Abstract
Background
Automated surveillance methods that re-use electronic health record data are considered an attractive alternative to traditional manual surveillance. However, surveillance algorithms need to be thoroughly validated before being implemented in a clinical setting. With semi-automated surveillance patients are classified as low or high probability of having developed infection, and only high probability patients subsequently undergo manual record review. The aim of this study was to externally validate two existing semi-automated surveillance algorithms for deep SSI after colorectal surgery, developed on Spanish and Dutch data, in a Swedish setting.
Methods
The algorithms were validated in 225 randomly selected surgeries from Karolinska University Hospital from the period January 1, 2015 until August 31, 2020. Both algorithms were based on (re)admission and discharge data, mortality, reoperations, radiology orders, and antibiotic prescriptions, while one additionally used microbiology cultures. SSI was based on ECDC definitions. Sensitivity, specificity, positive predictive value, negative predictive value, and workload reduction were assessed compared to manual surveillance.
Results
Both algorithms performed well, yet the algorithm not relying on microbiological culture data had highest sensitivity (97.6, 95%CI: 87.4–99.6), which was comparable to previously published results. The latter algorithm aligned best with clinical practice and would lead to 57% records less to review.
Conclusions
The results highlight the importance of thorough validation before implementation in other clinical settings than in which algorithms were originally developed: the algorithm excluding microbiology cultures had highest sensitivity in this new setting and has the potential to support large-scale semi-automated surveillance of SSI after colorectal surgery.
Similar content being viewed by others
Background
Healthcare-associated infections (HAIs) pose a major burden on the healthcare system, and result in increased morbidity, mortality, prolonged hospital stay, and additional costs [1,2,3]. HAIs yearly affect nearly four million patients in acute care hospitals in Europe and surgical site infections (SSIs) account for around 18% of all HAIs, annually affecting more than 500,000 patients [3]. After colorectal surgery, up to or more than 30% of patients develop an SSI [4].
Continuous surveillance with feedback to healthcare personnel and stakeholders is essential to allocate the required resources and assess the effect of interventions to prevent HAIs. Traditional HAI surveillance is often based on time-consuming and resource-intensive manual review of patient records, which is also prone to subjective interpretation and surveillance bias [5,6,7]. Automated surveillance methods that re-use electronic health record (EHR) data are being developed and considered an attractive alternative to this manual surveillance as it will reduce workload and generates standardised and continuous surveillance results [6, 7]. However, surveillance algorithms need to be thoroughly validated before being implemented in a clinical setting. Their transferability to other countries with different EHR systems and data management than the country of development needs to be assessed before implementation in new settings.
In this study, the aim was to externally validate two existing semi-automated surveillance algorithms for deep SSI after colorectal surgery, developed based on Spanish and Dutch data [8, 9], in a Swedish setting.
Methods
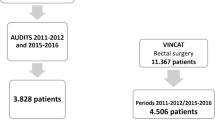
This retrospective study used EHR data from the Karolinska University Hospital (KUH) stored in the 2SPARE (2020 started Stockholm/Sweden Proactive Adverse Events REsearch) database. KUH is a tertiary care academic center with 1,100 beds divided between two hospitals (Huddinge and Solna), which serves the population of Region Stockholm (2.3 million inhabitants). The study was approved by the Regional Ethical Review Board in Stockholm (no. 2018/1030-31).
With semi-automated surveillance patients are divided in low- and high-probability cases where low-probability cases are automatically regarded as no SSI while high-probability cases undergo manual record review to determine SSI status [6]. Two existing semi-automated classification algorithms to assess deep SSI and/or organ/space SSI, from here on together referred to as deep SSI, were validated (Fig. 1):
-
1.
Original classification algorithm of van Rooden et al., i.e., probability of having a deep SSI based on (re)admission and discharge data, mortality, reoperations, radiology orders, antibiotic prescriptions, and microbiology cultures [8];
-
2.
Adapted classification algorithm according to Verberk et al., i.e., probability of having a deep SSI based on the original classification algorithm without the microbiology component [9].
The algorithms’ performance was assessed in a validation cohort of 225 colorectal surgeries selected via simple random sampling from the 2,675 performed surgeries in the period January 1, 2015 until August 31, 2020 (Fig. 1). Patient and surgery characteristics were recorded. The outcome of interest and gold standard was deep SSI versus no deep SSI (no SSI or only superficial SSI) within 30 days after colorectal surgery as annotated by two experienced infection control practitioners (ICPs) according to the European Centre for Disease Prevention and Control (ECDC) SSI definitions and guidelines [10]. Twenty cases were reviewed in overlap resulting in almost perfect agreement (95%) with a Cohen’s kappa of 0.87 for SSI classification. Both IPCs were blinded for the algorithm results.
