Abstract
Background
To assess the ability of procalcitonin (PCT) to distinguish between bacterial and nonbacterial causes of patients with severe acute exacerbation of COPD (AECOPD) admitted to the ICU, we conducted a retrospective analysis of two prospective studies including 375 patients with severe AECOPD with suspected lower respiratory tract infections. PCT levels were sequentially assessed at the time of inclusion, 6 h after and at day 1, using a sensitive immunoassay. The patients were classified according to the presence of a documented bacterial infection (including bacterial and viral coinfection) (BAC + group), or the absence of a documented bacterial infection (i.e., a documented viral infection alone or absence of a documented pathogen) (BAC- group). The accuracy of PCT levels in predicting bacterial infection (BAC + group) vs no bacterial infection (BAC- group) at different time points was evaluated by receiver operating characteristic (ROC) analysis.
Results
Regarding the entire cohort (n = 375), at any time, the PCT levels significantly differed between groups (Kruskal–Wallis test, p < 0.001). A pairwise comparison showed that PCT levels were significantly higher in patients with bacterial infection (n = 94) than in patients without documented pathogens (n = 218) (p < 0.001). No significant difference was observed between patients with bacterial and viral infection (n = 63). For example, the median PCT-H0 levels were 0.64 ng/ml [0.22–0.87] in the bacterial group vs 0.24 ng/ml [0.15–0.37] in the viral group and 0.16 ng/mL [0.11–0.22] in the group without documented pathogens. With a c-index of 0.64 (95% CI; 0.58–0.71) at H0, 0.64 [95% CI 0.57–0.70] at H6 and 0.63 (95% CI; 0.56–0.69) at H24, PCT had a low accuracy for predicting bacterial infection (BAC + group).
Conclusion
Despite higher PCT levels in severe AECOPD caused by bacterial infection, PCT had a poor accuracy to distinguish between bacterial and nonbacterial infection. Procalcitonin might not be sufficient as a standalone marker for initiating antibiotic treatment in this setting.
Similar content being viewed by others
Background
Procalcitonin (PCT) is considered useful for determining the likelihood that patients will develop bacterial infections, and several large randomized controlled clinical trials have investigated the ability of a PCT-based strategy to safely reduce antibiotic exposure in noncritically ill patients with lower respiratory tract infections [1,2,3,4,5,6,7]. Interestingly, two recent meta-analyses [8, 9], focusing on PCT-guided antibiotics in a mixed population of AECOPD (i.e., in the ICU and not in the ICU), suggested that a PCT-guided antibiotic strategy reduced antibiotic prescriptions compared with standard management without affecting clinical outcomes such as treatment failure, length of hospitalization and rate of re-exacerbation or overall mortality. These results suggested that PCT could be a useful biomarker to guide antibiotic therapy in this setting. However, due to the methodological limitations and the small overall study population, the authors underlined that the quality of the available evidence was considered low to moderate.
In addition, the safety of PCT-based strategies in critically ill medical patients is not clear. Despite PCT-based algorithms appear safe and reduce antibiotic exposure in critically ill patients [10,11,12,13,14,15,16,17,18], only few trials [11, 12] were designed to assess the impact on mortality. These studies reported contradictory results. In addition, in critically ill patients, the results of different meta-analyses assessing the effect of PCT-guided therapy on mortality are inconsistent [19,20,21,22].
Recently, a large multicenter randomized controlled clinical study from our group among AECOPD patients admitted in ICU showed that the use of PCT was not non-inferior to standard of care regarding 3-month mortality. Moreover, the PCT-guided strategy was significantly worse for patients not on antibiotics at baseline [23].
To explore this disappointing result, we hypothesized that PCT could fail to distinguish between bacterial and nonbacterial causes of severe AECOPD. Therefore, we aimed to assess the ability of PCT to distinguish between bacterial and nonbacterial causes of severe AECOPD, using all the data sets we previously published [23,24,25].
Methods
Patients
We conducted a retrospective analysis from two data sets of prospective studies (i.e., the PROCALCIVIR study, an observational cohort [24, 25] and the BPCTrea study, a randomized controlled trial [23]) including patients with severe AECOPD with suspected lower respiratory tract infections (detailed information provided in the Additional file 1: Appendix). The detailed study designs and main results have been published previously [23,24,25]. Briefly, a PROCALCIVIR study was conducted between September 2005 and September 2006 in one ICU in France. The BPCTrea study was conducted between October 2010 and March 2016 in 11 ICUs in France. PROCALCIVIR and BPCTrea studies allowed for antibiotics at the time of inclusion. These studies were conducted according to the principles of the Declaration of Helsinki.
