Abstract
Dysregulated circular RNAs (circRNAs) are implicated in prostate cancer (PCa) progression. Hsa_circ_0081234 (circTRRAP) has been revealed as a facilitator in PCa, but the mechanisms associated with circTRRAP in PCa progression are largely unclear. The present study was to explore the regulatory mechanism of circTRRAP-mediated PCa progression. A total of 50 PCa tissues and normal tissues were collected. RNA levels of circTRRAP, microRNA (miR)-515-5p and homeobox A1 (HOXA1) were detected by quantitative real-time polymerase chain reaction (qRT-PCR) or western blot. Cell viability, proliferation, migration, and invasion were estimated using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide, 5-ethynyl-2ʹ-deoxyuridine (EdU) and transwell assays, respectively. Cell glycolysis was assessed by measuring glucose uptake and lactate production. The target interaction between circTRRAP or HOXA1 and miR-515-5p was investigated by the dual-luciferase reporter assay. We observed the overt upregulaiton of circTRRAP in PCa samples and cells. Silencing of circTRRAP lowered tumor growth in vivo and restrained PCa cell viability, proliferation, migration, invasion, and glycolysis in vitro. miR-515-5p was negatively regulated by circTRRAP and its deficiency reversed the inhibiting effects of circTRRAP knockdown on PCa cell malignancy and glycolysis. HOXA1 was confirmed as a miR-515-5p target and miR-515-5p overexpression lessened PCa cell malignancy and glycolysis by decreasing HOXA1 expression. Importantly, circTRRAP mediated HOXA1 expression by functioning as a miR-515-5p sponge. In conclusion, circTRRAP took part in PCa progression and glycolysis through mediating the miR-515-5p/HOXA1 axis, suggesting that circTRRAP can serve as a potential therapeutic target for PCa patients.
Highlights
-
1.
CircTRRAP was overexpressed in PCa samples and cells.
-
2.
CircTRRAP silencing lowered PCa cell malignancy and glycolysis.
-
3.
CircTRRAP sponged miR-515-5p to mediate HOXA1 expression.
Similar content being viewed by others
Introduction
Prostate cancer is a common health issue in males contributing to the major cause of cancer-related deaths [1]. In the past decades, great progress has been made in research on the pathogenesis of PCa [2]. With the development of diagnosis and treatment of PCa, the outcomes of patients have gained much improvement [3, 4]. However, the challenge remains on the limited effective strategies. Therefore, improving the understanding of the underlying pathogenesis of PCa is crucial for investigating mechanism-based therapeutic strategies.
Circular RNAs (circRNAs) are a unique class of endogenous RNA transcripts, in principle generated using a back-splicing mechanism, with higher stability and tissue-dependent expression [5]. The biology of circRNAs has rapidly advanced and demonstrated their diverse roles, including regulation of gene expression and possible coding potential, and competitive interactions with proteins and microRNAs (miRNAs) under various pathological conditions [6]. Advances in RNA identification techniques have provided a large number of circRNAs exhibiting upregulation or downregulation in PCa tissues compared to normal adjacent tissues [7]. For instance, circ_0006156 expressed at low levels in PCa tissues, and circ_0006156 lowered PCa cell malignancy by binding to S100A9 [8]. Circ-PFKP facilitated cell proliferative capacity via activation of IMPDH2 in PCa [9]. Hsa_circ_0081234 (circTRRAP), derived from the TRRAP gene, was uncovered as a facilitator in PCa [10]. Currently, little is known on the underlying mechanism of circTRRAP in PCa progression.
MiRNAs participate in disease progression via regulating oncogenes or tumor-suppressive genes [11, 12]. Dysregulation of miRNA expression is associated with a variety of cancers, including PCa [13]. For instance, miR-29b was lowly expressed in PCa, and miR-29b upregulation lessened PCa cell invasion and migration [14]. Another example was that miR-128 targeted BMI-1 to restrain PCa progression [15]. Moreover, several circRNAs have been demonstrated to be involved in PCa progression by interacting with miRNAs [16,17,18]. For instance, circ-NOLC1 prompted PCa development via interaction with miR-647 [19]. Circ-0086722 drove PCa progression by mediating STAT5A expression by sponging miR-339-5p [20]. Existing evidence suggests a vital action of the circRNA/miRNA/mRNA competitive endogenous RNA (ceRNA) network pathway in PCa [21, 22]. Based on the important role of the ceRNA mechanism, we found that miR-515-5p might interact with circTRRAP through the circinteractome online prediction tool (https://circinteractome.irp.nia.nih.gov/). miR-515 has been demonstrated as a tumor repressor in PCa [23, 24], but the association between miR-515 and circTRRAP is indistinct in PCa. In addition, homeobox A1 (HOXA1), which is an oncogene in multiple tumors [25, 26], was predicted as a possible miR-515-5p molecular target using the Targetscan online prediction tool (https://www.targetscan.org/vert_80/). Currently, whether miR-515-5p is associated with HOXA1 in PCa needs to be verified.
