover, the Fritillaria species, known for their pharmacological effects on the respiratory system, possess antitussive, expectorant, and antiasthmatic properties. This genus has been included in the Ayurvedic (Fritillaria roylei), Korean (four species), and Chinese Pharmacopeia (ten species known as Bei Mu in Chinese), and are also widely used in Tibetan, Mongolian, Miao, Lisu, Tujia, Kazakh, Uighur, Jingpo and De’ang traditional medicine [240]. Other herbal products, Sumac, extracted from the Rhus genus, has demonstrated interactions with viral envelopes and host cell surfaces, exhibiting diverse antiviral activities against influenza A and B, HSV, and HIV. According to Korkmaz [241], it has been suggested as a potential treatment for COVID-19 infection.
Traditional medicines in the form of Jamu have been produced in Indonesia [242,243,244]. Popular Jamu formulations include wedhang jahe, jamu kunyit asam, jamu teulawak, and jamu beras kencur, which typically contain rhizomes from the ginger family (Zingiberaceae) such as Zingiber officinale and Curcuma longa. These formulations may also include additional ingredients like Cinnamomum verum bark, Citrus aurantifolia fruit, and starch fillers.
Although the use of Jamu is not officially recommended in treatment of COVID-19 patients, it has become a popular alternative among Indonesians as an immunostimulatory agents to prevent symptoms and promote speedy recovery from post-infection symptoms [245]. Another example is the application of virgin coconut oil (VCO) in the local communities as a therapeutic adjuvant to overcome inflammation caused by COVID-19. While no scientific evidence has been presented, clinical trials have been conducted in four hospitals in Yogyakarta caring for hospitalized COVID-19 patients [246]. It is important to note that traditional remedies should not be used as a substitute for medical treatment for COVID-19 and their effectiveness remains unproven.
According to the Committee on Herbal Medicinal Plant Products (EMEA/HMPC/892618/201), consuming the extract of Eucalyptus globulus Labill, in the form of dried leaves up to four times a day, is helpful in managing respiratory diseases such as bronchitis and rhinitis. This is because it contains active components such as 1,8-cineol and phenolic compounds. The British Herbal Pharmacopeia recommends the use of garlic products that contain sulfuric compounds, including allicin and mercaptan, amino acids, peptides, terpenes, minerals, and flavone glucosides to treat COVID-19, as it has traditionally been used to manage colds and whooping cough. However, it is important to note that Traditional Herbal Medicine Products (THMP) in Europe are not considered a treatment for COVID-19 as it is a severe, life-threatening illness [226].
The utilization of medicinal plant extracts mentioned earlier has gained widespread popularity worldwide as a recommendation to combat COVID-19, serving as an alternative to drugs provided by WHO and the Ministry of Health in each country. Zhang et al. [254], conducted in silico screening of Chinese herbal medicine and identified 13 active compounds effective against SARS-CoV-2. This method of analysis is beneficial in expediting drugs discovery process based on ethnobotanical reasoning. Exploring traditional medicine and ethnopharmacology presents a potential alternative for drugs discovery in combating COVID-19 pandemic. However, it is crucial to exercise caution regarding the preparation, dosage, and individual health considerations associated with traditional treatment prior to their application. Therefore, conducting a detailed investigation into the extraction of active compounds from traditional herbal medicines would prove advantageous as it focuses on the specific or known combination of active molecules responsible for their bioactivity, eliminating unnecessary or unsafe components that may be consumed.
Polyphenol-based secondary metabolites
Polyphenols are a prominent group of naturally occurring bioactive compounds found in plants that contain at least one substituted phenol ring or several hydroxyl groups on aromatic ring compounds. This group comprises four classes, namely phenolic acids, flavonoids, stilbenes, and lignans. The flavonoid class includes several derivatives such as chalcones, flavones, flavanones, flavonols, isoflavones, anthocyanins, and flavan-3-ols. Polyphenols are known for their broad antiviral activities against various viruses, including influenza A virus (H1N1), HBV/HCV, HSV, HIV, and Epstein-Barr virus (EBV) [255]. Table 5 shows some phenolic compounds that have been explored as antiviral agents, particularly against SARS-CoV-2. In silico and in vitro approaches have been used to study subsites of the virus, including proteins and enzymes related to SARS-CoV-2 and cell receptors in the human body.
Polyphenolic compounds can also act as antioxidants due to their hydroxyl groups, which react with radicals and oxidizing compounds. Resveratrol, a biflavonoid compound with the IUPAC name 3,5,4-trihydroxy-trans-stilbene, is a potent antioxidant that scavenges for reactive oxygen species, such as O2− and OH−, and lipid hydroperoxyl free radicals. Although it has poor oral bioavailability and water miscibility, resveratrol is rapidly metabolized in the body. Abba et al. [282] have stated the role of resveratrol and its action mechanisms in combating viral infections in human and animal cells. Therefore, resveratrol is presumed to have potential therapeutic benefits in treating COVID-19 by enhancing the immunity of infected patients. Quercetin is another popular phenolic compound that has been combined with N-acetylcysteine in the formulation of Quercinex to be directly administered to the deep lung tissue through a nebulizer to treat respiratory problems and multifocal pneumonia in COVID-19 patients [283]. Rutin, another phenolic compound, has been studied for its in-silico binding affinity to interact with the main protease of SARS-CoV-2 in the three-dimensional structures of PDB IDs 6LU7 ([170] and 6YNQ [169].
The polyphenols present in ethanol PPE have exhibited positive in vitro activity in reducing the interaction between SARS-CoV-2 spike glycoprotein and human ACE2, along with the activity of SARS-CoV-2 Mpro. ACE2 and TMPRSS2 gene expression levels were reduced by 30 and 70%, respectively, by applying PPE at 0.04 mg/mL on human kidney-2 cells infected by SARS-CoV-2 Spike pseudotyped lentivirus. Furthermore, PPE displayed the inhibition of Mpro activity by relatively 80% when used at 0.2 mg/mL [223]. It has also been evaluated in three commercial forms, namely pomegranate juice, a concentrated liquid extract, and 93% PP powder extract, to demonstrate its anti-influenza activity against PR8 (H1N1), X31 (H3N2), and H5N1 [221].
Dietary intake of polyphenols at high concentrations also regulates ACE2 expression and function, by acting as an antioxidant. Calceolarioside B is an active compound found in Akebia trifoliata fruit, which has been suggested as a potential dietary treatment for COVID-19 patients due to its various health benefits, including antimicrobial and anti-inflammatory effects [284]. This caffeic acid derivative is also present in other plants such as Stauntonia hexaphylla (leaves), Scutellaria galericulata L. (aerial parts), Forsythiae Fructus (fruit), and Mimulus guttatus (seeds) [285,286,287,288]. Figure 8 shows that active phenolic compounds, such as quercetin and vitamin C, have a synergistic effect as adjuncts in treating COVID-19 [289].
Based on this reasoning, it becomes evident that exploring the antioxidant potential of natural phenolic extracts, as well as other forms such as food-based extracts and polyphenol-containing functional foods or nutraceuticals [290,291,292], can be valuable in managing severe COVID-19 cases with inflammatory conditions like cytokine storm. Further discussion about the correlation between the activities of polyphenolic compounds, especially those with flavonoid structures, against the Mpro of SARS-CoV-2 has been elaborated in subchapter 8.
Alkaloid-based secondary metabolites
Alkaloids are a class of organic nitrogenous base compounds that occur naturally in plants, microorganisms, and animals. Their basicity depends on the form of nitrogen they contain, which can be primary, secondary, or tertiary amines. Alkaloids are categorized based on the amino acids that make up their nitrogen content and the structure of their alkaloid skeleton. These secondary metabolites have been found to exhibit a diverse range of biological and pharmacological activities, including antimicrobial, antiparasitic, antiasthma, analgesic, antihyperglycemic, anticancer, psychotropic, and stimulant properties. Some alkaloids have been identified as potential antiviral agents against SARS-CoV-2 through both in silico and in vitro assays, as outlined in Fig. 9, Table 6.
In 1818, quinine was discovered and isolated from Chincona bark, which prompted the exploration of other plant alkaloids due to their bioactivities [304]. Recent studies found that quinine exhibited promising activity against SARS-CoV-2 with an effective concentration of EC90 at 38.8 mM [305]. In Northern Chile, Schizanthus porrigens Graham, herbaceous species, contains a tropane-derived alkaloid called Schizanthine Z that actively binds to PLpro with docking affinity − 7.5 kcal/mol [168]. Another promising bis-benzylisoquinoline alkaloid, cepharanthine, showed strong activity against SARS-CoV-2 by blocking host Ca + channels and inhibiting virus fusion and entry [306]. Additionally, Cryptolepis sanguinolenta, a plant found in West Africa, contains antipathogenic-based alkaloids that could be a promising candidate for SARS-CoV-2 inhibitors [307]. Focus has also been directed to marine products in the search for secondary metabolite contents, particularly alkaloids. Some marine organisms such as sponges from Cryptotethya crypta, Dysidea avara, Crambe crambe, a cyanobacterium from Nostoc ellipsosporum, and starfishes from Fromia monilis and Celerina heffernani, produce a polycyclic guanidine alkaloid skeleton in their secondary metabolites, which act as antiviral agents [190].
The study conducted by Quimque et al. [308] focused on examining 97 antiviral secondary metabolites from fungi. They utilized computational modelling to screen these metabolites and identified Quinadoline B as the top-scoring compound, predicted to exhibit high binding affinity to various proteins associated with SARS-CoV-2, including PLpro, 3CLpro, RdRp, non-structural protein 15 (nsp15), and the spike binding domain to GRP78. Additionally, the ADMET value analysis indicated that quinadoline B is a favourable compound with high absorptive probability in the gastrointestinal tract and low capacity for crossing the blood–brain barrier.
Chowdhury [300] conducted a molecular docking study of five secondary metabolites from Tinospora cordifolia (Willd.) Hook.f. & Thomson (Menispermaceae) and found that berberine showed the best binding affinity of − 7.3 kcal/mol to 3CLpro of SARS-CoV-2, leading to an inhibition constant of 4.4 μM. Berberine has previously been shown to have antiviral activity against influenza, with a comparable IC50 to the standard drugs oseltamivir [309]. Garg and Roy [302] identified the four best molecules out of twenty antiviral alkaloids for potential scrutiny using Lipinski’s rule and docking study based on maximum negative binding energy with Mpro of SARS-CoV-2. Thalimonine, emetine, sophaline D, and tomatidine exhibited binding energies of − 8.39 kcal/mol, − 10.17 kcal/mol, − 8.79 kcal/mol, and − 9.58 kcal/mol, respectively. Thalimonine and sophaline D were recommended for further in vitro studies based on filters, parameters, and mechanisms of virtual bioactivity against Mpro of COVID-19. Various alkaloids derived from plants have been shown to have potent antiviral activity against various viruses, including coronavirus, as listed by Majnooni et al. [305] and Topcu et al. [304].
Terpenoid and its derivatives
Terpenoids are a diverse group of natural products that are derived from isoprene (1,3-butadiene) units. They are formed by combining carbon skeletons from other acetate and shikimate pathways such as steroidal saponins, cardioactive glycosides, and phytosterols. Terpenoids have many essential applications in the fields of medicine, cosmetics, and food industries. This group of secondary metabolites exhibits biological activities, including antitumor, anti-inflammatory, antibacterial, antiviral, antimalarial, and antidiabetic activities [310]. Some terpenoids have also been studied for their potential bioactivity against SARS-CoV-2 and illustrated in Fig. 10, Table 7.
Tetraterpenes, particularly astaxanthin from the carotenoid class, have been extensively discussed for their potential as an adjunctive supplement in COVID-19 [314]. Triterpene glycosides, such as saikosaponins A, B, C, and D, which can be isolated from Heteromorpha spp., Bupleurum spp., and Scrophularia scorodonia, have demonstrated antiviral activity against HCoV-22E9 [93]. Another saponin, 3-beta-O-(alpha-L-rhamnopyranosyl-(1- > 2)alpha-L-arabino pyranosyl)olean-12-ene-28-O-(alpha-L-rhamnopyranosyl-(1- > 4)-beta-D-glucopyranosyl-(1- > 6)-beta-D-glucopyranosyl) ester, isolated from leaves and stems of Oreopanax guatemalensis [315], exhibited the highest binding energy to interact with SARS-CoV-2 S-RBD compared to other terpenes using computer-based molecular simulation [316]. Li et al. [296] reported that extracts from Artemisia annua, Lycoris radiate, Pyrrosia lingua, and Lindera agregata were practical for anti-SARS-CoV screening analysis. The aqueous extract of Houttuynia cordata showed inhibition of viral 3CL protease and blockade activity of viral RNA-dependent RNA polymerase in SARS-CoV [317].
Glycyrrhizin, also known as glycyrrhizic acid, is a type of terpenoid saponin that is commonly extracted from the roots (Glycyrrhizae radix rhizoma) of glycyrrhiza plants, including Glycyrrhiza glabra L. and licorice root [251, 328]. The primary compound of glycyrrhizin is a triterpene glycoside, with its aglycone being 18b-glycyrrhetinic acid [329]. Due to its mode of action and characteristics, glycyrrhizin has the potential to be utilized as an anti-SARS-CoV-2 agents [251, 330]. In a case report of a non-hospitalized COVID-19 patient who took diammonium glycyrrhizinate, Ding et al. [331] stated that immune regulation against cytokine storm had improved, and inflammation was reduced.
Derivatives of glycyrrhizin, including the amide form, have been shown to exhibit higher anti-SARS-CoV activity than glycyrrhizin itself. This is due to the addition of amino acid residues on the glycoside part, while preserving the free -COOH function in C30, which appears to be crucial for the anti-SARS-CoV effect [332]. In vitro experiments on antiviral activity of aqueous licorice root extract containing glycyrrhizin against SARS-CoV-2 in Vero E6 cells have demonstrated that glycyrrhizin blocks SARS-CoV-2 replication by inhibiting Mpro, [252].
Terpenoids, specifically monoterpenes and sesquiterpenes, are highly active compounds that may interact more rapidly with the primary site of infection in COVID-19 patients when delivered via aerosol delivery systems such as nebulizers and inhalers. This form of drugs delivery is preferred over oral administration as it allows for direct entry of the terpenoids through the respiratory tract, which increases their bioavailability [255]. The aerosol form of terpenoids contains essential oils, which are volatile oils obtained from different parts of plants including leaves, fruit, flowers, bark, and roots using various extraction methods. Essential oils are classified as terpenoids because they predominantly contain monoterpenes and sesquiterpenes, rather than derivatives of phenyl, propanoid, and aromatic compounds.
According to Javed et al. [333], carvacrol, a phenolic monoterpene found in thyme and oregano, has demonstrated therapeutic properties against various viral diseases such as HSV type 1, bovine diarrhea virus, respiratory syncytial virus, and murine norovirus in vitro. It was proposed that carvacrol could also have potential mechanisms of action against SARS-CoV-2, the virus responsible for COVID-19. Specifically, carvacrol may interfere with ACE2 receptors in host cells, leading to protective effects against inflammation, and potentially hinder the virus's interaction with viral proteases during infection.
Essential oils derived from medicinal plants and their food matrices contain volatile compounds that can be quickly released, making them highly therapeutically potent compared to the original plants or herbs [334]. These essential oils can easily enter the body through inhalation and reach the bloodstream due to their high volatility. However, it is important to assess the duration of essential oil diffusion to ensure that it is safe to inhale and maintain indoor air quality [335].
Boukhatem (2020) conducted a literature review of published study articles and reported antiviral activities of essential oils and isolated compounds. The potential of essential oils from aromatic plants as antiviral compounds against coronavirus have also been explored [3, 207, 336]. Adorjan and Buchbauer [337], as well as Ojah [338], have listed essential oils with antiviral activities against human-targeting viruses. Using essential oils as a therapeutic antiviral intervention is a safe alternative due to their natural extract origin. Essential oils directly act on enveloped viruses, such as HSV type-1 and type-2, by binding to viral envelopes and glycoproteins. The plaque development assay supported this statement and showed that essential oils reduced viral load significantly during contact with virions before the adsorption process or during the pre-treatment step, but not when used before HSV-1 and HSV-2 adsorption and attachment [339, 340]. Time-of-addition experiments concluded that essential oils blocked virus adsorption [341]. Thymoquinone and black seed fixed oil were also found to be positively active against avian influenza virus (H9N2) and MCMV infection model 36. Pelargonium sidoides, extracted herbal products, has been licensed and marketed for patients with acute bronchitis, reducing rhinovirus infection and interfering with the reproduction of multiple respiratory viruses [163,164,165].