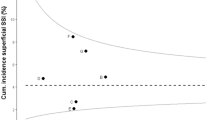
Flow chart of study and flow diagram of classification algorithms for deep surgical site infection. (SSI: surgical site infection; ECDC: European Centre for Disease Prevention and Control. Admissions: Length of stay ≥ 14 days or 1 readmission to original department or in-hospital mortality within follow-up (FU) time (= 45 days after surgery). Reoperations: Any reoperation by original surgery specialty within FU time. Radiology: ≥1 orders for CT scan within FU time. Antibiotics: ≥3 consecutive days of antibiotics (ATC J01) within FU time, starting after day 1. Microbiology: ≥1 culture taken from relevant body sites within FU time, excluding cultures taken on any day prior to day 1. Original classification algorithm: Figure originally published in van Rooden et al. [8], adapted and used with permission. Adapted classification algorithm: Figure originally published in Verberk et al. [9], adapted and used with permission)
Data acquisition, management and analysis were performed using R statistical software (version 3.6.1) and Python (version 3.7), and in accordance with current regulations concerning privacy and ethics. For algorithm performance, the sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were assessed. The confidence interval (CI) for these estimates were calculated using the asymptotic variance with Wilson score method.
Results
Within the validation cohort the median age was 66 year (IQR 55–75), 48.9% (n = 110) were female and the median body mass index was 25.6 (IQR 22.3–29.4). Most surgeries were primary (63.6%, n = 143), open procedures (77.3%, n = 174) and related to malignancy (76.9%, n = 173). The surgeries had a median duration of 316 min (IQR 206–427) and in 41.8% (n = 94) of surgeries a stoma was created. In 36.5% (n = 82) of surgeries the patient had an ASA class ≥ 3 and 24.0% (n = 54) of surgeries had a contaminated or dirty-infected wound classification. Within 30-days after surgery 18.2% (n = 41) of patients developed a deep SSI and 2.2% (n = 5) of the patients died.
Both semi-automated algorithms were applied to the validation cohort. Ordered radiology (47.6%, n = 107) and receiving antibiotic therapy for ≥ 3 consecutive days (41.3%, n = 93) were the most common components. In 38.7% (n = 87) of surgeries ≥ 14 days length-of-stay, readmission and/or mortality was present and in 31.1% (n = 70) a microbiological culture was taken. A reoperation (17.8%, n = 40) was the least common component.
Of the 41 patients with a deep SSI, 34 were classified as high probability by the original algorithm (sensitivity 82.9, 95%CI: 68.7–91.5) while 40 were classified as high probability by the adapted algorithm (sensitivity 97.6, 95%CI: 87.4–99.6) (Table 1). The six deep SSI cases only missed by the original algorithm all scored 2–3 components, but were lacking the microbiology component (in other words, no cultures were obtained). The one deep SSI case missed by both algorithms had none of the algorithm components: this deep SSI was manually assessed based on a clinical note describing pus from the rectal stump. The algorithms would lead to workload reduction for manual surveillance of 74% and 57%, respectively.
Discussion
External validation of two existing semi-automated surveillance algorithms after colorectal surgery in EHR data of a Swedish academic hospital center showed that the adapted classification algorithm, indicating high SSI probability based on (re)admission and discharge dates, mortality, reoperations, radiology orders and antibiotic prescriptions, performed best and outperformed the original classification algorithm which also included microbiology culture data. Only one mild case of deep SSI was missed, which would be hard to detect using only structured data as it was assessed merely based on a free-text medical note.
The original algorithm using microbiology culture data was less useful for this specific clinical setting as microbiological culture practices are not standard in suspected deep SSIs after colorectal surgery. However, this might be different in other settings and highlights the importance of thoroughly validation before implementation and pre-emptively investigating if algorithms correspond with clinical practices [9]. It should be emphasised that checking the algorithm periodically against clinical practice after implementation also remains important [6, 7, 9].
Although the original semi-automated classification algorithm was developed based on Spanish and Dutch data, and validated and adapted in the Netherlands, also within Sweden the sensitivity was high and comparable with previous results [8, 9]. These results confirm the potential of large-scale implementation of both, where especially the adapted algorithm, without microbiology data, has demonstrated robustness within Europe.
Strengths of our study were the independent external validation with the extensive availability of EHR data through which the performance of epidemiological surveillance using real-world, real-time data could be mimicked. Limitations were the usage of data from only one center in Sweden, focus of algorithms on only deep SSI and that absence of active post-discharge surveillance of SSI could result in missed SSI.