Hence, the research was to characterize the action of circTRRAP in PCa and the molecular mechanism of the circTRRAP/miR-515-5p/HOXA1 axis. All findings come together to confirm that circTRRAP might function as a miR-515-5p sponge to increase HOXA1 expression and consequently promote PCa progression. In conclusion, targeting circTRRAP to explore methods for PCa therapy is very promising.
Materials and methods
Patient and tissue collection
PCa patients (n = 50) who have signed the informed consent were recruited from Seventh Medical Center, PLA General Hospital. This study was performed under the approval of the Ethics Committee of Seventh Medical Center, PLA General Hospital.
Cell culture
Human prostatic epithelial cell line RWPE-1 (#165641, Mingzhoubio., Ningbo, China) and PCa cell lines DU145 (MZ-0058, Mingzhoubio.) and PC3 (MZ-0145, Mingzhoubio.) were cultured at 37 °C with 5% CO2. RWPE-1, DU145, and PC3 cell lines were cultured with the Keratinocyte Serum-Free Medium (K-SFM) Kit (#17005-042, Thermo, Waltham, MA, USA), MEM (Thermo) and Ham’s F-12K (Thermo), respectively.
Cell transfection
The overexpression plasmids of circTRRAP (circTRRAP) and HOXA1 (pcDNA-HOXA1) were generated with the pCD5-ciR (Geneseed, Guangzhou, China) or pcDNA3.1 (YouBio, Changsha, China) vectors with their respective empty vectors as negative controls (circ-NC and pcDNA-NC). Three small interfering RNAs (siRNAs) for circTRRAP circTRRAP [si-circTRRAP#1 (5ʹ-UGACGUGACAUUUGUUCGAGU-3ʹ), si-circTRRAP#2 (5ʹ-ACGUGACAUUUGUUCGAGUCU-3ʹ), and si-circTRRAP#3 (5ʹ-UGACAUUUGUUCGAGUCUCAC-3ʹ)] and non-target siRNA (si-NC, 5ʹ-AUAUCAUAGGUUGAUAAUGGU-3ʹ), miR-515-5p inhibitor (5ʹ-CAGAAAGUGCUUUCUUUUGGAGAA-3ʹ) and its control (inhibitor-NC, 5ʹ-CAGUACUUUUGUGUAGUACAA-3ʹ), as well as miR-515-5p mimic (miR-515-5p, 5ʹ-UUCUCCAAAAGAAAGCACUUUCUG-3ʹ) and its control (miR-NC, 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ) were generated by Fulengen (Guangzhou, China). The lipofecter liposomal transfection reagent (Beyotime, Shanghai, China) was used for transfection of DU145 and PC3 cells.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Trizol reagent (Solarbio, Beijing, China) was used for isolation of total RNA from PCa samples and cells. The PARIS Kit (Thermo) was utilized for isolation of nuclear and cytoplasmic fractions. RNase R (Epicentre Technologies) was utilized for digestion of total RNA (2 μg). RNA was reversely transcribed to complementary DNA (cDNA) using the reverse transcription PrimeScript RT Master Mix (Takara, Dalian, China) or RiboBio reverse transcription kit (Guangzhou, China). Quantification was done with SYBR (Takara) and specific primers (Table 1). The 2−ΔΔCt method was utilized for the calculation of the fold change [27].
Estimation of cell viability
PCa cells (5 × 103 cells) were plated into 96-well plates. After the incubation for 0, 24, 48 and 72 h, 10 μL of the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) solution (Beyotime) was added into each well. The cells were cultured for another 4 h and then each well was filled with 100 μL of dimethyl sulfoxide (Solarbio) for dissolving the crystal. The measurement of the optical density value was conducted with a microplate reader (Bio-Rad, Hercules, CA, USA).
Assessment of cell proliferation
PCa cells (5 × 103 cells) were seeded into 96-well plates and their proliferative capacity was measured using the Cell-Light™ 5-ethynyl-2ʹ-deoxyuridine (EdU) Apollo 488 Kit (RiboBio) following the manufacturer’s protocol. Briefly, cells were labeled with 50 μM EdU for 2 h, followed by fixation in 4% paraformaldehyde for 30 min. After staining with Apollo 488 and Hoechst, the number of positive cells exhibiting green fluorescence was recorded under a fluorescence microscope (Olympus, Tokyo, Japan).
Transwell assay
Estimation of cell migratory and invaded capacities was conducted with transwell inserts (Corning, Corning, NY, USA). For the invasion assay, the inserts were pre-coated with Matrigel. Meanwhile, the inserts without Matrigel were used for the migration assay. PCa cells (1 × 104) resuspended in serum-free medium were plated into the upper chambers. The lower chambers were filled with fresh medium containing 10% serum. After the incubation for 24 h, cells that crossed the membrane were stained with 0.5% crystal violet (Solarbio). Observation of migrating or invasive cells was done with a microscope (Olympus) at 100× magnification.