Essential oils are edible, but their potential toxicity requires caution when ingesting orally. The non-polar properties of essential oils make them easily permeable through skin membranes, leading to whole-body healing. As a result, essential oils can activate specific brain regions, influencing the hypothalamus and providing pain relief, mood enhancement, and improved cognitive function [334]. Due to concerns about their bioavailability, essential oils are recommended for topical application to the skin. Their lipophilic nature enables them to easily penetrate the skin and disrupt the virion envelope, inhibiting host cell attachment. Several essential oils with virucidal activity, such as lemongrass (Cymbopogon citratus) [342], lemon balm (Melissa officinalis) [343], peppermint [341], dwarf lavender cotton (Santolina insularis) [339], ginger (Zingiber officinale), thyme (Thymus vulgaris), hyssop (Hyssopus officinalis), and sandalwood (Santalum album) [340], have been identified. Table 7 highlights some potential essential oils and common terpenoids with activity against SARS-CoV-2.
The efficacy of natural plant essential oils in reducing virus titers (TCID50) against non-enveloped viruses at different temperatures and times was found to be insignificant. Several previous studies have investigated the impact of essential oils on non-enveloped viruses, such as norovirus, rotavirus, adenovirus, and HPV. For instance, Kovac et al. [344] examined the effect of Hyssopus officinalis and Thymus mastichina essential oils against murine norovirus (MNV-1) and human adenovirus serotype 2 (HAdV-2). Garozzo et al. [345] investigated Melaleuca alternifolia essential oil (tea tree oil, TTO) against polio type 1, ECHO 9, and Coxsackie B1. While Cermelli et al. Evaluated eucalyptus oil against adenovirus. In all of these [344, 346], essential oils were unable to mask non-enveloped viruses, indicating that they may not be a viable option for reducing foodborne viruses in the food industry. Conversely, essential oils have shown significant virucidal activity against enveloped viruses due to their ability to disrupt the virus's enveloped proteins and interaction with host cells. Jackwood et al. [347] reported that QR448(a), a blend of botanical oleoresins and essential oils developed by Quigley Pharma, Inc., exhibited virucidal effects against avian infectious bronchitis virus (IBV) in Vero E6 cells, embryonated eggs, and chickens by reacting before virus attachment and entry.
Antiviral mechanism of essential oils may be useful in inhibiting SARS-CoV-2 when the virus has structural similarities to other viruses. One such virus is HSV, an enveloped virus similar to SARS-CoV-2. Eugenol, extracted from Eugenia caryophyllus (Spreng.) Bullock & S.G. Harrison, was found to inhibit the replication of HSV standard strains [84] in HSV-1 and HSV-2 viruses [348] and delay the development of herpetic keratitis in HSV-1-infected mice. However, due to its stability concerns, eugenol is better suited for topical treatment rather than internal use. Eugenol in Syzygium aromaticum extract has also been evaluated for its ability to inhibit the replication of hepatitis C virus [220]. Another essential oil component, isoborneol from Salvia fruticosa, has dual viricidal activity against HSV-1, and inhibits virus replication and viral glycosylation at a concentration of 0.06%. Inactivation of the virus by isoborneol may lead to the interaction of the alcoholic moiety of isoborneol and the lipid in the virus envelope. Clove (S. aromaticum L.) has been reviewed as a potential therapeutic agents for anti-COVID-19 due to its essential oil content, with eugenol being a major component [349]. Clove extract has been shown to inhibit HCV replication [220] and exhibit chemopreventive activity [350]. This dried flower bud contains approximately 11–20% of the essential oil, while its dried leaves comprise less than 5% of the oil, with eugenol being a major component (70–90%) [350,351,352,353]. Eugenine, an isolated compound from Syzygum aromaticum extract, also exhibited anti-HSV potential activity [354]. Manuka oil was found to inactivate HSV before entering the cell, and its virucidal activity is believed to be due to the interference of β-triketones and other terpenes in adsorption and entry into host cells [337].
In an evaluation of coronavirus inhibition in HeLa-CEACAM1a cells, ethanol extracts of Nigella sativa, Anthemis hyalina, and peel of Citrus sinensis, which were presumed to contain essential oil compounds, were. Ulasli et al. [79] reported that A. hyaline extract molecules have the potential to treat CoV infections. Additionally, Salem and Hossain [99] noted that BSO from N. sativa exhibited a remarkable antiviral effect against MCMV infection.
According to Tkachenko's [102], essential oil extracted from the fruit and roots of Heracleum L. species (Apiaceae) demonstrated a toxicity LD50 of 0.2–0.4 mL against both Influenza Types A and B. The main constituents of these essential oils were found to be octyl acetate and octyl isobutyrate in the seeds, while the fruits contained monoterpenes such as pinene and limonene, as well as complexes of ethers of octyl and hexyl alcohols. The roots, on the other hand, were found to contain pinene, ocimene, and sesquiterpene derivatives. Hayashi et al. [88] reported that cinnamaldehyde, the primary constituent in Cinnamomi cortex (Cinnamomum cassia Blume), was able to reduce virus yields in the lungs during an infection with lethal influenza virus-induced pneumonia in the airways of mice.
CBD is a compound belonging to the class of cannabinoids and is typically found in Cannabis sativa. CBD has a unique chemical structure consisting of terpenes and phenols, which is commonly referred to as a terpenophenolic compound. Recent study by Anil et al. and Raj et al. [257, 311] suggests that CBD, along with other terpenoid and phenolic compounds, may possess potential activity against SARS-CoV-2. CBD has been detected in the blood plasma of healthy patients at concentrations in the nanomolar range when using approved CBD drugs. Conversely, CBD metabolite, 7-hydroxy-cannabidiol (7-OH-CBD), was found to be in the micromolar range. A study conducted on A549 human lung carcinoma cells expressing exogenous human ACE-2 receptor (A549-ACE2 cells) showed that CBD inhibited the replication of SARS-CoV-2 with an EC50 of 1.24, while 7-OH-CBD was 3.6 μM. Additionally, oral administration of CBD with a high-fat meal has been shown to increase the presence of 7-OH-CBD in the blood, which could effectively inhibit SARS-CoV-2 infection. Nguyen et al. [355] also found that CBD therapy resulted in a lower rate of testing positive for COVID-19 in patients. Furthermore, Chatow et al. [356] reported that CBD exhibited a synergistic effect on HCoV-229E-infected human lung fibroblasts when combined with the NT-VRL-1 terpene formulation. Current, CBD is available as a mouth spray under the licensed name Nabiximols, which is intended to reduce and relieve respiratory disease-related pain.
A study by Amparo et al. [357] on molecular docking of certain terpenoids and essential oils against SARS-CoV-2 showed satisfactory results. AutoDock Vina was used to investigate some compounds and the results revealed that (E)-α-atlantone, 14-hydroxy-α-muurolene, allo-aromadendrene epoxide, amorpha-4,9-dien-2-ol, aristolochene, azulenol, germacrene A, guaia-6,9-diene, hedycaryol, humulene epoxide II, α-amorphene, α-cadinene, α-calacorene, and α-muurolene have the highest binding energy values for PLpro, 3CLpro, S protein, and RdRp, respectively. Similarly, plant secondary metabolites such as bismahanine, eriodictyol-7-O-rutinoside, glycyrrhizic acid, and hypericin showed the highest binding energy against S protein, RdRp, TMPRSS2, and Mpro, respectively.
Antiviral activity of plant secondary metabolites against SARS-CoV-2 has been discussed, along with the various drugs discovery approaches that can be employed. Polyphenols, alkaloids, and terpenoids are some of the secondary metabolite classes that can serve as antiviral agents. Additionally, Machado et al. [358] and Pisoschi et al. [359] have suggested other secondary metabolites such as polysaccharides, lipids, vitamins, and animal-derived compounds for the modulation of early inflammatory responses in COVID-19 patients.
Structure–activity relationship (SAR)
SAR is a theoretical concept that links chemical molecules structure or structural-related properties to its biological activity or target properties. This model enables the modification of a moleculular structure to alter its bioactivity. Essentially, molecules with identical chemical properties that interact and bind with targets similarly will have similar activities. Therefore, SAR approach involves identifying the properties of molecules, such as geometric and electronic properties, solubility, and certain chemical groups, to predict its physicochemical and biological properties in targeting biological targets. SAR model reduces costs, time, and concerns related to toxicity bioassays.
Earlier studies by Mehaney et al. and Mengist et al. [366, 367] have discussed the detailed mechanism for designing inhibitors for SARS-CoV-2. Ye et al.[199] have also proposed a mechanism for inhibiting SARS-CoV-2 Mpro. The focus of this study is on the impact of flavonol structures on the binding affinities of Mpro to SARS-CoV-2, with Fig. 11 providing illustrations of three forms of flavonols with varying hydroxy substituents in the B ring skeleton of flavonol and glycosides.
Correlation of flavonol-based derivatives with their in-silico bioactivities with Mpro of SARS-CoV-2. All docking scores data presented here were obtained from different references, which will be influenced by their own use on (1) docking program, (2) docking algorithm, and (3) system set-up. Refs.: *[164]; **[281]; ***[265]; ****[262]; *****[360]; ******[361]; *******[362]; ********[363]; *********[364]; **********[365]
Quercetin derivatives, with mono- or di-substituents of the glycoside, exhibited high activity against Mpro. Quercetin-3-O-glucuronide 9, quercetin-3-O-rutinoside (rutin) 12, and quercetin-3,5-digalactoside 11 had binding affinities to Mpro of -9.4 kcal/mol, − 9.16 kcal/mol, and − 9.6 kcal/mol, respectively, which were higher than quercetin 2 (− 8.47 kcal/mol). This suggests that the presence of glycoside as a substituent on the quercetin skeleton is important for increasing binding affinity to Mpro. Most glycoside compounds have higher bioavailability in the body than aglycon [164]. However, isoquercitrin or isoquercetin 9, a monosubstituted quercetin glycoside, showed less binding energy than quercetin. The presence of glucose as the glycoside did not improve binding access to Mpro. Similar to quercetin, myricetin derivatives in a glycoside form also showed high binding affinity to Mpro compared to the lead compound, myricetin 3. Myricetin-3-O-rhamnoside (myricitrin) 13 and myricetin-3-O-rutinoside 14 are two examples of this. However, myricetin itself did not show a good binding affinity to Mpro, and was not ranked highly in previously reported study, with a binding energy of − 7.311 kcal/mol [164].
Myricetin derivatives have also been tested for their binding energy against the RdRp of SARS-CoV-2, and it has been found that myricetin-3-O-rutinoside (− 9.5 kcal/mol) has a lower binding energy compared to myricetin (− 8.4 kcal/mol). Myricetin has shown promising activity against Mpro in vitro, with an IC50 of 3.684 μM [262]. Therefore, it is expected that myricetin derivatives would exhibit better bioactivity against Mpro of SARS-CoV-2 based on in vitro and in vivo analysis.
Kaempferol 1, quercetin 2, and myricetin 3 are known structurally similar flavonols, differing only by the presence of hydroxy substituents at positions 3′ and 5′. Based on in silico analysis of their binding energy with Mpro, their relative binding energy values are comparable. The number of hydroxyl substituents on the B ring does not significantly affect their binding activity against Mpro, as they show similar binding activity values of around − 8.4 to − 8.5 kcal/mol, with critical energy data from other literature ranging from − 7.307 kcal/mol [362] to − 9.5 kcal/mol [164]. Quercetin 2 has been identified as crucial molecules [155] in the prophylaxis and treatment of COVID-19 patients due to its anti-inflammatory activity against cytokine storm during severe inflammation. Meanwhile, kaempferol derivatives, such as rhamnoside 4 and glucuronide 5 glycosides, exhibit higher binding affinities to Mpro (− 8.8 and − 9.1 kcal/mol, respectively) than the lead compound kaempferol (− 8.58 kcal/mol). The presence of two di-rhamnosides in the kaempferol skeleton at positions 3 and 7, as kaempferitrin 7, shows lower binding affinity to Mpro compared to kaempferol.
Based on the previous discussion, the optimal position for the hydroxy group on the B ring of the flavonol skeleton is on the para-substituted benzene ring, as shown in Fig. 12. The meta-position of the hydroxyl substituent does not significantly impact the binding energy, especially when comparing kaempferol 1, quercetin 2, and myricetin 3. When a glycosylated hydroxyl position is present on carbon number 3, compounds 4, 5, 6, 9, 10, 12–15 exhibit better binding affinity against Mpro than the original compounds or aglycon (1, 2, 3). An interesting observation is that compounds 7 and 11 have di-glycosides in their flavonol skeleton, but at different positions. While kaempferitrin 7 has two sugar moieties at positions 3 and 7, which reduces its binding energy against Mpro compared to the aglycon, kaempferol, quercetin-3,5-digalactoside 11 comprises two sugar moieties at positions 3 and 5, enabling it to bind better to the active site of Mpro than its aglycon, quercetin 2. The type of sugar presented as glycosides in molecules is presumed to influence molecules bioactivity against Mpro, with glucuronic acid, rhamnose, and rutinose enhancing and facilitating the binding affinity against Mpro, while glucose does not. This SAR between flavonol derivatives and Mpro may serve as the foundation for the development of a novel drugs compound against Mpro of SARS-CoV-2.
Current challenges against COVID-19
New variants of SARS-CoV-2 have emerged through mutations that increase their transmissibility, severity, and mortality. In the future, significant attention and funding will be given to the study of drugs, with a shift towards vaccine redesign when repurposing drugs is not enough. However, an effective vaccine cannot entirely prevent future mutant attacks, and not all vaccines are suitable for worldwide application due to factors such as environment, geography, and genetic diversity. Each SARS-CoV-2 variant has unique characteristics and infection roles, and current antibodies may not always be effective in neutralizing these variants. Therefore, specific vaccines are needed to enhance human immunity against each variant. The effectiveness of vaccines, such as NVX-CoV2373, varies across different regions and variants. In the UK, it showed 95.6% effectiveness against the original strain of SARS-CoV-2, but its efficacy in South Africa was 60%, and it was only 49.4% effective against the beta variant. Additionally, vaccine effectiveness diminishes over time, with up to a 50% reduction in efficacy observed after ten weeks of a booster dose [2].
After COVID-19 pandemic subsides, a major challenge that remains is increasing immunity to prevent unexpected virus mutations, thereby leading to questions, such as Should people receive annual vaccinations to prevent unexpected mutants? Further investigation on vaccine production is ongoing, but the long-term impact of immunization on the human body must also be investigated. Concerns have been raised about the potential carcinogenic effects of vaccination, which may not be detected quickly. Identifying adverse events after vaccination is important, especially for children, who have a longer expected future than older individuals. In some countries, vaccinations are mandatory for children over eight years old to improve their immunity. Therefore, it is important to track the progress in the muscular, cardiovascular, respiratory, and reproductive systems. In the coming years, study on COVID-19 will provide valuable scientific insights into preventing future unexpected viral infections.
Regarding the development of natural products into antiviral drugs, there are some considerations about what kind of challenges and problems researchers facing nowadays. Firstly, the complexity of natural products leads to time consuming and labour-intensive problems due to the long way of isolation, characterization and synthesis process [4, 6]. Secondly, standardization on the manufacture of the isolate or compound from natural products is difficult to be applied since a broad variation on bioactivity potency from different sources and batches [8]. In addition, formula stability and safety on the use of natural product extracts require specific strategies on the preservation of its efficacy and safety assessments which also to avoid any occurred resistances [10, 12]. The limited sources of natural products are also being an ethical and ecological concerns nowadays. Therefore, interdisciplinary collaborations among researchers in chemistry, biology, pharmacy, and medicine are required to commit in addressing the various scientific, regulatory and ethical challenges that arise.
Availability of data and materials
Not applicable.