Procedures
Circulating PCT levels were sequentially assessed at inclusion (PCT-H0), at 6 h after inclusion (PCT-H6), and on day 1 after inclusion (PCT-H24). PCT levels were measured by Elecsys BRAHMS PCT immunoassay (Roche Diagnostics GmbH, Mannheim, Germany) using a Cobas e411 analyzer, according to the manufacturer’s instructions.
In both, the PROCALCIVIR and BPCTrea studies, the microbiological investigation was encouraged, but left to the discretion of the attending physicians, according to the usual practice in each center.
Patients were classified according to the presence of a documented bacterial infection (including bacterial and viral coinfection) (BAC + group), or absence of a documented bacterial infection (i.e., a documented viral infection alone or absence of documented pathogens) (BAC− group).
Statistical analysis
Data are expressed as the mean ± SD or median (interquartile range [IQR]) and percentage, depending on the type of variable of interest. PCT levels were compared between the three groups (i,e., documented bacterial infection, documented viral infection alone and group without documented pathogens) using the Kruskal–Wallis test. In addition, a pairwise comparison between groups was performed using the Wilcoxon test. The discriminative ability of PCT in predicting documented bacterial infection (BAC +) vs nondocumented bacterial infection (BAC−) was evaluated by the receiver operating characteristic (ROC) analysis at each time point (i.e., H0, H6 and H24 after inclusion) in the overall cohort and in predefined subgroups (i.e., AECOPD with and without pneumonia and patients with or without antibiotics at inclusion). The area under the curve (AUC) and 95% confidence intervals are provided. Prism 8.3.1 (GraphPad software, LLC, San Diego, USA) was used for the data analysis. All tests were 2-sided, and a p-value < 0.05 was considered statistically significant.
Results
Patient characteristics
The baseline characteristics of the two studied cohorts are shown Table 1. Of 375 included patients, 94 (25%) patients had a documented bacterial infection (BAC + group) (including 76 (20%) patients with bacterial infection alone and 18 (5%) patients with bacterial and viral coinfection). Two hundred eighty one patients (75%) had nondocumented bacterial infection (BAC− group) (including 63 (17%) patients with documented viral infection alone and 218 patients without documented pathogens). Overall, 159 patients had severe AECOPD with pneumonia, and 202 patients received antibiotics at the time of inclusion.
Microbiological findings
Lower respiratory samples were taken from 268 (71%) patients, including sputum (n = 176), tracheobronchial aspirate (n = 35), bronchoalveolar lavage (n = 12), and distal protected specimens (n = 73). In addition, blood culture was performed in 93 (25%) patients. Additionally, serological diagnosis for antibodies to Legionella pneumophila or detection of Legionella pneumophila serogroup 1 urinary antigen test was performed in 297 (79%) patients. A polymerase chain reaction (PCR) assay for viral study was performed in 258 (69%) patients. The test included the following viruses: V. influenza A/H1/H3/H1N1v/B, parainfluenza 1, 2, 3, 4, VRS A/B, metapneumovirus, entero-rhinovirus, coronavirus OC43/229E/NL63/HKU1/MERS, adenovirus and bocavirus. Mycoplasma pneumoniae, Legionella pneumophila, Bordetella pertussis, and Chlamydophila pneumoniae were also detected by PCR assay. The most frequently isolated bacteria were Haemophilus influenza (n = 23), Pseudomonas spp. (n = 16), Streptococcus pneumonia (n = 8), Streptococcus spp. (n = 6) and Staphylococcus aureus (n = 6). The most frequently isolated viruses were rhinovirus (n = 23), syncytial respiratory virus (n = 16), parainfluenzae virus (n = 10), coronavirus (n = 9) and influenzae A/B virus (n = 9/1).
PCT levels
The circulating PCT levels were sequentially assessed at inclusion (PCT-H0) in 355 (95%) patients, at six hours after inclusion (PCT-H6) in 341 (91%) patients, and on day 1 after inclusion (PCT-H24) in 331 (89%) patients. The PCT levels of the documented infection group are shown Fig. 1a–c. At any time, PCT levels significantly differed between groups (Kruskal–Wallis test, p < 0.001). Using a Wilcoxon test, the PCT levels were significantly higher among patients with bacterial infection (n = 94) than among patients without documented pathogens (n = 218) (p < 0.001) but no significant difference was observed between patients with bacterial infection and patients with viral infection (n = 63). The median PCT-H0 levels were 0.64 ng/mL [0.22–0.87] in the bacterial group vs 0.24 ng/mL [0.15–0.37] in the viral group and 0.16 ng/mL [0.11–0.22] in the group without documented pathogens. PCT-H6 levels were 0.76 ng/mL [0.22–1.11] in the bacterial group vs 0.27 ng/mL [0.20–0.45] in the viral group and 0.18 ng/mL [0.12–0.27] in the group without documented pathogens. PCT-H24 levels were 0.59 ng/mL [0.20–1.17] in the bacterial group vs 0.26 ng/mL [0.16–0.40] in the viral group and 0.18 ng/mL [0.13–0.26] in the group without documented pathogens.