Western blot
Extraction of total protein was done with the total protein extraction kit (#BC3710, Solarbio). The protein samples (20 μg) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The membranes were blocked with the western blocking buffer (Solarbio) after transferring the membrane to a nitrocellulose membrane (Bio-Rad). Following incubation with specific primary antibodies, the membranes were incubated with a secondary antibody, the bands were visualized by SuperSignal West Pico Stable Peroxide Solution (Thermo). Primary antibodies were against PCNA (ab92552, 1:5000, Abcam, Cambridge, UK), MMP2 (ab181286, 1:1000, Abcam), MMP9 (ab76003, 1:5000, Abcam), HOXA1 (ab230513, 1:500, Abcam), and β-actin (ab115777, 1:200, Abcam).
Glucose and lactate level
PCa cells (1 × 104) were cultured for 24 h and then the culture medium was collected. Measurement of glucose uptake and lactate production was done with the Glucose Colorimetric Fluorometric Assay Kit (#K606-100, BioVision, USA) and the Lactate Colorimetric Assay Kit (#K627-100, BioVision) following manufacturer’s instructions, respectively.
Dual-luciferase reporter assay
PCa cells were transfected with a constructed luciferase reporter vector (circTRRAP-WT, circTRRAP-MUT, HOXA1 3ʹUTR-WT, or HOXA1 3ʹUTR-MUT) along with miR-515-5p or miR-NC. Cells were collected for measurement of the luciferase activity with a dual-luciferase assay kit (Promega, Madison, WI, USA) after 48 h of transfection. The luciferase plasmids used above were generated by inserting the wild-type and mutant sequences of circTRRAP and HOXA1 into the psiCHECK™-2 vector (Promega).
Xenograft models
The animal experiment permission was granted by the Animal Care Committee of the Seventh Medical Center, PLA General Hospital. Male BALB/c nude mice (4 weeks old) (Vital River, Beijing, China) were reared in conditions deprived of specific pathogens. PC3 cells (2 × 106) with Lenti-sh-NC or Lenti-sh-circTRRAP were injected into the flank of nude mice (n = 6). Tumor volume was monitored and calculated every 5 days (volume = (width2 × length)/2). After 30 days of injection, mice were anesthetized and then sacrificed for subsequent analysis.
IHC staining
Paraffin-embedded xenograft tumor sections were deparaffinized with xylene, rehydrated with graded ethanol, antigen retrieved with citrate buffer (10 mM, pH 6.0), and blocked for endogenous peroxidase with 3% H2O2 in TBS. Samples were incubated with primary antibodies against ki-67 (ab243878, 1:100, Abcam), MMP2 (ab97779, 1:500, Abcam), and MMP9 (ab76003, 1:1000, Abcam). After removal of excess antibody, tissue samples were reacted with DAB chromogen and substrate mixture (Thermo), followed by counterstaining with hematoxylin.
Statistical analysis
Bar plots were generated by GraphPad Prism 7 software (GraphPad, La Jolla, CA, USA). Each data represented the mean ± standard deviation (SD) of three biological replicates, with each biological replicate in triplicate. The difference was assessed by Student’s t-test or analysis of variance. Results were considered statistically significant when the P-value was less than 0.05.
Results
Validation of circTRRAP as an upregulated circRNA in PCa samples and cell lines
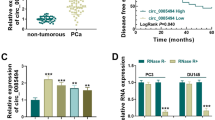
Hsa_circ_0081234 (circTRRAP) is derived from the TRRAP gene (exons 4 to 8) (Fig. 1A) and has been reported as a promoting regulator in PCa [10]. To explain the action of circTRRAP in PCa, the alteration in circTRRAP expression was validated. Results of qRT-PCR showed that circTRRAP expression was significantly elevated in PCa samples (n = 50) relative to matching normal samples (n = 50) (Fig. 1B). Similar circTRRAP trends were also seen in PCa cell lines when compared with the RWPE-1 cell line (Fig. 1C). Subcellular fractionation combined with qRT-PCR analysis exhibited that circTRRAP had a higher proportion in the cytoplasmic fraction of PCa cell lines (Fig. 1D). The combination of RNase R digestion and qRT-PCR analysis confirmed that circTRRAP has a more stable structure than the linear mRNA of the parental gene TRRAP (Fig. 1E). Collectively, high circTRRAP expression might be associated with PCa progression.