References
Ahmad A, Rehman MU, Alkharfy KM (2020) An alternative approach to minimize the risk of coronavirus (Covid-19) and similar infections. Eur Rev Med Pharmacol Sci 24:4030–4034. https://doi.org/10.2355/EURREV_202004_20873
El-Shabasy RM, Nayel MA, Taher MM, Abdelmonem R, Shoueir KR, Kenawy ER (2022) Three waves changes, new variant strains, and vaccination effect against COVID-19 pandemic. Int J Biol Macromol 204:161–168. https://doi.org/10.1016/j.ijbiomac.2022.01.118
Mazraedoost S, Behbudi G, Mousavi SM, Hashemi SA (2020) Covid-19 treatment by plant compounds. J Adv Appl NanoBio Tech 2:23–33. https://doi.org/10.4277/AANBT/2(1)33
Zhang QW, Lin LG, Ye WC (2018) Techniques for extraction and isolation of natural products: a comprehensive review. Chinese Med 13:1–26. https://doi.org/10.1186/s13020-018-0177-x
Panyod S, Ho CT, Sheen LY (2020) Dietary therapy and herbal medicine for COVID-19 prevention: a review and perspective. J Tradit Complement Med 10:420–427. https://doi.org/10.1016/j.jtcme.2020.05.004
Park D, Swayambhu G, Lyga T, Pfeifer BA (2021) Complex natural product production methods and options. Synth Syst Biotechnol 6:1–11. https://doi.org/10.1016/j.synbio.2020.12.001
Peiris JSM, Lai ST, Poon LLM, Guan Y, Yam LYC, Lim W, Nicholls J, Yee WKS, Yan WW, Cheung MT et al (2003) Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319–1325. https://doi.org/10.1016/S0140-6736(03)13077-2
Atanasov AG, Zotchev SB, Dirsch VM, Taskforce (2021) Natural products in drug discovery: advances and opportunities. Nat Rev Drug Discov 20:200–2016. https://doi.org/10.5772/intechopen.82860
Drosten C, Günther S, Preiser W, van der Werf S, Brodt H-R, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RAM et al (2003) Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348:1967–1976. https://doi.org/10.1056/nejmoa030747
Thomas E, Stewart LE, Darley BA, Pham AM, Esteban I, Panda SS (2021) Develop new antiviral drug candidates. Molecules 26:1–56. https://doi.org/10.3390/molecules26206197
Lee N, Hui D, Wu A, Chan P, Cameron P, Joynt G, Ahuja A, Yung M, Leung C, To K et al (2003) A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med 348:1986–1994. https://doi.org/10.5222/terh.2003.09293
Ma J, Gu Y, Xu P (2020) A roadmap to engineering antiviral natural products synthesis in microbes. Curr Opin Biotechnol 66:140–149. https://doi.org/10.1016/j.copbio.2020.07.008
Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Peñaranda S, Bankamp B, Maher K, Chen M (2003) Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394–1399. https://doi.org/10.1126/science.1085952
Cunha CB, Opal SM (2014) Middle East respiratory syndrome (MERS): a new zoonotic viral pneumonia. Virulence 5:650–654. https://doi.org/10.4161/viru.32077
WHO (2021) WHO lists two additional COVID-19 vaccines for emergency use and COVAX roll-out AstraZeneca/Oxford-developed vaccines to reach countries in the coming weeks. World Heal. Organ. 3–5. https://www.who.int/news/item/15-02-2021-who-lists-two-additional-covid-19-vaccines-for-emergency-use-and-covax-roll-out.
Raj VS, Osterhaus ADME, Fouchier RAM, Haagmans BL (2014) MERS: emergence of a novel human coronavirus. Curr Opin Virol 5:58–62
Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM (2012) Isolation of a novel coronavirus from a man with Pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. https://doi.org/10.1056/nejmoa1211721
Lam TT, Jia N, Zhang Y, Shum MH, Jiang J, Zhu H, Tong Y, Shi Y, Ni X, Liao Y et al (2020) Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583:282–285
Chellapandi P, Saranya S (2020) Genomics insights of SARS-CoV-2 (COVID-19) into target-based drug discovery. Med Chem Res 29:1777–1791. https://doi.org/10.1007/s00044-020-02610-8
Hu D, Zhu C, Ai L, He T, Wang Y, Ye F, Yang L, Ding C, Zhu X, Lv R et al (2018) Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bats. Emerg Microbes Infect 7:1–10. https://doi.org/10.1038/s41426-018-0155-5
Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N et al (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395:565–574. https://doi.org/10.1016/S0140-6736(20)30251-8
Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL et al (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. https://doi.org/10.1038/s41586-020-2012-7
Lau SKP, Luk HKH, Wong ACP, Li KSM, Zhu L, He Z, Fung J, Chan TTY, Fung KSC, Woo PCY (2020) Possible bat origin of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 26:1542–1547. https://doi.org/10.3201/eid2607.200092
Vasireddy D, Vanaparthy R, Mohan G, Malayala SV, Atluri P (2021) Review of COVID-19 Variants and COVID-19 Vaccine Efficacy: What the Clinician Should Know? J Clin Med Res 13:317–325. https://doi.org/10.1740/jocmr4518
Nicolete VC, Rodrigues PT, Fernandes ARJ, Corder RM, Tonini J, Buss LF, Sales FC, Faria NR, Sabino EC, Castro MC et al (2022) Epidemiology of COVID-19 after emergence of SARS-CoV-2 gamma variant, Brazilian Amazon, 2020–2021. Emerg Infect Dis 28:709–712. https://doi.org/10.3201/eid2803.211993
Shiehzadegan S, Alaghemand N, Fox M, Venketaraman V (2021) Analysis of the delta variant B.1.617.2 COVID-19. Clin Pract 11:778–784. https://doi.org/10.3390/clinpract11040093
Torjesen I (2021) Covid-19: delta variant is now UK’s most dominant strain and spreading through schools. BMJ 373:n1445. https://doi.org/10.1136/bmj.n1445
Akash K, Sharma A, Kumar D, Singh SK, Gupta G, Chellappan DK, Dua K, Nagraik R (2022) Molecular aspects of Omicron, vaccine development, and recombinant strain XE: a review. J Med Virol 94:4628–4643. https://doi.org/10.1002/jmv.27936
Goldsmith CS, Tatti KM, Ksiazek TG, Rollin PE, Comer JA, Lee WW, Rota PA, Bankamp B, Bellini WJ, Zaki SR (2004) Ultrastructural characterization of SARS coronavirus. Emerg Infect Dis 10:320–326. https://doi.org/10.3201/eid1002.030913
Tshibangu DST, Matondo A, Lengbiye EM, Inkoto CL, Ngoyi EM, Kabengele CN, Bongo GN, Gbolo BZ, Kilembe JT, Mwanangombo DT et al (2020) Possible effect of aromatic plants and essential oils against COVID-19: review of their antiviral activity. J Complement Altern Med Res 11:10–22. https://doi.org/10.9734/jocamr/2020/v11i130175
Wang HM, Liang PH (2010) Picornaviral 3C protease inhibitors and the dual 3C protease/coronaviral 3C-like protease inhibitors. Expert Opin Ther Pat 20:59–71. https://doi.org/10.1517/13543770903460323
Feuillet V, Canard B, Trautmann A (2021) Combining antivirals and immunomodulators to fight COVID-19. Trends Immunol 42:31–44. https://doi.org/10.1016/j.it.2020.11.003
Wang M, Zhao R, Gao L, Gao X, Wang D, Cao J (2020) SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front Cell Infect Microbiol 10:1–18
Goc A, Sumera W, Rath M, Niedzwiecki A (2021) Phenolic compounds disrupt spike-mediated receptor-binding and entry of SARS-CoV-2 pseudo-virions. PLoS ONE 16:1–20. https://doi.org/10.1371/journal.pone.0253489
Park A, Iwasaki A (2020) Type I and type iii interferons - induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe 27:870–878
Speerstra S, Chistov AA, Proskurin GV, Aralov AV, Ulashchik EA, Streshnev PP, Shmanai VV, Korshun VA, Schang LM (2018) Antivirals acting on viral envelopes via biophysical mechanisms of action. Antiviral Res 149:164–173. https://doi.org/10.1016/j.antiviral.2017.11.018
Wisskirchen K, Lucifora J, Michler T, Protzer U (2014) New pharmacological strategies to fight enveloped viruses. Trends Pharmacol Sci 35:470–478. https://doi.org/10.1016/j.tips.2014.06.004
Antonelli G, Turriziani O (2012) Antiviral therapy: old and current issues. Int J Antimicrob Agents 40:95–102. https://doi.org/10.1016/j.ijantimicag.2012.04.005
Gioia M, Ciaccio C, Calligari P, Simone G, Sbardella D, Tundo G, Fasciglione G, Masi A, Pierro D, Bocedi A et al (2020) Role of proteolytic enzymes in the COVID-19 infection and promising therapeutic approaches. Biochem Pharmacol 182:1–21. https://doi.org/10.1016/j.bcp.2020.114225
Pommier Y, Johnson AA, Marchand C (2005) Integrase inhibitors to treat HIV/AIDS. Nat Rev Drug Discov 4:236–248. https://doi.org/10.1038/nrd1660
Pour PM, Fakhri S, Asgary S, Farzaei MH, Echeverría J (2019) The signaling pathways, and therapeutic targets of antiviral agents: focusing on the antiviral approaches and clinical perspectives of anthocyanins in the management of viral diseases. Front Pharmacol. https://doi.org/10.3389/fphar.2019.01207
McKinlay MA, Pevear DC, Rossmann MG (1992) Treatment of the picorna virus common cold by inhibitors of viral uncoating and attachment. Annu Rev Microbiol 46:635–656. https://doi.org/10.1146/annurev.mi.46.100192.003223
Pathania S, Rawal RK, Singh PK (2022) RdRp (RNA-dependent RNA polymerase): a key target providing anti-virals for the management of various viral diseases. J Mol Struct 1250:131756. https://doi.org/10.1016/j.molstruc.2021.131756
Ghosh K, Amin SA, Gayen S, Jha T (2021) Unmasking of crucial structural fragments for coronavirus protease inhibitors and its implications in COVID-19 drug discovery. J Mol Struct. https://doi.org/10.1016/j.molstruc.2021.130366
Su HX, Yao S, Zhao W, Li M, Liu J, Shang W, Xie H, Ke C, Hu H, Gao M et al (2020) Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol Sin 41:1167–1177
Ma L, Liu H, Liu X, Yuan X, Xu C, Wang F, Lin J, Xu R, Zhang D (2021) Screening S protein ACE2 blockers from natural products: Strategies and advances in the discovery of potential inhibitors of COVID-19. Eur J Med Chem 226:1–23. https://doi.org/10.1016/j.ejmech.2021.113857
WHO (2021) Interim recommendations for use of the Pfizer–BioNTech COVID-19 vaccine, BNT162b2, under Emergency Use Listing. World Heal. Organ., 1–7.
WHO (2021) Interim recommendations for use of the Janssen Ad26.COV2S (COVID-19) vaccine. World Heal. Organ. 1–6.
Li CX, Noreen S, Zhang LX, Saeed M, Wu PF, Ijaz M, Dai DF, Maqbool I, Madni A, Akram F et al (2022) A critical analysis of SARS-CoV-2 (COVID-19) complexities, emerging variants, and therapeutic interventions and vaccination strategies. Biomed Pharmacother 146:112550. https://doi.org/10.1016/j.biopha.2021.112550
Rehman SU, Rehman SU, Yoo HH (2021) COVID-19 challenges and its therapeutics. Biomed Pharmacother 142:112015. https://doi.org/10.1016/j.biopha.2021.112015
Kezia V, Ramatillah DL (2022) Intensive monitroing of sinovac vaccine for safety and efficacy among Indonesian population. Int J Appl Pharm 14:44–48
Darbar S, Agarwal S, Saha S (2021) COVID19 Vaccine: COVAXIN ®-India’s first indigenous effective weapon to fight against coronavirus (a review). Parana J Sci Educ 7:1–9
Deshpande GR, Yadav PD, Abraham P, Nyayanit DA, Sapkal GN, Shete AM, Gupta N, Vadrevu KM, Ella R, Panda S et al (2022) Booster dose of the inactivated COVID-19 vaccine BBV152 (Covaxin) enhances the neutralizing antibody response against alpha, beta, delta and omicron variants of concern. J Travel Med 29:1–3. https://doi.org/10.1093/jtm/taac039
Emary KRW, Golubchik T, Aley PK, Ariani CV, Angus B, Bibi S, Blane B, Bonsall D, Cicconi P, Charlton S et al (2021) Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): an exploratory analysis of a randomised controlled trial. Lancet 397:1351–1362. https://doi.org/10.1016/S0140-6736(21)00628-0
Mallapaty S, Callaway E (2021) What scientists do and don’t know about the Oxford-AstraZeneca COVID vaccine. Nature 592:15–17. https://doi.org/10.1038/d41586-021-00785-7
van der Lubbe JEM, Rosendahl Huber SK, Vijayan A, Dekking L, van Huizen E, Vreugdenhil J, Choi Y, Baert MRM, Feddes-de Boer K, Izquierdo Gil A et al (2021) Ad26.COV2.S protects Syrian hamsters against G614 spike variant SARS-CoV-2 and does not enhance respiratory disease. npj Vaccines 6:1–12. https://doi.org/10.1038/s41541-021-00301-y
Botton J, Semenzato L, Jabagi MJ, Baricault B, Weill A, Dray-Spira R, Zureik M (2022) Effectiveness of Ad26.COV2.S Vaccine vs BNT162b2 vaccine for COVID-19 hospitalizations. JAMA Netw Open 5:2–7. https://doi.org/10.1001/jamanetworkopen.2022.0868
Mahase E (2021) Covid-19: Novavax vaccine efficacy is 86% against UK variant and 60% against South African variant. BMJ 372:2021. https://doi.org/10.1136/bmj.n296
Cromer D, Reynaldi A, Steain M, Triccas JA, Davenport MP, Khoury DS (2022) Relating in vitro neutralization level and protecztion in the CVnCoV (CUREVAC) trial. Clin Infect Dis 75:E878–E879. https://doi.org/10.1093/cid/ciac075
Cohen J (2021) What went wrong with CureVac’s mRNA vaccine pdf. Science. https://doi.org/10.1126/science.372.6549.1381
Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB et al (2021) Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med 384:403–416. https://doi.org/10.1056/nejmoa2035389
Chagla Z (2021) The BNT162b2 (BioNTech/Pfizer) vaccine had 95% efficacy against COVID-19 ≥7 days after the 2nd dose Ann. Intern Med. https://doi.org/10.7326/ACPJ202102160-015
Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C et al (2020) Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med 383:2603–2615. https://doi.org/10.1056/nejmoa2034577
WHO (2020) Timeline: WHO’s Covid-19 response. World Heal. Organ. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/interactive-timeline#!