In conclusion, the results from this study in Sweden, in conjunction with previous studies in the Netherlands and Spain, indicate that a classification algorithm based on (re)admission and discharge dates, mortality, reoperations, radiology orders and antibiotic prescriptions, could be widely implemented for semi-automated surveillance of SSI after colorectal surgery.
Data availability
The datasets generated and/or analysed in this study are not publicly available due to ethical limitations with sharing patient information, but are available from the corresponding author on reasonable request.
Abbreviations
- CI:
-
confidence interval
- ECDC:
-
European Centre for Disease Prevention and Control
- EHR:
-
electronic health record
- HAI:
-
healthcare-associated infection
- ICP:
-
infection control practitioner
- IQR:
-
interquartile range
- KUH:
-
Karolinska University Hospital
- NPV:
-
negative predictive value
- PPV:
-
positive predictive value
- SSI:
-
surgical site infection
References
Umscheid CA, Mitchell MD, Doshi JA, Agarwal R, Williams K, Brennan PJ. Estimating the proportion of healthcare-associated infections that are reasonably preventable and the related mortality and costs. Infect Control Hosp Epidemiol. 2011;32(2):101–14.
Cassini A, Plachouras D, Eckmanns T, Abu Sin M, Blank H-P, Ducomble T et al. Burden of six healthcare-associated infections on European population health: Estimating incidence-based disability-adjusted life years through a population prevalence-based modelling study. PLoS Med. 2016;13(10):e1002150.
Suetens C, Latour K, Kärki T, Ricchizzi E, Kinross P, Moro ML, et al. Prevalence of healthcare-associated infections, estimated incidence and composite antimicrobial resistance index in acute care hospitals and long-term care facilities: results from two european point prevalence surveys, 2016 to 2017. Eurosurveillance. 2018;23(46):pii=1800516.
Zywot A, Lau CSM, Stephen Fletcher H, Paul S. Bundles prevent surgical site infections after colorectal surgery: Meta-analysis and systematic review. J Gastrointest Surg. 2017;21(11):1915–30.
Mitchell BG, Hall L, Halton K, MacBeth D, Gardner A. Time spent by infection control professionals undertaking healthcare associated infection surveillance: A multi-centred cross sectional study. Infect Dis Health. 2016;21(1):36–40.
Sips ME, Bonten MJM, van Mourik MSM. Automated surveillance of healthcare-associated infections: state of the art. Curr Opin Infect Dis. 2017;30(4):425–31.
van Mourik MSM, van Rooden SM, Abbas M, Aspevall O, Astagneau P, Bonten MJM, et al. PRAISE: providing a roadmap for automated infection surveillance in Europe. Clin Microbiol Infect. 2021;27(Suppl 1):3–19.
van Rooden SM, Tacconelli E, Pujol M, Gomila A, Kluytmans JAJW, Romme J, et al. A framework to develop semiautomated surveillance of surgical site infections: An international multicenter study. Infect Control Hosp Epidemiol. 2020;41(2):194–201.
Verberk JDM, van der Kooi TII, Hetem DJ, Oostdam NEWM, Noordergraaf M, de Greeff SC et al. Semiautomated surveillance of deep surgical site infections after colorectal surgeries: A multicenter external validation of two surveillance algorithms. Infect Control Hosp Epidemiol. 2023;44(4):616–23.
European Centre for Disease Prevention and Control. Surveillance of surgical site infections and prevention indicators in european hospitals: HAI-Net SSI protocol, version 2.2. Stockholm: ECDC; 2017.
Acknowledgements
We would like to thank Anna Frej for her help with the manual annotation of medical records.
Funding
Open access funding provided by Karolinska Institute. The work was supported by Sweden´s Innovation Agency (Vinnova grant 2018–03350). PN was supported by Region Stockholm (clinical research appointment).
Author information
Authors and Affiliations
Contributions
SvdW, JV, MvM and PN conceptualised this study. CB gave valuable input for adapting the algorithms to the Swedish context. SvdW extracted, annotated, and analysed the data. All authors interpreted the data. MvM and PN supervised the study. SvdW and JV prepared and drafted the manuscript with the input of all other authors. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Regional Ethical Review Board in Stockholm (no. 2018/1030-31).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
van der Werff, S.D., Verberk, J.D., Buchli, C. et al. External validation of semi-automated surveillance algorithms for deep surgical site infections after colorectal surgery in an independent country. Antimicrob Resist Infect Control 12, 96 (2023). https://doi.org/10.1186/s13756-023-01288-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-023-01288-y