CircTRRAP was overexpressed in PCa. A Schematic illustration of the structure of circTRRAP. B CircTRRAP expression was measured in PCa tissues (n = 50) and corresponding normal tissues by qRT-PCR. C CircTRRAP abundance was detected in PC cell lines by qRT-PCR. D CircTRRAP abundance in the nuclear and cytoplasm fractions was assessed by qRT-PCR. E RNase R was administrated to digest the extracted RNA for analysis of the stability of circTRRAP. *P < 0.05
Knockdown of circTRRAP restrained PCa cell malignant phenotypes and lowered PCa cell glycolysis
To assess the effect of circTRRAP silencing on PCa cell malignant phenotypes, we transfected PCa cells with circTRRAP-specific siRNAs or a non-target control si-NC. PCa cells transfected with circTRRAP-specific siRNAs displayed substantially lower circTRRAP expression validated by qRT-PCR, and si-circTRRAP#1, which caused the greatest change in circTRRAP expression, was used for subsequent functional analysis (Fig. 2A). Functionally, circTRRAP-deficient PCa cell lines had low cell viability and proliferative capacity, as confirmed by MTT assay and EdU assay (Fig. 2B and C). Transwell assays showed that depletion of circTRRAP resulted in reduced migratory and invasive abilities of PCa cell lines (Fig. 2D and E). Also, circTRRAP knockdown decreased PCNA, MMP2, and MMP9 protein levels (Fig. 2F). Insufficiency of circTRRAP resulted in a significant reduction in both glucose uptake and lactate production in PCa cell lines (Fig. 2G and H). Together, circTRRAP prompted malignant phenotypes and glycolysis of PCa cells.
Knockdown of circTRRAP reduced PCa cell malignancy and glycolysis. A Transfection efficiencies of three siRNAs targeting circTRRAP were determined by qRT-PCR. B–E Cell viability, proliferation, migration, and invasion in si-circTRRAP#1 or si-NC-transfected PCa cells were estimated by MTT, EdU, and transwell assays. F and G PCNA, MMP2, and MMP9 protein levels in si-circTRRAP#1 or si-NC-transfected PCa cells were detected by western blot. G and H The glucose uptake and lactate production of si-circTRRAP#1 or si-NC-transfected PCa cells were measured. *P < 0.05
MiR-515-5p was sponged by circTRRAP
Because cytoplasmic circRNAs can affect miRNAs and corresponding downstream genes through a ceRNA mechanism, we explored the function of circTRRAP as a miRNA molecular sponge. An online prediction tool Circular RNA Interactome showed that circTRRAP possessed bases that may adsorb miR-515-5p (Fig. 3A). For validation of this association, the luciferase reporter vectors circTRRAP-WT and circTRRAP-MUT were constructed. The data of dual-luciferase reporter assay showed that the luciferase reporter circTRRAP-WT had a lower luciferase activity in the miR-515-5p-overexpression group, but the luciferase reporter circTRRAP-MUT did not change significantly (Fig. 3B and C). Moreover, circTRRAP insufficiency led to an elevation in miR-515-5p expression, but circTRRAP overexpression had the opposite function (Fig. 3D and E). Furthermore, the abundance of miR-515-5p was significantly decreased in PCa samples and cells (Fig. 3F and G). Meanwhile, there was a negative correlation between miR-515-5p and circTRRAP expression levels in PCa samples (r = − 0.455, P = 0.0009) (Fig. 3H). These findings suggested circTRRAP as a miR-515-5p molecular sponge.
CircTRRAP functioned as a miR-515-5p decoy. A The complementary binding sites of circTRRAP with miR-515-5p. B and C Luciferase activity was measured in PCa cells transfected with circTRRAP-WT or circTRRAP-MUT and miR-NC or miR-515-5p. D and E miR-515-5p abundance was measured in PCa cells transfected with si-NC, si-circTRRAP#1, circ-NC or circTRRAP by qRT-PCR. F and G miR-515-5p abundance was detected in PCa tissues and cell lines by qRT-PCR. H Pearson’s correlation analysis determined the correlation between the levels of circTRRAP and miR-515-5p in PCa tissues. *P < 0.05
Knockdown of miR-515-5p alleviated circTRRAP insufficiency-mediated impacts on PCa cell malignant phenotypes and glycolysis
Whether circTRRAP-mediated regulation of PCa progression is linked to miR-515-5p was further elucidated. As shown in Fig. 4A, the increased expression of miR-515-5p urged by circTRRAP knockdown was weakened by the introduction of miR-515-5p inhibitor. Furthermore, circTRRAP knockdown-mediated repression of PCa cell viability, proliferation, migration, and invasion was whittled by downregulation of miR-515-5p (Fig. 4B–F). Additionally, interference of circTRRAP-induced downregulation of MMP2 and MMP9 was mitigated by miR-515-5p deficiency (Fig. 4G). Moreover, the decreased levels of glucose uptake and lactate production in PCa cells mediated by circTRRAP inhibition were impaired after miR-515-5p silencing (Fig. 4H and I). These results uncovered that circTRRAP interacted with miR-515-5p to mediate PCa progression.