Jahan R, Paul AK, Jannat K, Rahmatullah M (2021) Plant essential oils: possible COVID-19 therapeutics. Nat Prod Commun 16:2021. https://doi.org/10.1177/1934578X21996149
Hameed S, Khan AF, Khan S, Wasay M (2022) First report of cerebral venous thrombosis following inactivated-virus covid vaccination (Sinopharm and Sinovac). J Stroke Cerebrovasc Dis 31:10–13. https://doi.org/10.1016/j.jstrokecerebrovasdis.2021.106298
Zhao H, Li Y, Wang Z (2022) Adverse event of sinovac coronavirus vaccine: deafness. Vaccine 40:521–523. https://doi.org/10.1016/j.vaccine.2021.11.091
Hardt K, Vandebosch A, Sadoff J, Gars MLe, Truyers C, Lowson D, Dromme I Van, Vingerhoets J, Kamphuis T, Scheper G et al (2022) Efficacy and safety of a booster regimen of Ad26COV2S vaccine against covid-19. Medrxiv. https://doi.org/10.1101/2022.01.28.22270043
Banerjee S, Sandhu M, Tonzi E, Tambe A, Gambhir H (2021) Immune-mediated thrombocytopenia associated with Ad26.COV2.S (Janssen; Johnson & Johnson) vaccine. Am J Ther 28:e604–e605. https://doi.org/10.1111/bjh
Sulemankhil I, Abdelrahman M, Negi SI (2022) Temporal association between the COVID-19 Ad26.COV2.S vaccine and acute myocarditis: a case report and literature review. Cardiovasc Revascularization Med 38:117–123. https://doi.org/10.1016/j.carrev.2021.08.012
Tobaiqy M, Elkout H, Maclure K (2021) Analysis of thrombotic adverse reactions of covid-19 astrazeneca vaccine reported to eudravigilance database. Vaccines 9:1–8. https://doi.org/10.3390/vaccines9040393
Oldenburg J, Klamroth R, Langer F, Albisetti M, Von Auer C, Ay C, Korte W, Scharf RE, Pötzsch B, Greinacher A (2021) Diagnosis and management of vaccine-related thrombosis following AstraZeneca COVID-19 Vaccination: guidance statement from the GTH. Hamostaseologie 41:E1. https://doi.org/10.1055/s-0041-1729135
Wise J (2021) Covid-19: European countries suspend use of Oxford-AstraZeneca vaccine after reports of blood clots. BMJ 372:n699. https://doi.org/10.1136/bmj.n699
Nagy A, Alhatlani B (2021) An overview of current COVID-19 vaccine platforms. Comput Struct Biotechnol J 19:2508–2517. https://doi.org/10.1016/j.csbj.2021.04.061
Park JW, Lagniton PNP, Liu Y, Xu RH (2021) mRNA vaccines for covid-19: What, why and how. Int J Biol Sci 17:1446–1460. https://doi.org/10.7150/ijbs.59233
Ouled Aitouna A, Belghiti ME, Eşme A, Anouar E, Ouled Aitouna A, Zeroual A, Salah M, Chekroun A, El Alaoui El Abdallaoui H, Benharref A et al (2021) Chemical reactivities and molecular docking studies of parthenolide with the main protease of HEP-G2 and SARS-CoV-2. J Mol Struct 1243:1–9. https://doi.org/10.1016/j.molstruc.2021.130705
Zhang J, Xiong Y, Wang F, Zhang F, Yang X, Lin G, Tian P, Ge G, Gao D (2022) Discovery of 9,10-dihydrophenanthrene derivatives as SARS-CoV-2 3CLpro inhibitors for treating COVID-19. Eur J Med Chem. 228:1–16. https://doi.org/10.1016/j.ejmech.2021.114030
Shen L, Niu J, Wang CBH, Wang W, Zhu N, Deng Y, Wang H, Ye F, Chen S et al (2019) High-throughput screening and identification of potent broad-spectrum inhibitors of coronaviruses. J Virol 93(1):15
Ulasli M, Gurses SA, Bayraktar R, Yumrutas O, Oztuzcu S, Igci M, Igci YZ, Cakmak EA, Arslan A (2014) The effects of Nigella sativa (Ns), anthemis hyalina (Ah) and Citrus sinensis (Cs) extracts on the replication of coronavirus and the expression of TRP genes family. Mol Biol Rep 41:1703–1711. https://doi.org/10.1007/s11033-014-3019-7
Park JY, Yuk HJ, Ryu HW, Lim SH, Kim KS, Park KH, Ryu YB, Lee WS (2017) Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J Enzyme Inhib Med Chem 32:504–512. https://doi.org/10.1080/14756366.2016.1265519
Lin SC, Ho CT, Chuo WH, Li S, Wang TT, Lin CC (2017) Effective inhibition of MERS-CoV infection by resveratrol. BMC Infect Dis 17:1–10. https://doi.org/10.1186/s12879-017-2253-8
Park JY, Ko JA, Kim DW, Kim YM, Kwon HJ, Jeong HJ, Kim CY, Park KH, Lee WS, Ryu YB (2016) Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV. J Enzyme Inhib Med Chem 31:23–30. https://doi.org/10.3109/14756366.2014.1003215
Park JY, Kim JH, Kwon JM, Kwon HJ, Jeong HJ, Kim YM, Kim D, Lee WS, Ryu YB (2013) Dieckol, a SARS-CoV 3CLpro inhibitor, isolated from the edible brown algae Ecklonia cava. Bioorganic Med Chem 21:3730–3737. https://doi.org/10.1016/j.bmc.2013.04.026
Tragoolpua Y, Jatisatienr A (2007) Anti-herpes Simplex Virus activities of Eugenia caryophyllus (Spreng.) Bullock & S. G. Harrison and essential oil. Eugenol Phyther Res 21:1153–1158. https://doi.org/10.1002/ptr
Armaka M, Papanikolaou E, Sivropoulou A, Arsenakis M (1999) Antiviral properties of isoborneol, a potent inhibitor of herpes simplex virus type 1. Antiviral Res 43:79–92. https://doi.org/10.1016/S0166-3542(99)00036-4
Astani A, Reichling J, Schnitzler P (2011) Screening for antiviral activities of isolated compounds from essential oils. Evidence-based Compl Altern Med 2011:1–8. https://doi.org/10.1093/ecam/nep187
Serkedjieva J, Velcheva M (2003) In vitro anti-influenza virus activity of the pavine alkaloid (-)-thalimonine isolated from Thalictrum simplex L. Antivir Chem Chemother 14:75–80. https://doi.org/10.1177/095632020301400202
Hayashi K, Imanishi N, Kashiwayama Y, Kawano A, Terasawa K, Shimada Y, Ochiai H (2007) Inhibitory effect of cinnamaldehyde, derived from Cinnamomi cortex, on the growth of influenza A/PR/8 virus in vitro and in vivo. Antiviral Res 74:1–8. https://doi.org/10.1016/j.antiviral.2007.01.003
Xie XH, Zang N, Li S (2012) Resveratrol Inhibits respiratory syncytial virus-induced IL-6 production, decreases viral replication, and downregulates TRIF expression in airway epithelial cells. Inflammation 35:1392–1401. https://doi.org/10.1007/s10753-012-9452-7
Liu T, Zang N, Zhou N, Li W, Xie X, Deng Y, Ren L, Long X, Li S, Zhou L et al (2014) Resveratrol Inhibits the TRIF-dependent pathway by upregulating sterile alpha and armadillo motif protein, contributing to anti-inflammatory effects after respiratory syncytial virus infection. J Virol 88:4229–4236. https://doi.org/10.1128/jvi.03637-13
Kim D, Min J, Jang M, Lee J, Shin Y, Park C, Song J, Kim H, Kim S, Jin Y et al (2019) Natural bis-benzylisoquinoline alkaloids-tetrandrine, fangchinoline, and cepharanthine, inhibit human coronavirus OC43 infection of MRS-5 human lung cells. Biomolecules 9:1–16
Diniz LR, Perez-Castillo Y, Elshabrawy H, da Filho C, SM, and Sousa, D. (2021) Bioactive terpenes and their derivatives as potential sars-cov-2 proteases inhibitors from molecular modeling studies. Biomolecules 11:1–19
Cheng PW, Ng LT, Chiang LC, Lin CC (2006) Antiviral effects of saikosaponins on human coronavirus 229E in vitro. Clin Exp Pharmacol Physiol 33:612–616. https://doi.org/10.1111/j.1440-1681.2006.04415.x
Chen F, Chan KH, Jiang Y, Kao RYT, Lu HT, Fan KW, Cheng VCC, Tsui WHW, Hung IFN, Lee TSW et al (2004) In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J Clin Virol 31:69–75. https://doi.org/10.1016/j.jcv.2004.03.003
Wen CC, Kuo YH, Jan JT, Liang PH, Wang SY, Liu HG, Lee CK, Chang ST, Kuo CJ, Lee SS et al (2007) Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J Med Chem 50:4087–4095. https://doi.org/10.1021/jm070295s
Kim DW, Seo KH, Curtis-Long MJ, Oh KY, Oh JW, Cho JK, Lee KH, Park KH (2014) Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. J Enzyme Inhib Med Chem 29:59–63. https://doi.org/10.3109/14756366.2012.753591
Kitamura K, Honda M, Yoshizaki H, Yamamoto S, Nakane H, Fukushima M, Ono K, Tokunaga T (1998) Baicalin, an inhibitor of HIV-1 production in vitro. Antiviral Res 37:131–140. https://doi.org/10.1016/S0166-3542(97)00069-7
Li BQ, Fu T, Dongyan Y, Mikovits JA, Ruscetti FW, Wang JM (2000) Flavonoid baicalin inhibits HIV-1 infection at the level of viral entry. Biochem Biophys Res Commun 276:534–538. https://doi.org/10.1006/bbrc.2000.3485
Salem ML, Hossain MS (2000) Protective effect of black seed oil from Nigella sativa against murine cytomegalovirus infection. Int J Immunopharmacol 22:729–740. https://doi.org/10.1016/S0192-0561(00)00036-9
Gu C, Yin A, Yuan H, Yang K, Luo J, Zhan Y (2019) New anti-HBV norbisabolane sesquiterpenes from Phyllantus acidus. Fitoterapia 137:1–7. https://doi.org/10.1016/j.fitote.2019.04.006
Loizzo MR, Saab AM, Tundis R, Statti GA, Menichimi F, Lampronti D, Gambari R, Cinatl J, Doerr HW (2008) Phytochemical analysis and in vitro antiviral activities of the essential oils of seven Lebanon species. Chem Biodivers 5:461–470. https://doi.org/10.1002/cbdv.200890045
Tkachenko KG (2006) Antiviral activity of the essential oils of some Heracleum L. species. J. Herbs. Spices Med Plants 12:1–12. https://doi.org/10.1300/J044v12n03_01
Su H, Yao S, Zhao W, Li M, Liu J, Shang W, Xie H, Ke C, Gao M, Yu K et al (2020) Discovery of baicalin and baicalein as novel, natural product inhibitors of SARS-CoV-2 3CL protease in vitro. bioRxiv. https://doi.org/10.1101/2020.04.13.038687
Lammers AJJ, Brohet RM, Theunissen REP, Koster C, Rood R, Verhagen DWM, Brinkman K, Hassing RJ, Dofferhoff A, el Moussaoui R et al (2021) Early hydroxychloroquine but not chloroquine use reduces ICU admission in COVID-19 patients. Int J Infect Dis 101:283–289. https://doi.org/10.1016/j.ijid.2020.12.008
Omokhua-Uyi AG, Van Staden J (2021) Natural product remedies for COVID-19: a focus on safety. South African J Bot 139:386–398. https://doi.org/10.1016/j.sajb.2021.03.012
de Almeida S, Soares J, dos Santos K, Alves J, Ribeiro A, Jacob I, Ferreira Cj, dos Santos J, de Oliveira J, Junior Lb et al (2020) COVID-19 therapy: What weapons do we bring into battle? Bioorganic Med. Chem 28:1–26
Murtuja S, Shilkar D, Sarkar B, Sinha BN, Jayaprakash V (2021) A short survey of dengue protease inhibitor development in the past 6 years (2015–2020) with an emphasis on similarities between DENV and SARS-CoV-2 proteases. Bioorganic Med Chem 49:1–24. https://doi.org/10.1016/j.bmc.2021.116415
Hashem A, Alghamdi B, Algaissi A, Alshehri F, Bukhari A, Alfaleh M, Memish Z (2020) Therapeutic use of chloroquine and hydroxychloroquine in Covid-19 and other viral infections: a narrative review. Travel Med Infect Dis 35:1–15
Prodromos C, Rumschlag T (2020) Hydroxychloroquine is effective, and consistently so when provided early, for COVID-19: a systematic review. New Microbes New Infect 38:1–11. https://doi.org/10.1016/j.nmni.2020.100776
Roy A, Das R, Roy D, Saha S, Ghosh NN, Bhattacharyya S, Roy MN (2022) Encapsulated hydroxychloroquine and chloroquine into cyclic oligosaccharides are the potential therapeutics for COVID-19: insights from first-principles calculations. J Mol Struct 1247:131371. https://doi.org/10.1016/j.molstruc.2021.131371
Singh AK, Singh A, Shaikh A, Singh R, Misra A (2020) Chloroquine and hydroxychloroquine in the treatment of COVID-19 with or without diabetes: A systematic search and a narrative review with a special reference to India and other developing countries. Diabetes Metab Syndr Clin Res Rev 14(3):241
Lamback EB, de Oliveira MA, Haddad AF, Vieira AFM, Neto ALF, da Maia T (2021) Hydroxychloroquine with azithromycin in patients hospitalized for mild and moderate COVID-19. Brazilian J Infect Dis 25:1–6. https://doi.org/10.1016/j.bjid.2021.101549
Gies V, Bekaddour N, Dieudonne Y, Guffroy A, Frenger Q, Gros F, Rodero M, Herbeuval J, Korganow A (2020) Beyond anti-viral effects of chloroquine/hydroxychloroquine. Front Immunol 11:1–8
Omrani AS, Pathan SA, Thomas SA, Harris TRE, Coyle PV, Thomas CE, Qureshi I, Bhutta ZA, Mawlawi NA, Kahlout RA et al (2020) Randomized double-blinded placebo-controlled trial of hydroxychloroquine with or without azithromycin for virologic cure of non-severe Covid-19. EClinicalMedicine 29–30:1–13. https://doi.org/10.1016/j.eclinm.2020.100645
Ni Y, Liao J, Qian Z, Wu C, Zhang X, Zhang J, Xie Y, Jiang S (2022) Synthesis and evaluation of enantiomers of hydroxychloroquine against SARS-CoV-2 in vitro. Bioorganic Med Chem 53:1–8. https://doi.org/10.1016/j.bmc.2021.116523
Purwati M, Nasronudin A, Hendrianto E, Karsari D, Dinaryanti A, Ertanti N, Ihsan IS, Purnama DS, Asmarawati TP et al (2021) An in vitro study of dual drug combinations of anti-viral agents, antibiotics, and/or hydroxychloroquine against the SARS-CoV-2 virus isolated from hospitalized patients in surabaya Indonesia. PLoS ONE 16(1):27. https://doi.org/10.1371/journal.pone.0252302
Szymkuć S, Gajewska EP, Molga K, Wołos A, Roszak R, Beker W, Moskal M, Dittwald P, Grzybowski BA (2020) Computer-generated “synthetic contingency” plans at times of logistics and supply problems: scenarios for hydroxychloroquine and remdesivir. Chem Sci 11:6736–6744. https://doi.org/10.1039/d0sc01799j
Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM (2020) The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res 178:3–6. https://doi.org/10.1016/j.antiviral.2020.104787
Zaheer T, Pal K, Abbas RZ, del Torres M (2021) COVID-19 and ivermectin: potential threats associated with human use. J Mol Struct 1243:130808. https://doi.org/10.1016/j.molstruc.2021.130808
Chaccour C, Casellas A, Blanco-Di Matteo A, Pineda I, Fernandez-Montero A, Ruiz-Castillo P, Richardson MA, Rodríguez-Mateos M, Jordán-Iborra C, Brew J et al (2021) The effect of early treatment with ivermectin on viral load, symptoms and humoral response in patients with non-severe COVID-19: a pilot, double-blind, placebo-controlled, randomized clinical trial. EClinicalMedicine 32:1–9. https://doi.org/10.1016/j.eclinm.2020.100720
WHO (2021) WHO advises that ivermectin only be used to treat COVID-19 within clinical trials. World Heal Orga 1:1–14
Gendrot M, Andreani J, Jardot P, Hutter S, Delandre O, Boxberger M, Mosnier J, Bideau M, Duflot I, Fonta I et al (2020) In vitro antiviral activity of doxycycline against SARS-CoV-2. Molecules 25:1–10
Lu H (2020) Drug treatment options for the 2019-new coronavirus (2019-nCoV). Biosci Trends 14:69–71. https://doi.org/10.1002/jmv.25678.4
Chakraborty R, Parvez S (2020) COVID-19: An overview of the current pharmacological interventions, vaccines, and clinical trials biochem. Pharmacol 180(1):16. https://doi.org/10.1016/j.bcp.2020.114184
Yousefi H, Mashouri L, Okpechi S, Alahari N, Alahari S (2021) Repurposing existing drugs for the treatment of COVID-19/SARS-CoV-2 infection: a review describing drug mechanisms of action. Biochem Pharmacol 183:1–14. https://doi.org/10.1016/j.bcp.2020.114296
Kucukoglu K, Faydalı N, Bul D (2020) What are the drugs having potential against COVID-19? Med Chem Res 29:1935–1955. https://doi.org/10.1007/s00044-020-02625-1
Frediansyah A, Nainu F, Dhama K, Mudatsir M, Harapan H (2021) Remdesivir and its antiviral activity against COVID-19: a systematic review. Clin Epidemiol Glob Heal 9:123–127. https://doi.org/10.1016/j.cegh.2020.07.011
Tian L, Qiang T, Liang C, Ren X, Jia M, Zhang J, Li J, Wan M, YuWen X, Li H et al (2021) RNA-dependent RNA polymerase (RdRp) inhibitors: the current landscape and repurposing for the COVID-19 pandemic. Eur. J. Med. Chem. 213:1–19. https://doi.org/10.1016/j.ejmech.2021.113201
Hsu WS, Yen JH, Wang YS (2013) Formulas of components of citronella oil against mosquitoes (Aedes aegypti). J. Environ. Sci. Heal. —Part B Pestic. Food Contam Agric Wastes 48:1014–1019. https://doi.org/10.1080/03601234.2013.816613
Hung Y, Lee J, Chiu C, Lee C, Tsai P, Hsu I, Ko W (2022) Oral nirmatrelvir/ritonavir therapy for COVID-19: the dawn in the dark? Antibiotic 11:1–7. https://doi.org/10.3390/antibiotics11020220
Eng H, Dantonio AL, Kadar EP, Obach RS, Di L, Lin J, Patel NC, Boras B, Walker GS, Novak JJ et al (2022) Disposition of PF-07321332 (Nirmatrelvir), an orally bioavailable inhibitor of SARS-CoV-2 3CL protease, across animals and humans. Drug Metab Dispos. https://doi.org/10.1124/dmd.121.000801
Rai DK, Yurgelonis I, McMonagle P, Rothan HA, Hao L, Gribenko A, Titova E, Kreiswirth B, White KM, Zhu Y et al (2022) Nirmatrelvir, an orally active Mpro inhibitor, is a potent inhibitor of SARS-CoV-2 Variants of Concern. biorxiv 35:1–22
Hammond J, Leister-Tebbe H, Gardner A, Abreu P, Bao W, Wisemandle W, Baniecki M, Hendrick VM, Damle B, Simón-Campos A et al (2022) Oral nirmatrelvir for high-risk, nonhospitalized adults with covid-19. N Engl J Med. https://doi.org/10.1056/nejmoa2118542
Owen D, Allerton CM, Anderson A, Aschenbrenner L, Avery M, Berritt S, Boras B, Cardin R, Carlo A, Coffman K et al (2021) An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of Covid-19. Science 374:1586–1593
Ullrich S, Ekanayake KB, Otting G, Nitsche C (2022) Main protease mutants of SARS-CoV-2 variants remain susceptible to nirmatrelvir. Bioorg Med Chem Lett 62:1–4. https://doi.org/10.1016/j.bmcl.2022.128629
Ramos-Guzmán CA, Ruiz-Pernía JJ, Tuñón I (2021) Computational simulations on the binding and reactivity of a nitrile inhibitor of the SARS-CoV-2 main protease. Chem Commun 57:9096–9099. https://doi.org/10.1039/d1cc03953a
Ngo ST, Nguyen TH, Tung NT, Mai BK (2022) Insights into the binding and covalent inhibition mechanism of PF-07321332 to SARS-CoV-2 Mpro. RSC Adv 12:3729–3737. https://doi.org/10.1039/d1ra08752e
Rosales R, Rodriguez ML, Rai DK, Cardin RD, Anderson AS, Sordillo EM, van Bakel H, Simon V, García-Sastre A, White KM (2022) Nirmatrelvir, molnupiravir, and remdesivir maintain potent in vitro activity against the SARS-CoV-2 Omicron variant. BioRxiv. https://doi.org/10.1101/2022.01.17.476685
Sacco MD, Hu Y, Gongora MV, Meilleur F, Kemp MT, Zhang X, Wang J, Chen Y (2022) The P132H mutation in the main protease of Omicron SARS-CoV-2 decreases thermal stability without compromising catalysis or small-molecule drug inhibition. biorxiv. https://doi.org/10.1101/2022.01.26.477774
Zarenezhad E, Marzi M (2022) Review on molnupiravir as a promising oral drug for the treatment of COVID-19. Med Chem Res 31:232–243. https://doi.org/10.1007/s00044-021-02841-3
WHO (2022) WHO recommends two new drugs to treat COVID-19. World Heal. Organ. https://www.who.int/news/item/14-01-2022-who-recommends-two-new-drugs-to-treat-covid-19.
Reyes, E. This treatment can protect vulnerable people from COVID . But many don’t know about it. Los Angeles Times. 2022.
Ramajayam R, Tan KP, Liang PH (2011) Recent development of 3C and 3CL protease inhibitors for anti-coronavirus and anti-picornavirus drug discovery. Biochem Soc Trans 39:1371–1375. https://doi.org/10.1042/BST0391371
Binford SL, Maldonado F, Brothers MA, Weady PT, Zalman LS, Meador JW III, Matthews DA, Patick A (2005) Conservation of amino acids in human rhinovirus 3C protease correlates with broad-spectrum antiviral activity of rupintrivir, a novel human rhinovirus 3C protease inhibitor. Antimicrob Agents Chemother 49:619–626
Fink K, Nitsche A, Neumann M, Grossegesse M, Eisele KH, Danysz W (2021) Amantadine inhibits sars-cov-2 in vitro. Viruses 13:1–10. https://doi.org/10.3390/v13040539
Hou Y, Okuda K, Edwards C, Martinez D, Asakura T, Dinnon K III, Kato T, Lee R, Yount B, Mascenik T et al (2020) SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 182:429–446
Sallard E, Lescure FX, Yazdanpanah Y, Mentre F, Peiffer-Smadja N (2020) Type 1 interferons as a potential treatment against COVID-19. Antiviral Res 178:1–4. https://doi.org/10.1016/j.antiviral.2020.104791
Mantlo E, Bukreyeva N, Maruyama J, Paessler S, Huang C (2020) Antiviral activities of type I interferons to SARS-CoV-2 infection. Antiviral Res 179:1–4. https://doi.org/10.1016/j.antiviral.2020.104811
Elmaaty AA, Alnajjar R, Hamed MIA, Khattab M, Khalifa MM, Al-Karmalawy AA (2021) Revisiting activity of some glucocorticoids as a potential inhibitor of SARS-CoV-2 main protease: theoretical study. RSC Adv 11:10027–10042. https://doi.org/10.1039/d0ra10674g
Wang S, Li W, Hui H, Tiwari SK, Zhang Q, Croker BA, Rawlings S, Smith D, Carlin AF, Rana TM (2020) Cholesterol 25-Hydroxylase inhibits SARS -CoV-2 and other coronaviruses by depleting membrane cholesterol. EMBO J 39:1–13. https://doi.org/10.1252/embj.2020106057
Zu S, Deng Y, Zhou C, Li J, Li L, Chen Q, Li X, Zhao H, Gold S, He J et al (2020) 25-Hydroxycholesterol is a potent SARS-CoV-2 inhibitor. Cell Res 30:1043–1045
Marcello A, Civra A, Bonotto R, Alves L, Rajasekharan S, Giacobone C, Caccia C, Cavalli R, Adami M, Brambilla P et al (2020) The cholesterol metabolite 27-hydroxycholesterol inhibits SARS-CoV-2 and is markedly decreased in Covid-19 patients. Redox Biol 36:1–11
Naggie S, Boulware DR, Lindsell CJ, Stewart TG, Slandzicki AJ, Lim SC, Cohen J, Kavtaradze D, Amon AP, Gabriel A et al (2023) Effect of higher-dose ivermectin for 6 days vs placebo on time to sustained recovery in outpatients with COVID-19. JAMA 329:888–897. https://doi.org/10.1001/jama.2023.1650
Chakraborty C, Sharma AR, Bhattacharya M, Agoramoorthy G, Lee SS (2021) The drug repurposing for COVID-19 clinical trials provide very effective therapeutic combinations: lessons learned from major clinical studies. Front Pharmacol 12:1–15. https://doi.org/10.3389/fphar.2021.704205
Takahashi JA, Barbosa BVR, Lima MTNS, Cardoso PG, Contigli C, Pimenta LPS (2021) Antiviral fungal metabolites and some insights into their contribution to the current COVID-19 pandemic. Bioorganic Med Chem 46:116366. https://doi.org/10.1016/j.bmc.2021.116366
Elzupir AO (2020) Inhibition of SARS-CoV-2 main protease 3CLpro by means of α-ketoamide and pyridone-containing pharmaceuticals using in silico molecular docking. J Mol Struct 1222:128878. https://doi.org/10.1016/j.molstruc.2020.128878
Silva LR, da Silva Santos-Júnior PF, de Andrade Brandão J, Anderson L, Bassi ÊJ, Xavier de Araújo-Júnior J, Cardoso SH, da Silva-Júnior EF (2020) Druggable targets from coronaviruses for designing new antiviral drugs. Bioorganic Med Chem 28:1–23. https://doi.org/10.1016/j.bmc.2020.115745
Ashraf S, Ashraf S, Ashraf M, Imran M, Kalsoom L, Siddiqui U, Farooq I, Akmal R, Akram M (2023) Honey and Nigella sativa against COVID-19 in Pakistan (HNS-COVID-PK): a multicenter placebo-controlled randomized clinical trial. Phyther Res 37:627–644. https://doi.org/10.1002/ptr.7640
Chopra A, Tillu G, Chuadhary K, Reddy G, Srivastava A, Lakdawala M, Gode D, Reddy H, Tamboli S, Saluja M et al (2023) Co-administration of AYUSH 64 as an adjunct to standard of care in mild and moderate COVID-19: a randomized, controlled, multicentric clinical trial. PLoS ONE 18:e0282688. https://doi.org/10.1371/journal.pone.0282688
Ghosh K, Amin SA, Gayen S, Jha T (2021) Chemical-informatics approach to COVID-19 drug discovery: Exploration of important fragments and data mining based prediction of some hits from natural origins as main protease (Mpro) inhibitors. J Mol Struct 1224:129026. https://doi.org/10.1016/j.molstruc.2020.129026
Rashid H, Ahmad N, Abdalla M, Khan K, Martines MAU, Shabana S (2021) Molecular docking and dynamic simulations of cefixime, etoposide and nebrodenside a against the pathogenic proteins of SARS-CoV-2. J Mol Struct 1247:131296. https://doi.org/10.1016/j.molstruc.2021.131296
Erdogan T (2021) DFT, molecular docking and molecular dynamics simulation studies on some newly introduced natural products for their potential use against SARS-CoV-2. J Mol Struct 1242:1–14. https://doi.org/10.1016/j.molstruc.2021.130733
Esam Z, Akhavan M, lotfi, M, and Bekhradnia, A, (2022) Molecular docking and dynamics studies of nicotinamide riboside as a potential multi-target nutraceutical against SARS-CoV-2 entry, replication, and transcription: A new insight. J Mol Struct 1247:131394. https://doi.org/10.1016/j.molstruc.2021.131394
Puttaswamy H, Gowtham HG, Ojha MD, Yadav A, Choudhir G, Raguraman V, Kongkham B, Selvaraju K, Shareef S, Gehlot P et al (2020) In silico studies evidenced the role of structurally diverse plant secondary metabolites in reducing SARS-CoV-2 pathogenesis. Sci Rep 10:1–24. https://doi.org/10.1038/s41598-020-77602-0
Sepay N, Sekar A, Halder UC, Alarifi A, Afzal M (2021) Anti-COVID-19 terpenoid from marine sources: a docking, admet and molecular dynamics study. J Mol Struct 1228:129433. https://doi.org/10.1016/j.molstruc.2020.129433
Khalifa I, Zhu W, Mohammed HHH, Dutta K, Li C (2020) Tannins inhibit SARS-CoV-2 through binding with catalytic dyad residues of 3CLpro: an in silico approach with 19 structural different hydrolysable tannins. J Food Biochem 44:1–19. https://doi.org/10.1111/jfbc.13432
Peele KA, Potla Durthi C, Srihansa T, Krupanidhi S, Ayyagari VS, Babu DJ, Indira M, Reddy AR, Venkateswarulu TC (2020) Molecular docking and dynamic simulations for antiviral compounds against SARS-CoV-2: a computational study. Inform Med Unlocked 19:1–6. https://doi.org/10.1016/j.imu.2020.100345
Alfaro M, Alfaro I, Angel C (2020) Identification of potential inhibitors of SARS-CoV-2 papain-like protease from tropane alkaloids from Schizanthus porrigens: a molecular docking study. Chem Phys Lett 761:1–10. https://doi.org/10.1016/j.cplett.2020.138068
Arora S, Lohiya G, Moharir K, SYShah S (2020) Identification of potential flavonoid inhibitors of the SARS-CoV-2 main protease 6YNQ A molecular docking study. Digit Chinese Med 3:239–248
Xu Z, Yang L, Zhang X, Zhang Q, Yang Z, Liu Y, Wei S, Liu W (2020) Discovery of potential flavonoid inhibitors against COVID-19 3CL proteinase based on virtual screening strategy. Front Mol Biosci 7:1–8. https://doi.org/10.3389/fmolb.2020.556481
Adem S, Eyupoglu V, Sarfraz I, Rasul A, Ali M (2020) Identification of potent COVID-19 main protease (Mpro) inhibitors from natural polyphenols: an in silico strategy unveils a hope against CORONA. Preprints. https://doi.org/10.2944/preprints202003.0333.v1
Vijayakumar B, Ramesh D, Joji A, Prakasan J, Kannan T (2020) In silico pharmacokinetic and molecular docking studies of natural flavonoids and synthetic indole chalcones against essential proteins of SARS-CoV-2. Eur J Pharmacol 886:1–11
Rameshkumar M, Indu P, Arunagirinathan N, Venkatadri B, El-Serehy H, Ahmad A (2021) Computational selection of flavonoid compounds as inhibitors against SARS-CoV-2 main protease, RNA-dependent RNA polymerase and spike proteins. a molecular docking study. Saudi J Biol Sci 28:448–458
Zaki AA, Ashour A, Elhady SS, Darwish KM, Al-Karmalawy AA (2022) Calendulaglycoside a showing potential activity against SARS-CoV-2 main protease: molecular docking, molecular dynamics, and SAR studies. J Tradit Complement Med 12:16–34. https://doi.org/10.1016/j.jtcme.2021.05.001
Giofrè SV, Napoli E, Iraci N, Speciale A, Cimino F, Muscarà C, Molonia MS, Ruberto G, Saija A (2021) Interaction of selected terpenoids with two SARS-CoV-2 key therapeutic targets: an in silico study through molecular docking and dynamics simulations. Comput Biol Med 134:1–13. https://doi.org/10.1016/j.compbiomed.2021.104538
Abdelmohsen UR, Albohy A, Abdulrazik BS, Bayoumi SAL, Malak LG, Khallaf ISA, Bringmann G, Farag SF (2021) Natural coumarins as potential anti-SARS-CoV-2 agents supported by docking analysis. RSC Adv 11:16970–16979. https://doi.org/10.1039/d1ra01989a
Thangavel N, Al Bratty M, Al Hazmi HA, Najmi A, Ali Alaqi RO (2021) Molecular docking and molecular dynamics aided virtual search of olivenet™ directory for secoiridoids to combat SARS-CoV-2 infection and associated hyperinflammatory responses. Front Mol Biosci 7:1–17. https://doi.org/10.3389/fmolb.2020.627767
Kumar V, Parate S, Yoon S, Lee G, Lee KW (2021) Computational simulations identified marine-derived natural bioactive compounds as replication inhibitors of SARS-CoV-2. Front Microbiol 12:1–15. https://doi.org/10.3389/fmicb.2021.647295
Gajjar ND, Dhameliya TM, Shah GB (2021) In search of RdRp and Mpro inhibitors against SARS CoV-2: Molecular docking, molecular dynamic simulations and ADMET analysis. J Mol Struct 1239:130488. https://doi.org/10.1016/j.molstruc.2021.130488
Adelusi TI, Oyedele AQK, Monday OE, Boyenle ID, Idris MO, Ogunlana AT, Ayoola AM, Fatoki JO, Kolawole OE, David KB et al (2022) Dietary polyphenols mitigate SARS-CoV-2 main protease (Mpro)–molecular dynamics, molecular mechanics, and density functional theory investigations. J Mol Struct 1250:131879. https://doi.org/10.1016/j.molstruc.2021.131879
Nebigil C, Moog C, Vagner S, Jessel N, Smith D, Desaubry L (2020) Flavaglines as natural products targeting eIF4A and prohibitins: From traditional Chinese medicine to antiviral activity against coronaviruses. Eur J Med Chem 203:1–9. https://doi.org/10.1016/j.ejmech.2020.112653
Ellinger B, Bojkova D, Zaliani A, Cinatl J, Claussen C, Westhaus S, Reinshagen J, Kuzikov M, Wolf M, Geisslinger G et al (2020) Identification of inhibitors of SARS-CoV-2 in-vitro cellular toxicity in human (Caco-2) cells using a large scale drug repurposing collection. Res Sq 23:1–19. https://doi.org/10.2203/rs.3.rs-23951/v1
Bojkova D, Klann K, Koch B, Widera M, Krause D, Ciesek S, Cinatl J, Münch C (2020) Proteomics of SARS-CoV-2 infected host cell proteomics reveal potential therapy targets. Nature 583:469–472. https://doi.org/10.1038/s41586-020-2332-7
Hoehl S, Rabenau H, Berger A, Kortenbusch M, Cinatl J, Bojkova D, Behrens P, Böddinghaus B, Götsch U, Naujoks F et al (2020) Evidence of SARS-CoV-2 infection in returning travelers from Wuhan. China N Engl J Med 382:1278–1280. https://doi.org/10.1056/nejmc2001899
Escalante DE, Ferguson DM (2021) Structural modeling and analysis of the SARS-CoV-2 cell entry inhibitor camostat bound to the trypsin-like protease TMPRSS2. Med Chem Res 30:399–409. https://doi.org/10.1007/s00044-021-02708-7
Zaporozhets TS, Besednova NN (2020) Biologically active compounds from marine organisms in the strategies for combating coronaviruses. AIMS Microbiol 6:470–494. https://doi.org/10.3934/microbiol.2020028
Khan MT, Ali A, Wang Q, Irfan M, Khan A, Zeb MT, Zhang YJ, Chinnasamy S, Wei DQ (2021) Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2—a molecular dynamic study. J Biomol Struct Dyn 39:3627–3637. https://doi.org/10.1080/07391102.2020.1769733
Singh R, Chauhan N, Kuddus M (2021) Exploring the therapeutic potential of marine-derived bioactive compounds against COVID-19. Environ Sci Pollut Res 28:52798–52809. https://doi.org/10.1007/s11356-021-16104-6
Omotuyi IO, Nash O, Ajiboye BO, Olumekun VO, Oyinloye BE, Osuntokun OT, Olonisakin A, Ajayi AO, Olusanya O, Akomolafe FS et al (2021) Aframomum melegueta secondary metabolites exhibit polypharmacology against SARS-CoV-2 drug targets: in vitro validation of furin inhibition. Phyther Res 35:908–919. https://doi.org/10.1002/ptr.6843
El-Demerdash A, Metwaly AM, Hassan A, El-Aziz TMA, Elkaeed EB, Eissa IH, Arafa RK, Stockand JD (2021) Comprehensive virtual screening of the antiviral potentialities of marine polycyclic guanidine alkaloids against sars-cov-2 (Covid-19). Biomolecules 11:1–25. https://doi.org/10.3390/biom11030460
Kar P, Kumar V, Vellingiri B, Sen A, Jaishee N, Anandraj A, Malhotra H, Bhattacharyya S, Mukhopadhyay S, Kinoshita M et al (2020) Anisotine and amarogentin as promising inhibitory candidates against SARS-CoV-2 proteins: a computatational investigation. J Biomol Struct Dyn 40:1–11
Vardhan S, Sahoo SK (2004) (2020) Searching inhibitors for three important proteins of COVID-19 through molecular docking studies. arXiv 08095:1–13
Falade VA, Adelusi TI, Adedotun IO, Abdul-Hammed M, Lawal TA, Agboluaje SA (2021) In silico investigation of saponins and tannins as potential inhibitors of SARS-CoV-2 main protease (Mpro). Silico Pharmacol 9:1–15. https://doi.org/10.1007/s40203-020-00071-w
Kandeil A, Mostafa A, Kutkat O, Moatasim Y, Al-Karmalawy AA, Rashad AA, Kayed AE, Kayed AE, El-Shesheny R, Kayali G et al (2021) Bioactive polyphenolic compounds showing strong antiviral activities against severe acute respiratory syndrome coronavirus 2. Pathogens 10:758. https://doi.org/10.3390/pathogens10060758
Carroll AR, Copp BR, Davis RA, Keyzers RA, Prinsep MR (2020) Marine natural products. Nat Prod Rep 37:175–223. https://doi.org/10.1039/c9np00069k