Deficiency of miR-515-5p attenuated circTRRAP knockdown-mediated effects on PCa cell malignancy and glycolysis. A miR-515-5p abundance was measured in PCa cells transfected with si-NC, si-circTRRAP#1, si-circTRRAP#1 + inhibitor-NC, or si-circTRRAP#1 + miR-515-5p inhibitor by qRT-PCR. B–F Cell viability, proliferation, migration and invasion were detected in the above cells by MTT, EdU and transwell assays. G PCNA, MMP2, and MMP9 protein levels were detected in the above cells by western blot. H and I The glucose uptake and lactate production of the above cells were measured. *P < 0.05
HOXA1 was a miR-515-5p target
TargetScan predicted that HOXA1 contained the complementary binding sites with miR-515-5p (Fig. 5A). To ascertain the impact of miR-515-5p on HOXA1, a fragment containing the 3ʹUTR of HOXA1 was spliced into the psiCHECK™-2 vector. The reduced luciferase activity was observed in cells co-transfected with the HOXA1 3ʹUTR-WT vector with miR-515-5p mimic (Fig. 5B and C). Moreover, HOXA1 mRNA and protein levels were remarkably repressed by miR-515-5p overexpression in PCa cells (Fig. 5D and E). In addition, the upregulation of HOXA1 mRNA and protein levels was got in PCa tissues (n = 50) compared with that in normal tissues (Fig. 5F and G). Similarly, the abundances of HOXA1 mRNA and protein were also significantly higher in PCa cells than that in RWPE-1 cells (Fig. 5H and I). Besides, the HOXA1 mRNA level in PCa tissues had a negative correlation with miR-515-5p (r = − 0.437, P = 0.0015) (Fig. 5J). Also, circTRRAP silencing led to an overt reduction in the mRNA and protein levels of HOXA1, but miR-515-5p inhibition partly reversed this reduction (Fig. 5K and L). Expectedly, the HOXA1 mRNA level in PCa tissues had a positive correlation with circTRRAP (Fig. 5M). The above results illustrated the targeting relationship between miR-515-5p and HOXA1 and that circTRRAP interacted with miR-515-5p to mediate HOXA1 expression.
HOXA1 was a miR-515-5p target. A The binding sites of miR-515-5p with HOXA1. B and C Luciferase activity was detected in PCa cells transfected with HOXA1 3ʹUTR-WT or HOXA1 3ʹUTR-MUT and miR-515-5p or miR-NC. D and E HOXA1 mRNA and protein levels were detected in PCa cells transfected with miR-NC or miR-515-5p. F–I The abundances of HOXA1 mRNA and protein were examined in PCa tissues and cells. J Pearson’s correlation analysis assessed the correlation between the levels of miR-515-5p and HOXA1 mRNA in PCa tissues. K and L HOXA1 mRNA and protein levels were estimated in PCa cells transfected with si-NC, si-circTRRAP#1, si-circTRRAP#1 + inhibitor-NC, or si-circTRRAP#1 + miR-515-5p inhibitor. M Pearson’s correlation analysis determined the correlation between the levels of circTRRAP and HOXA1 mRNA in PCa tissues. *P < 0.05
miR-515-5p targeted HOXA1 to lower PCa cell malignancy and glycolysis
To explore whether HOXA1 is associated with miR-515-5p-mediated effects on PCa cell malignancy and glycolysis, rescue experiments were conducted. The reduced mRNA and protein levels of HOXA1 promoted by miR-515-5p overexpression were whittled by introduction of the pcDNA-HOXA1 plasmid (Fig. 6A and B). Furthermore, miR-515-5p overexpression restrained PCa cell viability and reduced proliferative, migratory and invasive capacities of PCa cells, while HOXA1 overexpression weakened these impacts (Fig. 6C–F). Moreover, the decreased protein levels of PCNA, MMP2 and MMP9 caused by miR-515-5p upregulation were whittled by HOXA1 overexpression (Fig. 6G and H). In addition, miR-515-5p increase-induced reduction of glucose uptake and lactate production was impaired by the overexpression of HOXA1 (Fig. 6I and J). These data indicated that miR-515-5p targeted HOXA1 to inhibit PCa cell malignancy and glycolysis.
miR-515-5p lessened PCa cell malignancy and glycolysis by targeting HOXA1. A and B HOXA1 mRNA and protein levels were estimated in PCa cells transfected with miR-NC, miR-515-5p, miR-515-5p + pcDNA-NC, or miR-515-5p + pcDNA-HOXA1. C–F Cell viability, proliferation, migration and invasion were determined in the above cells. G and H Detection of PCNA, MMP2, and MMP9 protein levels was done in the above cells. I and J Measurement of glucose uptake and lactate production was conducted in the above cells. *P < 0.05
Interference of circTRRAP reduced PCa growth in vivo
To validate the action of circTRRAP in vivo, PC3 cells stably expression of Lenti-sh-NC or Lenti-sh-circTRRAP were constructed and injected into mice. In vivo experiments demonstrated that circTRRAP silencing impaired tumor growth (volume and weight) relative to the control group (Fig. 7A and B). Moreover, tumor sample-derived from mice with circTRRAP silencing had lower levels of circTRRAP, HOXA1, and PCNA, as well as higher levels of miR-515-5p (Fig. 7C and D). IHC analysis also showed that the number of ki-67/MMP2/MMP9-positive cells was lowered in tumor sample-derived from mice with circTRRAP silencing (Fig. 7E). Together, circTRRAP prompted PCa growth in vivo.