Rathinavel T, Meganathan B, Kumarasamy S, Ammashi S, Thangaswamy S, Ragunathan Y, Palanisamy S (2021) Potential COVID-19 drug from natural phenolic compounds through in silico virtual screening approach. Biointerf Res. Appl. Chem. 11:10161–10173. https://doi.org/10.33263/BRIAC113.1016110173
Davella R, Gurrapu S, Mamidala E (2021) Phenolic compounds as promising drug candidates against Covid-19 -an integrated molecular docking and dynamics simulation study. Mater Today Proc 51:1–6
Kowalczyk M, Golonko A, Świsłocka R, Kalinowska M, Parcheta M, Swiergiel A, Lewandowski W (2021) Drug design strategies for the treatment of viral disease. plant phenolic compounds and their derivatives. Front Pharmacol 12:1–21. https://doi.org/10.3389/fphar.2021.709104
Ye N, Caruso F, Rossi M (2021) Mechanistic insights into the inhibition of SARS-CoV-2 main protease by clovamide and its derivatives. Silico Stud Biophys 1:377–404. https://doi.org/10.3390/biophysica1040028
Abdusalam AAA, Murugaiyah V (2020) Identification of potential inhibitors of 3CL protease of SARS-CoV-2 From ZINC database by molecular docking-based virtual screening. Front Mol Biosci 7:1–11. https://doi.org/10.3389/fmolb.2020.603037
Barage S, Karthic A, Bavi R, Desai N, Kumar R, Kumar V, Lee KW (2022) Identification and characterization of novel RdRp and Nsp15 inhibitors for SARS-COV2 using computational approach. J Biomol Struct Dyn 40:2557–2574. https://doi.org/10.1080/07391102.2020.1841026
Kumar Verma A, Kumar V, Singh S, Goswami BC, Camps I, Sekar A, Yoon S, Lee KW (2021) Repurposing potential of Ayurvedic medicinal plants derived active principles against SARS-CoV-2 associated target proteins revealed by molecular docking, molecular dynamics and MM-PBSA studies. Biomed Pharmacother 137:1–17. https://doi.org/10.1016/j.biopha.2021.111356
Kumar R, Kumar V, Lee KW (2021) A computational drug repurposing approach in identifying the cephalosporin antibiotic and anti-hepatitis C drug derivatives for COVID-19 treatment. Comput Biol Med 130:1–13. https://doi.org/10.1016/j.compbiomed.2020.104186
Peele KA, Kumar V, Parate S, Srirama K, Lee KW, Venkateswarulu TC (2021) Insilico drug repurposing using FDA approved drugs against Membrane protein of SARS-CoV-2. J Pharm Sci 110:2346–2354. https://doi.org/10.1016/j.xphs.2021.03.004
Sharma AD, Kaur I (2020) Jensenone from eucalyptus essential oil as a potential inhibitor of COVID 19 corona virus infection. Res Rev Biotechnol Biosci 7:59–66. https://doi.org/10.5281/zenodo.3748477
Paul A, Sarkar A, Saha S, Maji A, Janah P, Kumar Maity T (2021) Synthetic and computational efforts towards the development of peptidomimetics and small-molecule SARS-CoV 3CLpro inhibitors. Bioorganic Med Chem 46:1–39. https://doi.org/10.1016/j.bmc.2021.116301
da Silva JKR, Figueiredo PLB, Byler KG, Setzer WN (2020) Essential oils as antiviral agents. Potential of essential oils to treat sars−cov−2 infection: an in−silico investigation. Int J Mol Sci 21:1–35. https://doi.org/10.3390/ijms21103426
Fiscon G, Conte F, Farina L, Paci P (2021) SAveRUNNER: a network-based algorithm for drug repurposing and its application to COVID-19. PLoS Comput Biol 17:1–30. https://doi.org/10.1371/JOURNAL.PCBI.1008686
Sibilio P, Bini S, Fiscon G, Sponziello M, Conte F, Farina L, Verrienti A (2021) In silico drug repurposing in COVID-19: A network-based analysis. Biomed Pharmacother 142:1–10. https://doi.org/10.1016/j.biopha.2021.111954
Gysi DM, Do Valle Í, Zitnik M, Ameli A, Gan X, Varol O, Ghiassian SD, Patten JJ, Davey RA, Loscalzo J et al (2021) Network medicine framework for identifying drug-repurposing opportunities for COVID-19. Proc Natl Acad Sci USA 118:1–11. https://doi.org/10.1073/pnas.2025581118
Vatansever EC, Yang KS, Drelich AK, Kratch KC, Cho CC, Kempaiah KR, Hsu JC, Mellott DM, Xu S, Tseng CTK et al (2021) Bepridil is potent against SARS-CoV-2 in vitro. Proc Natl Acad Sci USA 118:1–8. https://doi.org/10.1073/pnas.2012201118
Asdadi A, Hamdouch A, Gharby S, Hassani LM (2020) Chemical characterization of essential oil of Artemisia herba-alba asso and his possible potential against covid-19. J Anal Sci Appl Biotechnol 2:67–72. https://doi.org/10.4802/IMIST.PRSM/jasab-v2i2.21589
Romeo I, Mesiti F, Lupia A, Alcaro S (2021) Current updates on naturally occurring compounds recognizing SARS-CoV-2 druggable targets. Molecules 26:1–27. https://doi.org/10.3390/molecules26030632
Demeritte A, Wuest WM (2020) A look around the West Indies: The spices of life are secondary metabolites. Bioorganic Med Chem 28:1–11. https://doi.org/10.1016/j.bmc.2020.115792
Ademiluyi AO, Oyeniran OH, Oboh G (2020) tropical food spices: a promising panacea for the novel coronavirus disease (COVID-19). efood 1:347. https://doi.org/10.2991/efood.k.201022.001
Sengupta S, Bhattacharyya D, Kasle G, Karmakar S, Sahu O, Ganguly A, Addya S, Das Sarma J (2021) Potential immunomodulatory properties of biologically active components of spices against SARS-CoV-2 and Pan β-coronaviruses. Front Cell Infect Microbiol 11:1–12. https://doi.org/10.3389/fcimb.2021.729622
Singh NA, Kumar P (2021) Spices and herbs: potential antiviral preventives and immunity boosters during COVID-19. Phyther Res 35:2745–2757. https://doi.org/10.1002/ptr.7019
Bousquet J, Czarlewski W, Zuberbier T, Mullol J, Blain H, Cristol JP, De La Torre R, Le Moing V, Lozano NP, Bedbrook A et al (2021) Spices to control COVID-19 symptoms: yes, but not only. Int Arch Allergy Immunol 182:489–495. https://doi.org/10.1159/000513538
Parham S, Kharazi AZ, Bakhsheshi-Rad HR, Nur H, Ismail AF, Sharif S, Ramakrishna S, Berto F (2020) Antioxidant, antimicrobial and antiviral properties of herbal materials. Antioxidants 9:1–36. https://doi.org/10.3390/antiox9121309
Hussein G, Miyashiro H, Nakamura N, Hattori M, Kakiuchi N, Shimotohno K (2000) Inhibitory effects of Sudanese medicinal plant extracts on hepatitis C virus (HCV) protease. Phyther Res 14:510–516. https://doi.org/10.1002/1099-1573(200011)14:7%3c510::AID-PTR646%3e3.0.CO;2-B
Sundararajan A, Ganapathy R, Huan L, Dunlap JR, Webby RJ, Kotwal GJ, Sangster MY (2010) Influenza virus variation in susceptibility to inactivation by pomegranate polyphenols is determined by envelope glycoproteins. Antiviral Res 88:1–9. https://doi.org/10.1016/j.antiviral.2010.06.014
Surucic R, Travar M, Petkovic M, Tubic B, Stojilkovic M, Grabez M, Savikin K, Zdunic G, Skrbic R (2021) Pomegranate peel extract polyphenols attenuate the SARS-CoV-2 S-glycoprotein binding ability to ACE2 Receptor. In silico and in vitro studies. Bioorg Chem 114:1–7
Tito A, Colantuono A, Pirone L, Pedone E, Intartaglia D, Giamundo G, Conte I, Vitaglione P, Apone F (2021) Pomegranate peel extract as an inhibitor of SARS-CoV-2 spike binding to human ACE2 receptor (in vitro): a promising source of novel antiviral drugs. Front Chem 9:1–11. https://doi.org/10.3389/fchem.2021.638187
Ali S, Alam M, Khatoon F, Fatima U, Elasbali AM, Adnan M, Islam A, Hassan MI, Snoussi M, De Feo V (2022) Natural products can be used in therapeutic management of COVID-19: Probable mechanistic insights. Biomed Pharmacother 147:1–11. https://doi.org/10.1016/j.biopha.2022.112658
An X, Zhang YH, Duan L, Jin D, Zhao S, Zhou RR, Duan Y, Lian F, Tong X (2021) The direct evidence and mechanism of traditional Chinese medicine treatment of COVID-19. Biomed Pharmacother 137:1–17. https://doi.org/10.1016/j.biopha.2021.111267
Silveira D, Prieto-Garcia JM, Boylan F, Estrada O, Fonseca-Bazzo YM, Jamal CM, Magalhães PO, Pereira EO, Tomczyk M, Heinrich M (2020) COVID-19: is there evidence for the use of herbal medicines as adjuvant symptomatic therapy? Front Pharmacol 11:1–44. https://doi.org/10.3389/fphar.2020.581840
Yang Y, Islam MS, Wang J, Li Y, Chen X (2020) Traditional Chinese medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): a review and perspective. Int J Biol Sci 16:1708–1717. https://doi.org/10.7150/ijbs.45538
Yi L, Li Z, Yuan K, Qu X, Chen J, Wang G, Zhang H, Luo H, Zhu L, Jiang P et al (2004) Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J Virol 78:11334–11339. https://doi.org/10.1128/jvi.78.20.11334-11339.2004
Xi S, Li Y, Yue L, Gong Y, Qian L, Liang T, Ye Y (2020) Role of traditional Chinese medicine in the management of viral pneumonia. Front Pharmacol 11:1–26. https://doi.org/10.3389/fphar.2020.582322
Ni L, Zhou L, Zhou M, Zhao J, Wang DW (2020) Combination of the western medicine and the Chinese traditional patent medicine in treating a family case of CoVID-19. Front Med 14:210–214
Ng AWT, Poon SL, Huang MN, Lim JQ, Boot A, Yu W, Suzuki Y, Thangaraju S, Ng CCY, Tan P et al (2017) Aristolochic acids and their derivatives are widely implicated in liver cancers in Taiwan and throughout Asia. Sci Transl Med 9:1–12. https://doi.org/10.1126/scitranslmed.aan6446
Deng W, Xu Y, Kong Q, Xue J, Yu P, Liu J, Lv Q, Li F, Wei Q, Bao L (2020) Therapeutic efficacy of pudilan xiaoyan oral liquid (PDL) for COVID-19 in vitro and in vivo. Signal Transduct Target Ther 5:1–3. https://doi.org/10.1038/s41392-020-0176-0
Hetrick B, Yu D, Olanrewaju AA, Chilin LD, He S, Dabbagh D, Alluhaibi G, Ma YC, Hofmann LA, Hakami RM et al (2021) A traditional medicine, respiratory detox shot (RDS), inhibits the infection of SARS-CoV, SARS-CoV-2, and the influenza a virus in vitro. Cell Biosci 11:1–12. https://doi.org/10.1186/s13578-021-00609-1
Care H (2020) COVID - 19: opportunity for mainstreaming Ayurveda to transform Indian COVID - 19: opportunity for mainstreaming Ayurveda to transform indian health care. J Ayurveda 14:42–48. https://doi.org/10.4103/joa.joa
Maurya VK, Kumar S, Prasad AK, Bhatt MLB, Saxena SK (2020) Structure-based drug designing for potential antiviral activity of selected natural products from Ayurveda against SARS-CoV-2 spike glycoprotein and its cellular receptor. VirusDisease 31:179–193. https://doi.org/10.1007/s13337-020-00598-8
Borse S, Joshi M, Saggam A, Bhat V, Walia S, Marathe A, Sagar S, Chavan-Gautam P, Girme A, Hingorani L et al (2021) Ayurveda botanicals in COVID-19 management: an in silico multi-target approach. PLoS ONE 16:1–33. https://doi.org/10.1371/journal.pone.0248479
Ram T, Munikumar M, Raju V, Devaraj P, Boiroju N, Hemalatha R, Prasad P, Gundeti M, Sisodia B, Pawar S et al (2022) In silico evaluation of the compounds of the ayurvedic drug, AYUSH-64, for the action against the SARS-CoV-2 main protease. J Ayurveda Integr Med 13:100413. https://doi.org/10.1016/j.jaim.2021.02.004
Segneanu A, Cepan C, Grozescu I, Cziple F, Olariu S, Ratiu S, Lazar V, Murariu S, Velciov S, Marti T (2019) Therapeutic use of some Romanian medicinal plants. In: Perveen S, Al-Taweel A (eds) Pharmacognosy. Intech, London
Khanna K, Kohli SK, Kaur R, Bhardwaj A, Bhardwaj V, Ohri P, Sharma A, Ahmad A, Bhardwaj R, Ahmad P (2021) Herbal immune-boosters: Substantial warriors of pandemic Covid-19 battle. Phytomedicine 85:153361. https://doi.org/10.1016/j.phymed.2020.153361
Wang Y, Hou H, Ren Q, Hu H, Yang T, Li X (2021) Natural drug sources for respiratory diseases from Fritillaria: chemical and biological analyses. Chin Med 16:1–41. https://doi.org/10.1186/s13020-021-00450-1
Korkmaz H (2021) Could sumac be effective on COVID-19 treatment? J Med Food 24:563–568. https://doi.org/10.1089/jmf.2020.0104
Wijaya SH, Afendi FM, Batubara I, Huang M, Ono N, Kanaya S, Altaf-Ul-amin M (2021) Identification of targeted proteins by jamu formulas for different efficacies using machine learning approach. Life 11:1–11. https://doi.org/10.3390/life11080866
Wijaya SH, Batubara I, Nishioka T, Altaf-Ul-Amin M, Kanaya S (2017) Metabolomic studies of Indonesian Jamu medicines: prediction of Jamu efficacy and identification of important metabolites. Mol Inform 36:1–16. https://doi.org/10.1002/minf.201700050
Fadli A, Kusuma W, Annisa B, Heryanto IR (2021) Screening of potential indonesia herbal compounds based on multi-label classification for 2019 coronavirus disease _ enhanced reader.pdf. Big Data Cogn Comput 5:1–19
Hartanti D, Dhiani BA, Charisma SL, Wahyuningrum R (2020) The potential roles of Jamu for COVID-19: a learn from the traditional Chinese medicine. Pharm Sci Res 7:12–22. https://doi.org/10.7454/psr.v7i4.1083
CNN (2021) 4 RS di Yogya Kembangkan Obat Covid dari Minyak Kelapa Murni. CNN Indones, Mampang Prapatan
Lin CW, Tsai FJ, Tsai CH, Lai CC, Wan L, Ho TY, Hsieh CC, Chao PDL (2005) Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antiviral Res 68:36–42. https://doi.org/10.1016/j.antiviral.2005.07.002
Nguyen TT, Woo H, Kang H, Nguyen V, Kim Y, Kim D, Ahn S, Xia Y, Kim D (2012) Flavonoid-mediated inhibition of SARS coronavirus 3C-like protease expressed in Pichia pastoris. Biotechnol Lett 34:831–838
Schwarz S, Wang K, Yu W, Sun B, Schwarz W (2011) Emodin inhibits current through SARS-associated coronavirus 3a protein. Antiviral Res 90:64–69. https://doi.org/10.1016/j.antiviral.2011.02.008
Ho TY, Wu SL, Chen JC, Li CC, Hsiang CY (2007) Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antiviral Res 74:92–101. https://doi.org/10.1016/j.antiviral.2006.04.014
Bailly C, Vergoten G (2020) Glycyrrhizin: an alternative drug for the treatment of COVID-19 infection and the associated respiratory syndrome? Pharmacol Ther 214:1–16
van de Sand L, Bormann M, Alt M, Schipper L, Heilingloh CS, Steinmann E, Todt D, Dittmer U, Elsner C, Witzke O et al (2021) Glycyrrhizin effectively inhibits sars-cov-2 replication by inhibiting the viral main protease. Viruses 13:1–10. https://doi.org/10.3390/v13040609
Jo S, Kim S, Shin DH, Kim MS (2020) Inhibition of SARS-CoV 3CL protease by flavonoids. J Enzyme Inhib Med Chem 35:145–151. https://doi.org/10.1080/14756366.2019.1690480
Zhang D, Wu K, Zhang X, qiong S, Peng B (2020) In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. J Integr Med. https://doi.org/10.1016/j.joim.2020.02.005
Paraiso IL, Revel JS, Stevens JF (2020) Potential use of polyphenols in the battle against COVID-19. Curr Opin Food Sci 32:149–155. https://doi.org/10.1016/j.cofs.2020.08.004
Ma Q, Li R, Pan W, Huang W, Liu B, Xie Y, Wang Z, Li C, Jiang H, Huang J et al (2020) Phillyrin (KD-1) exerts anti-viral and anti-inflammatory activities against novel coronavirus (SARS-CoV-2) and human coronavirus 229E (HCoV-229E) by suppressing the nuclear factor kappa B (NF-κB) signaling pathway. Phytomedicine 78:1–9. https://doi.org/10.1016/j.phymed.2020.153296
Raj V, Park J, Cho K, Choi P, Kim T, Ham J, Lee J (2021) Assessment of antiviral potencies of cannabinoids against SARS-CoV-2 using computational and in vitro approaches. Int J Biol Macromol 168:474–485. https://doi.org/10.1016/j.ijbiomac.2020.12.020
Biagioli M, Marchiano S, Roselli R, Giorgio C, Bellini R, Bordoni M, Gidari A, Sabbatini S, Francisci D, Fiorillo B et al (2021) Discovery of a AHR pelargonidin agonist that counter-regulates Ace2 expression and attenuates ACE2-SARS-CoV-2 interaction. Biochem Pharmacol 188:1–15
Schwarz S, Sauter D, Wang K, Zhang R, Sun B, Karioti A, Bilia A, Efferth T, Schwarz W (2014) Kaempferol derivatives as antiviral drugs against the 3a channel protein of coronavirus. Planta Med 80:177–182
Bettuzzi S, Gabba L, Cataldo S (2021) Efficacy of a polyphenolic, standardized green tea extract for the treatment of COVID-19 syndrome: a proof-of-principle study. Covid 1:2–12. https://doi.org/10.3390/covid1010002
Bormann M, Alt M, Schipper L, van de Sand L, Le-Trilling VTK, Rink L, Heinen N, Madel RJ, Otte M, Wuensch K et al (2021) Turmeric root and its bioactive ingredient curcumin effectively neutralize sars-cov-2 in vitro. Viruses 13:1–12. https://doi.org/10.3390/v13101914
Xiao T, Cui M, Zheng C, Wang M, Sun R, Gao D, Bao J, Ren S, Yang B, Lin J et al (2021) Myricetin inhibits SARS-CoV-2 viral replication by targeting Mpro and ameliorates pulmonary inflammation. Front Pharmacol 12:1–9. https://doi.org/10.3389/fphar.2021.669642
Chen Z, Cui Q, Cooper L, Zhang P, Lee H, Chen Z, Wang Y, Liu X, Rong L, Du R (2021) Ginkgolic acid and anacardic acid are specific covalent inhibitors of SARS-CoV-2 cysteine proteases. Cell Biosci 11:1–8. https://doi.org/10.1186/s13578-021-00564-x
Ghosh R, Chakraborty A, Biswas A, Chowdhuri S (2021) Identification of polyphenols from Broussonetia papyrifera as SARS CoV-2 main protease inhibitors using in silico docking and molecular dynamics simulation approaches. J Biomol Struct Dyn 39:6747–6760. https://doi.org/10.1080/07391102.2020.1802347
Khaerunnisa S, Kurniawan H, Awaluddin R, Suhartati S (2020) Potential inhibitor of COVID-19 main protease (M pro) from several medicinal plant compounds by molecular docking study. Preprints 3:1–14. https://doi.org/10.2944/preprints202003.0226.v1
Ghosh R, Chakraborty A, Biswas A, Chowduri S (2020) Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors-an in silico docking and molecular dynamics simulation study. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2020.1779818
Maiti S, Banerjee A (2021) Epigallocatechin gallate and theaflavin gallate interaction in SARS-CoV-2 spike-protein central channel with reference to the hydroxychloroquine interaction: Bioinformatics and molecular docking study. Drug Dev Res 82:86–96. https://doi.org/10.1002/ddr.21730
Adem Ş, Eyupoglu V, Sarfraz I, Rasul A, Zahoor AF, Ali M, Abdalla M, Ibrahim IM, Elfiky AA (2021) Caffeic acid derivatives (CAFDs) as inhibitors of SARS-CoV-2: CAFDs-based functional foods as a potential alternative approach to combat COVID-19. Phytomedicine 85:1–17. https://doi.org/10.1016/j.phymed.2020.153310
Núñez MJ, Díaz-Eufracio BI, Medina-Franco JL, Olmedo DA (2021) Latin American databases of natural products: biodiversity and drug discovery against SARS-CoV-2. RSC Adv 11:16051–16064. https://doi.org/10.1039/d1ra01507a
Samy MN, Attia E, Shoman M, Khalil H, Sugimoto S, Matsunami K, Fahim J (2021) Phytochemical investigation of Amphilophium paniculatum an underexplored bignoniaceae species as a source of SARS-CoV-2 Mpro inhibitory metabolites. Isolation, identification, and molecular docking study. South African J Bot 141:421–430
Hasan A, Nahar N, Jannat K, Afroze T, Jahan R, Rahmatullah M (2020) In silico studies on phytochemicals of Pimpinella Anisum L. (Apiaceae) as potential inhibitors of SARS-CoV-2 3C-like protease. Int J Ayurveda 5:8–14
Chauhan, A, and Kalra, S (2020) Identification of potent COVID-19 main protease (MPRO) inhibitors from flavonoids. Res. Sq., 1–11.
Gentile D, Patamia V, Scala A, Sciortino MT, Piperno A, Rescifina A (2020) Putative inhibitors of SARS-COV-2 main protease from a library of marine natural products: a virtual screening and molecular modeling study. Mar Drugs 18:1–19. https://doi.org/10.3390/md18040225
Bhardwaj VK, Singh R, Sharma J, Rajendran V, Purohit R, Kumar S (2020) Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J Biomol Struct Dyn 39:1–10. https://doi.org/10.1080/07391102.2020.1766572
Khan MF, Khan MA, Khan ZA, Ahamad T, Ansari WA (2020) Identification of dietary molecules as therapeutic agents to combat COVID-19 using molecular docking studies. Res Sq. https://doi.org/10.21203/rs.3.rs-19560/v1
Zhang J-J, Shen X, Yan YM, WANG Y, Cheng Y-X (2020) Discovery of anti-SARS-CoV-2 agents from commercially available flavor via docking screening. OSFPreprint. https://doi.org/10.31219/osf.io/vjch2
Lung J, Lin Y, Yang Y, Chou Y, Chang G, Tsai M, Hsu C, Yeh R, Shu L, Cheng Y et al (2020) The potential SARS-CoV-2 entry inhibitor. biorxiv Prepr Serv Biol 55:1–22
ulQamar MT, Alqahtani SM, Alamri MA, Che LL (2020) Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J Pharm Anal 10:313–319. https://doi.org/10.1016/j.jpha.2020.03.009
Owis A, El-Hawary M, Amir D, Aly O, Abdelmohsen U, Kamel M (2020) Molecular docking reveals the potential of Salvadora persica flavonoids to inhibit COVID-19 virus main protease. RSC Adv 10:1–7
Vincent S, Arokiyaraj S, Saravanan M, Dhanraj M (2020) Molecular docking studies on the anti-viral effects of compounds from kabasura kudineer on SARS-CoV-2 3CLpro. Front Mol Biosci 7:1–12. https://doi.org/10.3389/fmolb.2020.613401
Murthy T, Joshi T, Gunnan S, Kulkarni N, Venkatesh P, Kumar B, Gowrishankar B (2021) In silico analysis of Phyllanthus amarus phytochemicals as potent drugs against SARS-CoV-2 main protease. Curr Res Green Sustain Chem 4:1–14. https://doi.org/10.1016/j.crgsc.2021.100159
Abba Y, Hassim H, Hamzah H, Noordin MM (2015) antiviral activity of resveratrol against human and animal viruses. Adv Virol 184241:1–7
Schettig R, Sears T, Klein M, Tan-Lim R, Matthias R Jr, Aussems C, Hummel M, Sears R, Poteet Z, Warren D et al (2020) COVID-19 patient with multifocal pneumonia and respiratory difficulty resolved quickly: possible antiviral and anti-inflammatory benefits of quercinex (nebulized quercetin-NAC) as adjuvant. Adv Infect Dis 10:45–55. https://doi.org/10.4236/aid.2020.103006
Wang X, Yu N, Peng H, Hu Z, Sun Y, Zhu X, Jiang L, Xiong H (2019) The profiling of bioactives in Akebia trifoliata pericarp and metabolites bioavailability and in vivo anti-inflammatory activities in DSS-induced colitis mice. Food Funct. https://doi.org/10.1039/c9fo00393b
Hwang SH, Kwon SH, Kim SB, Lim SS (2017) Inhibitory activities of stauntonia hexaphylla leaf constituents on rat lens aldose reductase and formation of advanced glycation end products and antioxidant. Biomed Res Int 4273257:1–9. https://doi.org/10.1155/2017/4273257
Ersöz T, Taşdemir D, Çaliş I, Ireland CM (2002) Phenylethanoid glycosides from Scutellaria galericulata. Turkish J Chem 26:465–471
Dong Z, Lu X, Tong X, Dong Y, Tang L, Liu M (2017) Forsythiae fructus: a review on its phytochemistry, quality control, pharmacology and pharmacokinetics. Molecules 22:1–49. https://doi.org/10.3390/molecules22091466
Keefover-Ring K, Holeski LM, Bowers MD, Clauss AD, Lindroth RL (2014) Phenylpropanoid glycosides of mimulus guttatus (yellow monkeyflower). Phytochem Lett 10:132–139. https://doi.org/10.1016/j.phytol.2014.08.016
Colunga Biancatelli RML, Berrill M, Catravas JD, Marik PE (2020) Quercetin and vitamin C: an experimental, synergistic therapy for the prevention and treatment of SARS-CoV-2 related disease (COVID-19). Front Immunol 11:1–11. https://doi.org/10.3389/fimmu.2020.01451
Liskova A, Samec M, Koklesova L, Samuel SM, Zhai K, Al-Ishaq RK, Abotaleb M, Nosal V, Kajo K, Ashrafizadeh M et al (2021) Flavonoids against the SARS-CoV-2 induced inflammatory storm. Biomed Pharmacother 138:111430. https://doi.org/10.1016/j.biopha.2021.111430
Annunziata G, Sanduzzi Zamparelli M, Santoro C, Ciampaglia R, Stornaiuolo M, Tenore GC, Sanduzzi A, Novellino E (2020) May polyphenols have a role against coronavirus infection? An overview of in vitro evidence. Front Med 7:1–8. https://doi.org/10.3389/fmed.2020.00240
Berretta A, Silveira M, Capcha J, Jong D (2020) Propolis and its potential against SARS-CoV-2 infection mechanisms and COVID-19 disease. running title: propolis against SARS-CoV-2 infection and COVID-19. Biomed Pharmacother 131:1–16. https://doi.org/10.1016/j.biopha.2020.110622
Huang L, Li H, Yuen TT-T, Ye Z, Fu Q, Sun W, Xu Q, Yang Y, Chan JF-W, Zhang G et al (2020) Berbamine inhibits the infection of SARS-CoV-2 and flaviviruses by compromising TPRMLs-mediated endolysosomal trafficking of viral receptors. Res Sq. https://doi.org/10.21203/rs.3.rs-30922/v1
Choy KT, Wong AYL, Kaewpreedee P, Sia SF, Chen D, Hui KPY, Chu DKW, Chan MCW, Cheung PPH, Huang X et al (2020) Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res 178:1–5. https://doi.org/10.1016/j.antiviral.2020.104786
Xia B, Shen X, He Y, Pan X, Wang Y, Yang F, Fang S, Wu Y, Zuo X, Xie Z et al (2021) SARS-CoV-2 envelope protein causes acute respiratory distress syndrome (ARDS)-like pathological damage and constitutes an antiviral target. Cell Res 31:847–860. https://doi.org/10.1038/s41422-021-00519-4
Li SY, Chen C, Zhang HQ, Guo HY, Wang H, Wang L, Zhang X, Hua SN, Yu J, Xiao PG et al (2005) Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antiviral Res 67:18–23. https://doi.org/10.1016/j.antiviral.2005.02.007
Stanic-Vucinic D, Minic S, Nikolic M, Velickovic T (2018) Spirulina phycobiliproteins as food components and complements. In: Jacob-Lopes E, Zepka LQ, Queiroz MI (eds) Microalgal Biotechnology. IntechOpen, London, pp 129–149
Pendyala B, Patras A, Dash C (2021) Phycobilins as Potent food bioactive broad-spectrum inhibitors against proteases of SARS-CoV-2 and other coronaviruses: a preliminary study. Front Microbiol. https://doi.org/10.3389/fmicb.2021.645713
Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G (2020) Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30:269–271. https://doi.org/10.1038/s41422-020-0282-0
Chowdhury P (2021) In silico investigation of phytoconstituents from indian medicinal herb “Tinospora Cordifolia (Giloy)” as potential inhibitors against SARS-CoV-2 (COVID-19). J Biomol Struct Dyn 39:6792–6809
Mohammadi S, Heidarizadeh M, Entesari M, Esmailpour A, Esmailpour M, Moradi R, Sakhaee N, Doustkhah E (2020) In silico investigation on the inhibiting role of nicotine/caffeine by blocking the s protein of sars-cov-2 versus ace2 receptor. Microorganisms 8:1–14. https://doi.org/10.3390/microorganisms8101600
Garg S, Roy A (2020) In silico analysis of selected alkaloids against main protease (Mpro) of SARS-CoV-2. Chem Biol Interact 332:1–12. https://doi.org/10.1016/j.cbi.2020.109309
Roza D, Selly R, Munsirwan R, Fadhilah G (2021) Molecular docking of quinine derivative as inhibitor in sars-Cov-2. J Phys Conf Ser 1819:1–8. https://doi.org/10.1088/1742-6596/1819/1/012053
Topcu G, Şenol H, Alim Toraman GÖ, Altan VM (2020) Natural alkaloids as potential anti-coronavirus compounds. Bezmialem Sci 8:131–139. https://doi.org/10.12435/bas.galenos.2020.5035
Majnooni MB, Fakhri S, Bahrami G, Naseri M, Farzaei MH, Echeverr J (2021) Alkaloids as potential phytochemicals against SARS-CoV-2: approaches to the associated pivotal mechanisms. Evidence-based Complement Altern Med 2021:1–21. https://doi.org/10.1155/2021/6632623
He CL, Huang LY, Wang K, Gu CJ, Hu J, Zhang GJ, Xu W, Xie YH, Tang N, Huang AL (2021) Identification of bis-benzylisoquinoline alkaloids as SARS-CoV-2 entry inhibitors from a library of natural products. Signal Transduct Target Ther 6:1–3. https://doi.org/10.1038/s41392-021-00531-5
Borquaye LS, Gasu EN, Ampomah GB, Kyei LK, Amarh MA, Mensah CN, Nartey D, Commodore M, Adomako AK, Acheampong P et al (2020) Alkaloids from cryptolepis sanguinolenta as potential inhibitors of SARS-CoV-2 viral proteins: an in silico study. Biomed Res Int 2020:1–14. https://doi.org/10.1155/2020/5324560
Quimque M, Notarte K, Fernandez R, Mendoza M, Liman R, Lim J, Pilapil L, Ong J, Pastrana A, Khan A et al (2021) Virtual screening-driven drug discovery of SARS-CoV2 enzyme inhibitors targeting viral attachment, replication, post-translational modification and host immunity evasion infection mechanisms. J Biomol Struct Dyn 39:1–18
Gaba S, Saini A, Singh G, Monga V (2021) An insight into the medicinal attributes of berberine derivatives: a review. Bioorganic Med Chem 38:1–20. https://doi.org/10.1016/j.bmc.2021.116143
Yang W, Chen X, Li Y, Guo S, Wang Z, Yu X (2020) Advances in pharmacological activities of terpenoids. Nat Prod Commun 15:1–13. https://doi.org/10.1177/1934578X20903555
Anil SM, Shalev N, Vinayaka AC, Nadarajan S, Namdar D, Belausov E, Shoval I, Mani KA, Mechrez G, Koltai H (2021) Cannabis compounds exhibit anti-inflammatory activity in vitro in COVID-19-related inflammation in lung epithelial cells and pro-inflammatory activity in macrophages. Sci Rep 11:1–15. https://doi.org/10.1038/s41598-021-81049-2
Wu Z, Chen X, Ni W, Zhou D, Chai S, Ye W, Zhang Z, Guo Y, Ren L, Zeng Y (2021) The inhibition of Mpro, the primary protease of COVID-19, by Poria cocos and its active compounds_ a network pharmacology and molecular docking study. RSC Adv 11:11821–11843