Knockdown of circTRRAP decreased PCa growth in vivo. A Tumor volume was measured every 5 days. B The mean weight of tumors in both groups was at the end point. C The abundances of circTRRAP and miR-515-5p were detected in tumors in both groups. D Western blot analyzed HOXA1 and PCNA protein levels in tumors in both groups. E IHC analysis of ki-67, MMP2 and MMP9 protein levels in tumors in both groups. *P < 0.05
Discussion
The unique molecular structure of circRNAs and their stage-specific and cell/tissue-specific expression characteristics make them more likely to be cancer therapeutic targets than linear transcripts [28]. Some scholars have confirmed the important role of some dysregulated circRNAs in PCa progression [29]. However, the molecular mechanism by which most circRNAs play an important role in PCa remains unclear.
A previous study indicated the upregulation of circTRRAP in PCa samples with spinal metastases, and exosomal circTRRAP facilitated cell epithelial-mesenchymal transition and invasion by binding to miR-1 and subsequently increasing MAP3K1 expression in PCa [10]. Our data validated the overexpression of circTRRAP in PCa samples in comparison to matching normal samples. Functional experiments manifested that circTRRAP knockdown lowered PCa cell viability and repressed PCa cell proliferative, migratory, and invasive capacities, as well as weakened tumor growth in mouse xenograft models. These results highlighted the promoting effect of circTRRAP on PCa growth. The novelty of our study lied in the discovery that circTRRAP was involved in glycolysis in PCa cells and circTRRAP mediated PCa cell malignancy and glycolysis by the miR-515-5p/HOXA1 axis.
Cancer cells have the ability to produce energy in an oxygen-independent manner, that is, their metabolic phenotype is preferentially dependent on glycolysis [30]. Targeting glycolysis has great potential to treat tumors based on the dependence of cancer cells on glycolysis to influence tumorigenesis [31]. Herein, circTRRAP silencing reduced glucose uptake and lactate production in PCa cells, manifesting that circTRRAP took part in glycolysis in PCa.
Existing evidence points to the importance of circRNAs in cancer progression by regulating the miRNA/mRNA axis [32]. Currently, circTRRAP had been shown to regulate the miR-1/MAP3K1 axis to participate in PCa progression [10]. Here, we discovered the possible association of circTRRAP with miR-515-5p by bioinformatics analysis and subsequently confirmed their targeting relationship by dual-luciferase reporter assays. miR-515-5p as a suppressor has been exposed in diverse cancers, such as gastric cancer [33] and breast cancer [34]. In PCa, the downregulation of miR-515-5p mediated by circ-0057553 overexpression facilitated cancer cell aerobic glycolysis, invasion, and migration [24]. Furthermore, circ-PAPPA [35] and circ-FOXM1 [36] prompted PCa cell malignancy by sequestering miR-515-5p. Our results displayed the downregulation of miR-515-5p in PCa samples and cells. Downregulation of miR-515-5p abated the suppressive effects of circTRRAP silencing on PCa cell malignancy and glycolysis. All results supported that circTRRAP mediated PCa cell malignancy and glycolysis by interacting with miR-515-5p.
Next, this research probed into the targets of miR-515-5p. Here, Bioinformatics analysis and validation experiments confirmed that HOXA1 was a miR-515-5p target. HOXA1 is a conserved member of a family of homeobox transcription factors that coordinately regulate early developmental patterns, organogenesis, and cell fate [37]. There is increasing evidence that dysregulation of HOXA1 affects tumorigenesis, including gastric cancer [25], glioblastoma [38], and melanoma [39]. In PCa, HOXA1 silencing suppressed tumor growth and metastasis in xenograft models [40]. Moreover, circ_0074032 sequestered miR-198 and subsequently elevated HOXA1 expression, thus leading to prompting PCa progression [41]. Here, HOXA1 had high levels in PCa samples and cells, and HOXA1 upregulation alleviated miR-515-5p mimic-mediated repression of cell glycolysis and malignancy in PCa cells. Additionally, circTRRAP regulated HOXA1 expression by competitively sponging miR-515-5p, highlighting the network mechanism of the circTRRAP/miR-515-5p/HOXA1 axis in PCa cells.
In conclusion, circTRRAP in PCa samples and cells was highly expressed. Furthermore, knockdown of circTRRAP lessened PCa cell malignancy and glycolysis via decreasing HOXA1 expression via sponging and sequestering miR-515-5p. This research disclosed a novel mechanism for PCa progression.
Availability of data and materials
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
References
Attard G, Parker C, Eeles RA, Schroder F, Tomlins SA, Tannock I, Drake CG, de Bono JS (2016) Prostate cancer. Lancet 387(10013):70–82
Howard N, Clementino M, Kim D, Wang L, Verma A, Shi X, Zhang Z, DiPaola RS (2019) New developments in mechanisms of prostate cancer progression. Semin Cancer Biol 57:111–116
Litwin MS, Tan HJ (2017) The diagnosis and treatment of prostate cancer: a review. JAMA 317(24):2532–2542
Ghosh S, Tvsvgk T, Somasundaram V, Deepti MJSJ (2021) The domino effect-treatment of superior vena cava obstruction triggering tumor lysis syndrome: a case report. SciMed J 3(1):44–50
Han B, Chao J, Yao H (2018) Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther 187:31–44
Li J, Sun D, Pu W, Wang J, Peng Y (2020) Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer 6(4):319–336
Taheri M, Najafi S, Basiri A, Hussen BM, Baniahmad A, Jamali E, Ghafouri-Fard S (2021) The role and clinical potentials of circular RNAs in prostate cancer. Front Oncol 11:781414
Zhang Y, Liu F, Feng Y, Xu X, Wang Y, Zhu S, Dong J, Zhao S, Xu B, Feng N (2022) CircRNA circ_0006156 inhibits the metastasis of prostate cancer by blocking the ubiquitination of S100A9. Cancer Gene Ther. https://doi.org/10.1038/s41417-022-00492-z
Wang S, Chao F, Zhang C, Han D, Xu G, Chen G (2022) Circular RNA circPFKP promotes cell proliferation by activating IMPDH2 in prostate cancer. Cancer Lett 524:109–120
Zhang G, Liu Y, Yang J, Wang H, Xing Z (2021) Inhibition of circ_0081234 reduces prostate cancer tumor growth and metastasis via miR-1/MAP3K1 axis. J Gene Med. https://doi.org/10.1002/jgm.3376
Cozar JM, Robles-Fernandez I, Rodriguez-Martinez A, Puche-Sanz I, Vazquez-Alonso F, Lorente JA, Martinez-Gonzalez LJ, Alvarez-Cubero MJ (2019) The role of miRNAs as biomarkers in prostate cancer. Mutat Res 781:165–174
Khorasgani MA, Nejad PM, Bashi MMM, Hedayati MJSJ (2019) Increased expression of miR-377–3p in patients with relapsing remitting multiple sclerosis. SciMed J 1(2):48–54
Fabris L, Ceder Y, Chinnaiyan AM, Jenster GW, Sorensen KD, Tomlins S, Visakorpi T, Calin GA (2016) The potential of MicroRNAs as prostate cancer biomarkers. Eur Urol 70(2):312–322
Ru P, Steele R, Newhall P, Phillips NJ, Toth K, Ray RB (2012) miRNA-29b suppresses prostate cancer metastasis by regulating epithelial-mesenchymal transition signaling. Mol Cancer Ther 11(5):1166–1173
Jin M, Zhang T, Liu C, Badeaux MA, Liu B, Liu R, Jeter C, Chen X, Vlassov AV, Tang DG (2014) miRNA-128 suppresses prostate cancer by inhibiting BMI-1 to inhibit tumor-initiating cells. Cancer Res 74(15):4183–4195
Chao F, Song Z, Wang S, Ma Z, Zhuo Z, Meng T, Xu G, Chen G (2021) Novel circular RNA circSOBP governs amoeboid migration through the regulation of the miR-141-3p/MYPT1/p-MLC2 axis in prostate cancer. Clin Transl Med 11(3):e360
Chen Y, Yang F, Fang E, Xiao W, Mei H, Li H, Li D, Song H, Wang J, Hong M et al (2019) Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ 26(7):1346–1364
Mao Y, Li W, Hua B, Gu X, Pan W, Chen Q, Xu B, Lu C, Wang Z (2020) Circular RNA_PDHX promotes the proliferation and invasion of prostate cancer by sponging MiR-378a-3p. Front Cell Dev Biol 8:602707
Chen W, Cen S, Zhou X, Yang T, Wu K, Zou L, Luo J, Li C, Lv D, Mao X (2020) Circular RNA CircNOLC1, upregulated by NF-KappaB, promotes the progression of prostate cancer via miR-647/PAQR4 axis. Front Cell Dev Biol 8:624764
Deng W, Zhou X, Zhu K, Chen R, Liu X, Chen L, Jiang H, Hu B, Zeng Z, Cheng X et al (2022) Novel circular RNA circ_0086722 drives tumor progression by regulating the miR-339-5p/STAT5A axis in prostate cancer. Cancer Lett 533:215606
Yang Z, Qu CB, Zhang Y, Zhang WF, Wang DD, Gao CC, Ma L, Chen JS, Liu KL, Zheng B et al (2019) Dysregulation of p53-RBM25-mediated circAMOTL1L biogenesis contributes to prostate cancer progression through the circAMOTL1L-miR-193a-5p-Pcdha pathway. Oncogene 38(14):2516–2532
Wu G, Sun Y, Xiang Z, Wang K, Liu B, Xiao G, Niu Y, Wu D, Chang C (2019) Preclinical study using circular RNA 17 and micro RNA 181c–5p to suppress the enzalutamide-resistant prostate cancer progression. Cell Death Dis 10(2):37
Zhang X, Zhou J, Xue D, Li Z, Liu Y, Dong L (2019) MiR-515-5p acts as a tumor suppressor via targeting TRIP13 in prostate cancer. Int J Biol Macromol 129:227–232