Nair M, Huang Y, Fidock DAP, Wagoner SJ, Towler J, Weathers MJ, P, (2021) Artemisia annua L. extracts inhibit the in vitro replication of SARS-CoV-2 and two of its variants. J Ethnopharmacol 274:1–8
Talukdar J, Bhadra B, Dattaroy T, Nagle V, Dasgupta S (2020) Potential of natural astaxanthin in alleviating the risk of cytokine storm in COVID-19. Biomed Pharmacother 132:110886. https://doi.org/10.1016/j.biopha.2020.110886
Melek FR, Miyase T, Abdel-Khalik SM, Hetta MH, Mahmoud II (2002) Triterpenoid saponins from Oreopanax guatemalensis. Phytochemistry 60:185–195. https://doi.org/10.1016/S0031-9422(02)00058-4
Muhseen ZT, Hameed AR, Al-Hasani HMH, Tahir ul Qamar M, Li G (2020) Promising terpenes as SARS-CoV-2 spike receptor-binding domain (RBD) attachment inhibitors to the human ACE2 receptor: Integrated computational approach. J Mol Liq 320:1–10. https://doi.org/10.1016/j.molliq.2020.114493
Yu MS, Lee J, Lee JM, Kim Y, Chin YW, Jee JG, Keum YS, Jeong YJ (2012) Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorganic Med Chem Lett 22:4049–4054. https://doi.org/10.1016/j.bmcl.2012.04.081
Reichling J, Koch C, Stahl-Biskup E, Sojka C, Schnitzler P (2005) Virucidal activity of a β-triketone-rich essential oil of Leptospermum scoparium (manuka oil) against HSV-1 and HSV-2 in cell culture. Planta Med 71:1123–1127. https://doi.org/10.1055/s-2005-873175
Snene A, El Mokni R, Jmii H, Jlassi I, Jaïdane H, Falconieri D, Piras A, Dhaouadi H, Porcedda S, Hammami S (2017) In vitro antimicrobial, antioxidant and antiviral activities of the essential oil and various extracts of wild (Daucus virgatus (Poir.) Maire) from Tunisia. Ind Crops Prod 109:109–115. https://doi.org/10.1016/j.indcrop.2017.08.015
Lelešius R, Karpovaite A, Mickiene R, Drevinskas T, Tiso N, Ragažinskiene O, Kubiliene L, Maruška A, Šalomskas A (2019) In vitro antiviral activity of fifteen plant extracts against avian infectious bronchitis virus. BMC Vet Res 15:1–10. https://doi.org/10.1186/s12917-019-1925-6
Park JY, Kim JH, Kim YM, Jeong HJ, Kim DW, Park KH, Kwon HJ, Park SJ, Lee WS, Ryu YB (2012) Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases. Bioorganic Med Chem 20:5928–5935. https://doi.org/10.1016/j.bmc.2012.07.038
Thuy BTP, My TTA, Hai NTT, Hieu LT, Hoa TT, Loan HTP, Triet NT, Van Anh TT, Quy PT, Van Tat P (2020) Investigation into SARS-CoV-2 resistance of compounds in garlic essential oil. ACS Omega. 5(8312):8320. https://doi.org/10.1021/acsomega.0c00772
Sharma AD, Kaur I (2020) Eucalyptol (1,8 cineole) from eucalyptus essential oil a potential inhibitor of Covid-19 corona virus infection by molecular docking studies. Preprints. https://doi.org/10.20944/preprints202003.0455.v1
Yadalam PK, Varatharajan K, Rajapandian K, Chopra P, Arumuganainar D, Nagarathnam T, Sohn H, Madhavan T (2021) Antiviral essential oil components against SARS-CoV-2 in pre-procedural mouth rinses for dental setting during Covid-19: a computational study. Front Chem 9:1–11
Roviello V, Roviello GN (2021) Lower COVID-19 mortality in Italian forested areas suggests immunoprotection by Mediterranean plants. Environ Chem Lett 19:699–710. https://doi.org/10.1007/s10311-020-01063-0
Kulkarni SA, Nagarajan SK, Ramesh V, Palaniyandi V, Selvam SP, Madhavan T (2020) Computational evaluation of major components from plant essential oils as potent inhibitors of SARS-CoV-2 spike protein. J Mol Struct 1221:128823. https://doi.org/10.1016/j.molstruc.2020.128823
Aanouz I, Belhassan A, El-Khatabi K, Lakhlifi T, El-ldrissi M, Bouachrine M (2021) Moroccan medicinal plants as inhibitors against SARS-CoV-2 main protease: computational investigations. J Biomol Struct Dyn 39:2971–2979. https://doi.org/10.1080/07391102.2020.1758790
Chapman RL, Andurkar SV (2022) A review of natural products, their effects on SARS-CoV-2 and their utility as lead compounds in the discovery of drugs for the treatment of COVID-19. Med Chem Res 31:40–51. https://doi.org/10.1007/s00044-021-02826-2
Shibata S (2000) A drug over the millennia: pharmacognosy, chemistry, and pharmacology of licorice. Yakugaku Zasshi 120:849–862. https://doi.org/10.1248/yakushi1947.120.10_849
Luo P, Liu D, Li J (2020) Pharmacological perspective: glycyrrhizin may be an efficacious therapeutic agent for COVID-19. Int J Antimicrob Agents 55:1–3. https://doi.org/10.1016/j.ijantimicag.2020.105995
Ding H, Deng W, Ding L, Ye X, Yin S, Huang W (2020) Glycyrrhetinic acid and its derivatives as potential alternative medicine to relieve symptoms in nonhospitalized COVID-19 patients. J Med Virol 92:2200–2204. https://doi.org/10.1002/jmv.26064
Hoever G, Baltina L, Michaelis M, Kondratenko R, Baltina L, Tolstikov GA, Doerr HW, Cinatl J (2005) Antiviral activity of glycyrrhizic acid derivatives against SARS-coronavirus. J Med Chem 48:1256–1259. https://doi.org/10.1021/jm0493008
Javed H, Meeran MFN, Jha NK, Ojha S (2021) Carvacrol, a plant metabolite targeting viral protease (Mpro) and ACE2 in host cells can be a possible candidate for COVID-19. Front Plant Sci 11:1–10. https://doi.org/10.3389/fpls.2020.601335
Halder D, Barik BB, Dasgupta RK, Roy SD (2018) Aroma therapy: an art of healing. Indian Res J Pharm Sci. 5:1540–1558. https://doi.org/10.21276/irjps.2018.5.3.2
Angulo-Milhem S, Verriele M, Nicolas M, Thevenet F (2021) Indoor use of essential oils: emission rates, exposure time and impact on air quality. Atmos Environ 244:1–13
Boukhatem MN (2020) Effective antiviral activity of essential oils and their characteristics terpenes against coronaviruses: an update. J Pharmacol Clin Toxicol 8:1138
Adorjan B, Buchbauer G (2010) Biological properties of essential oils: An updated review. Flavour Fragr J 25:407–426. https://doi.org/10.1002/ffj.2024
Ojah EO (2020) Exploring essential oils as prospective therapy against the ravaging Coronavirus (SARS-CoV-2). Iberoam J Med 04:322–330
De Logu A, Loy G, Pellerano ML, Bonsignore L, Schivo ML (2000) Inactivation of HSV-1 and HSV-2 and prevention of cell-to-cell virus spread by Santolina insularis essential oil. Antiviral Res 48:177–185. https://doi.org/10.1016/S0166-3542(00)00127-3
Schnitzler P, Koch C, Reichling J (2007) Susceptibility of drug-resistant clinical herpes simplex virus type 1 strains to essential oils of ginger, thyme, hyssop, and sandalwood. Antimicrob Agents Chemother 51:1859–1862. https://doi.org/10.1128/AAC.00426-06
Schuhmacher A, Reichling J, Schnitzler P (2003) Virucidal effect of peppermint oil on the enveloped viruses herpes simplex virus type 1 and type 2 in vitro. Phytomedicine 10:504–510. https://doi.org/10.1078/094471103322331467
Minami M, Kita M, Nakaya T, Yamamoto T, Kuriyama H, Imanishi J (2003) The inhibitory effect of essential oils on herpes simplex virus type-1 replication in vitro. Microbiol Immunol 47:681–684. https://doi.org/10.1111/j.1348-0421.2003.tb03431.x
Schnitzler P, Schuhmacher A, Astani A, Reichling J (2008) Melissa officinalis oil affects infectivity of enveloped herpesviruses. Phytomedicine 15:734–740. https://doi.org/10.1016/j.phymed.2008.04.018
Kovač K, Diez-Valcarce M, Raspor P, Hernández M, Rodríguez-Lázaro D (2012) Natural plant essential oils do not inactivate non-enveloped enteric viruses. Food Environ Virol 4:209–212. https://doi.org/10.1007/s12560-012-9088-7
Garozzo A, Timpanaro R, Stivala A, Bisignano G, Castro A (2011) Activity of Melaleuca alternifolia (tea tree) oil on Influenza virus A/PR/8: study on the mechanism of action. Antiviral Res 89:83–88. https://doi.org/10.1016/j.antiviral.2010.11.010
Cermelli C, Fabio A, Fabio G, Quaglio P (2008) Effect of eucalyptus essential oil on respiratory bacteria and viruses. Curr Microbiol 56:89–92. https://doi.org/10.1007/s00284-007-9045-0
Jackwood MW, Rosenbloom R, Petteruti M, Hilt DA, McCall AW, Williams SM (2010) Avian coronavirus infectious bronchitis virus susceptibility to botanical oleoresins and essential oils in vitro and in vivo. Virus Res 149:86–94. https://doi.org/10.1016/j.virusres.2010.01.006
Benencia F, Courrges MC (2000) In vitro and in vivo activity of eugenol on human herpesvirus. Phyther Res 14:495–500. https://doi.org/10.1002/1099-1573(200011)14:7%3c495::AID-PTR650%3e3.0.CO;2-8
Vicidomini C, Roviello V, Roviello GN (2021) molecular basis of the therapeutical potential of clove (Syzygium aromaticum L.) and clues to its anti-COVID-19 utility. Molecules 26:1–12. https://doi.org/10.3390/molecules26071880
Banerjee S, Panda C, Das KS (2006) Clove (Syzygium aromaticum L.), a potential chemopreventive agent for lung cancer. Carcinogenesis. 27:1645–1654
Chaieb K, Hajlaoui H, Zmantar T, Kahla-Nakbi AB, Rouabhia M, Mahdouani K, Bakhrouf A (2007) The chemical composition and biological activity of clove essential oil, eugenia caryophyllata (Syzigium aromaticum L. myrtaceae): a short review. Phyther Res 21:501–506. https://doi.org/10.1002/ptr
Raina VK, Srivastava SK, Aggarwal KK, Syamasundar KV, Kumar S (2001) Essential oil composition of Syzygium aromaticum leaf from Little Andaman. India Flavour Fragr J 16:334–336. https://doi.org/10.1002/ffj.1005
Pino JA, Marbot R, Agüero J, Fuentes V (2001) Essential oil from buds and leaves of clove (Syzygium aromaticum (L.) Merr. et Perry) grown in Cuba. J Essent Oil Res 13:278–279. https://doi.org/10.1080/10412905.2001.9699693
Kurokawa M, Hozumi T, Basnet P, Nakano M, Kadota S, Namba T, Kawana T, Shiraki K (1998) Purification and characterization of eugeniin as an anti-herpesvirus compound from Geum japonicum and Syzygium aromaticum. J Pharmacol Exp Ther 284:728–735. https://doi.org/10.1146/annurev-immunol-032713-120231
Nguyen LC, Yang D, Nicolaescu V, Best TJ, Ohtsuki T, Chen S, Friesen JB, Drayman N, Mohamed A, Dann C et al (2021) Cannabidiol inhibits SARS-CoV-2 replication and promotes the host innate immune response. biorxiv 70:1–15
Chatow L, Nudel A, Nesher I, Hemo DH, Rozenberg P, Voropaev H, Winkler I, Levy R, Kerem Z, Yaniv Z et al (2021) In vitro evaluation of the activity of terpenes and cannabidiol against human coronavirus E229. Life 11:1–10. https://doi.org/10.3390/life11040290
Amparo TR, Seibert JB, Silveira BM, Costa FSF, Almeida TC, Braga SFP, da Silva GN, dos Santos ODH, de Souza GHB (2021) Brazilian essential oils as source for the discovery of new anti-COVID-19 drug: a review guided by in silico study. Phytochem Rev 20:1013–1032. https://doi.org/10.1007/s11101-021-09754-4
Amaral-Machado L, Oliveira WN, Rodrigues VM, Albuquerque NA, Alencar ÉN, Egito EST (2021) Could natural products modulate early inflammatory responses, preventing acute respiratory distress syndrome in COVID-19-confirmed patients? Biomed Pharmacother 134:1–20. https://doi.org/10.1016/j.biopha.2020.111143
Pisoschi A, Pop A, Iordache F, Stanca L, Geicu O, Bilteanu L, Serban A (2022) Antioxidant, anti-inflammatory and immunomodulatory roles of vitamins in COVID-19 therapy. Eur J Med Chem 232:1–24. https://doi.org/10.1016/j.ejmech.2022.114175
da Silva FMA, da Silva KPA, de Oliveira LPM, Costa EV, Koolen HHF, Pinheiro MLB, de Souza AQL, de Souza ADL (2020) Flavonoid glycosides and their putative human metabolites as potential inhibitors of the sars-cov-2 main protease (Mpro) and RNA-dependent RNA polymerase (RdRp). Mem Inst Oswaldo Cruz 115:1–8. https://doi.org/10.1590/0074-02760200207
Antonio AD, Wiedemann LS, Junior VF (2020) Natural products’ role against COVID-19. RSC Adv 10:23379–23393
Teli DM, Shah MB, Chhabria MT (2021) In silico screening of natural compounds as potential inhibitors of SARS-CoV-2 main protease and spike RBD: targets for COVID-19. Front Mol Biosci 7:1–25. https://doi.org/10.3389/fmolb.2020.599079
Contreras-Puentes N, Alvíz-Amador A (2020) Virtual screening of natural metabolites and antiviral drugs with potential inhibitory activity against 3CL-PRO and PL-PRO. Biomed Pharmacol J 13:933–941. https://doi.org/10.13005/BPJ/1962
Joshi RS, Jagdale SS, Bansode SB, Shankar SS, Tellis MB, Pandya VK, Chugh A, Giri AP, Kulkarni MJ (2020) Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J Biomol Struct Dyn 1:1–16. https://doi.org/10.1080/07391102.2020.1760137
Huynh T, Wang H, Luan B (2020) Structure-based lead optimization of herbal medicine rutin for inhibiting SARS-CoV-2’s main protease. Phys Chem Chem Phys 22:25335–25343. https://doi.org/10.1039/d0cp03867a
Mengist HM, Dilnessa T, Jin T (2021) Structural basis of potential inhibitors targeting SARS-CoV-2 main protease. Front Chem 9:1–19. https://doi.org/10.3389/fchem.2021.622898
Mehany T, Khalifa I, Barakat H, Althwab SA, Alharbi YM, El-Sohaimy S (2021) Polyphenols as promising biologically active substances for preventing SARS-CoV-2: a review with research evidence and underlying mechanisms. Food Biosci 40:1–11. https://doi.org/10.1016/j.fbio.2021.100891
Verma S, Twilley D, Esmear T, Oosthuizen CB, Reid AM, Nel M, Lall N (2020) Anti-SARS-CoV natural products with the potential to inhibit SARS-CoV-2 (COVID-19). Front Pharmacol 11:1–20. https://doi.org/10.3389/fphar.2020.561334
Acknowledgements
The authors also acknowledge Indonesia Endowment Fund for Education (Lembaga Pengelola Dana Pendidikan, LPDP) for supporting the present work.
Funding
This work is supported by Indonesia Endowment Fund for Education (Lembaga Pengelola Dana Pendidikan, LPDP) under-Grant Number KET-235/LPDP.4/2021 and LOG-3382/LPDP/LPDP.3/2023.
Author information
Authors and Affiliations
Contributions
IO wrote the first draft of the manuscript, wrote additional sections, and created the figures. IO, MS, MFAB, YUK, and SF created the concept, edited, and revised the manuscript together. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Oktavianawati, I., Santoso, M., Bakar, M.F.A. et al. Recent progress on drugs discovery study for treatment of COVID-19: repurposing existing drugs and current natural bioactive molecules. Appl Biol Chem 66, 89 (2023). https://doi.org/10.1186/s13765-023-00842-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13765-023-00842-x