Zhang Y, Shi Z, Li Z, Wang X, Zheng P, Li H (2020) Circ_0057553/miR-515-5p regulates prostate cancer cell proliferation, apoptosis, migration, invasion and aerobic glycolysis by targeting YES1. Onco Targets Ther 13:11289–11299
Yuan C, Zhu X, Han Y, Song C, Liu C, Lu S, Zhang M, Yu F, Peng Z, Zhou C (2016) Elevated HOXA1 expression correlates with accelerated tumor cell proliferation and poor prognosis in gastric cancer partly via cyclin D1. J Exp Clin Cancer Res 35:15
Zhang Y, Pan Q, Shao Z (2020) Tumor-suppressive role of microRNA-202-3p in hepatocellular carcinoma through the KDM3A/HOXA1/MEIS3 pathway. Front Cell Dev Biol 8:556004
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4):402–408
Kristensen LS, Hansen TB, Venø MT, Kjems J (2018) Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 37(5):555–565
Greene J, Baird AM, Lim M, Flynn J, McNevin C, Brady L, Sheils O, Gray SG, McDermott R, Finn SP (2021) Differential CircRNA expression signatures may serve as potential novel biomarkers in prostate cancer. Front Cell Dev Biol 9:605686
Ganapathy-Kanniappan S, Geschwind JF (2013) Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer 12:152
Pelicano H, Martin DS, Xu RH, Huang P (2006) Glycolysis inhibition for anticancer treatment. Oncogene 25(34):4633–4646
Khan S, Jha A, Panda AC, Dixit A (2021) Cancer-associated circRNA-miRNA-mRNA regulatory networks: a meta-analysis. Front Mol Biosci 8:671309
Pu Z, Xu M, Yuan X, Xie H, Zhao J (2020) Circular RNA circCUL3 accelerates the warburg effect progression of gastric cancer through regulating the STAT3/HK2 Axis. Mol Ther Nucleic Acids 22:310–318
Qiao K, Ning S, Wan L, Wu H, Wang Q, Zhang X, Xu S, Pang D (2019) LINC00673 is activated by YY1 and promotes the proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo signaling pathway. J Exp Clin Cancer Res CR 38(1):418
Wang G, Zhao H, Duan X, Ren Z (2021) CircRNA pappalysin 1 facilitates prostate cancer development through miR-515-5p/FKBP1A axis. Andrologia 53(11):e14227
Liu GX, Zheng T, Zhang Y, Hao P (2022) CircFOXM1 silencing represses cell proliferation, migration and invasion by regulating miR-515-5p/ADAM10 axis in prostate cancer. Anticancer Drugs 33(1):e573–e583
Tischfield MA, Bosley TM, Salih MA, Alorainy IA, Sener EC, Nester MJ, Oystreck DT, Chan WM, Andrews C, Erickson RP et al (2005) Homozygous HOXA1 mutations disrupt human brainstem, inner ear, cardiovascular and cognitive development. Nat Genet 37(10):1035–1037
Li Q, Dong C, Cui J, Wang Y, Hong X (2018) Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J Exp Clin Cancer Res CR 37(1):265
Wardwell-Ozgo J, Dogruluk T, Gifford A, Zhang Y, Heffernan TP, van Doorn R, Creighton CJ, Chin L, Scott KL (2014) HOXA1 drives melanoma tumor growth and metastasis and elicits an invasion gene expression signature that prognosticates clinical outcome. Oncogene 33(8):1017–1026
Wang H, Liu G, Shen D, Ye H, Huang J, Jiao L, Sun Y (2015) HOXA1 enhances the cell proliferation, invasion and metastasis of prostate cancer cells. Oncol Rep 34(3):1203–1210
Feng C, Wang Q, Deng L, Peng N, Yang M, Wang X (2022) Hsa_circ_0074032 promotes prostate cancer progression through elevating homeobox A1 expression by serving as a microRNA-198 decoy. Andrologia 54(1):e14312
Acknowledgements
Not applicable.
Funding
No funding was received.
Author information
Authors and Affiliations
Contributions
Conceptualization and Methodology: YG and JT; Formal analysis and Data curation: ZJ, GZ and XA; Validation and Investigation: ZL and YG; Writing - original draft preparation and Writing - review and editing: ZL, YG, JT and ZJ. All authors read and approve the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The present study was approved by the ethical review committee of Seventh Medical Center, PLA General Hospital. Written informed consent was obtained from all enrolled patients.
Consent for publication
Patients agree to participate in this work.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, Z., Guan, Y., Teng, J. et al. CircTRRAP (hsa_circ_0081234) participates in prostate cancer progression and glycolysis by HOXA1 via functioning as a miR-515-5p sponge. Appl Biol Chem 65, 58 (2022). https://doi.org/10.1186/s13765-022-00722-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13765-022-00722-w