We conducted subgroup analyses among patients with (ATB+) and without (ATB−) antibiotics and with (PNP+) or without (PNP−) pneumonia at the time of inclusion. In the subgroup of patients with antibiotics, PCT levels significantly differed between groups. PCT levels were significantly higher in patients with documented bacterial infection compared to the other groups (detailed information regarding each group may be found in the Additional file 1: Appendix). No difference in PCT levels was observed in the subgroup of patients without antibiotics at the time of inclusion (detailed information regarding each group may be found in the Additional file 1: Appendix). In the subgroup with pneumonia, we observed no difference in PCT levels between the different groups (detailed information regarding each group may be found in the Additional file 1: Appendix). In contrast, in the subgroup of patients without pneumonia, PCT levels significantly differed between groups. PCT levels were significantly higher in patients with documented infection (i.e., bacterial and viral groups) compared to the group without documented pathogens (detailed information regarding each group may be found in the Additional file 1: Appendix).
Ability of PCT to distinguish between bacterial (BAC + group) and nonbacterial infections (BAC−group)
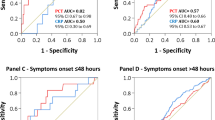
The ROC curves at any time (i.e., H0, H6 and H24 after inclusion) for the prediction of documented bacterial infection (including bacterial and viral coinfection) (BAC + group) vs nondocumented bacterial infection (i.e., documented viral infection alone or absence documented pathogens) (BAC− group) for the PCT levels are shown Fig. 2. With a c-index of 0.64 (95% CI 0.58–0.71) at H0, 0.64 [95% CI 0.57–0.70] at H6 and 0.63 (95% CI 0.56–0.69) at H24, PCT had a low accuracy for predicting bacterial infection.
Receiver operating characteristic (ROC) curves at any time (i.e., H0, H6 and H24 after inclusion) for the prediction of documented bacterial infection (including bacterial and viral coinfection) (BAC + group) vs nondocumented bacterial infection (i.e., documented viral infection alone or absence of documented pathogen) (BAC− group) for the PCT levels
Similar results were observed in subgroups of patients with (ATB+) and without (ATB−) antibiotics and with (PNP+) or without (PNP−) pneumonia at the time of inclusion (detailed information regarding each group may be found in the Additional file 1: Appendix).
Discussion
In this large cohort of patients with severe AECOPD admitted to the ICU, PCT had a low ability to discriminate those patients with and without bacterial documented infection in the overall cohort and in all predefined subgroups (i.e., patients with and without pneumonia and patients with or without antibiotics at the time of inclusion). This poor performance of PCT may contribute to explain the excess mortality observed in the subgroup of patients without antibiotics at baseline in the BPCTrea trial [23]. This result suggests that a PCT-based strategy to initiate antibiotic treatment should be considered with cautious in this setting.
To our knowledge, we provided the largest cohort ever assembled investigating the ability of PCT to predict bacterial infection in a homogenous population of critically ill patients with severe AECOPD and suspected lower respiratory tract infection who need noninvasive or invasive mechanical ventilation. PCT failed to predict bacterial infection in the overall cohort and all predefined subgroups. This result contrasts with previous meta-analyses reporting the good diagnostic accuracy of PCT among mixed patients hospitalized for suspected bacterial infections (including patients admitted to the ICU) both to differentiate bacterial infections from viral infections and to differentiate bacterial infections from other noninfective causes of systemic inflammation [26,27,28,29]. However, although PCT could be considered an accurate marker for diagnosing bacterial infection, the result of this test needs to be interpreted with caution and might not be sufficient as a standalone marker for initiating antibiotic treatment, specifically in ICU patients [27, 30]. This result is supported by a recent prospective multicenter trial in ICU patients addressing a large panel of circulating biomarkers previously tested in a sepsis setting to differentiate sepsis from nonseptic SIRS [31]. In that study [31], no biomarker, alone or in combination, was able to detect infection. With a ROC-AUC of 0.55 [0.47–0.62], PCT poorly discriminated sepsis from nonseptic SIRS [31], a result consistent with those reported here. In addition, a recent meta-analysis [8] focusing on the potential of PCT in predicting bacterial exacerbation in severe COPD exacerbation, reported an area under the receiver operator characteristic curve of 0.77 [0.73–0.80], indicating moderate accuracy. A subgroup analysis revealed that the pooled sensitivity and specificity of PCT for patients admitted in ICU was lower than for other patients.
Algorithms using PCT in critically ill patients suspected of bacterial infection can lead to two opposite clinical decisions: (i) initiate antibiotics; (ii) discontinue antibiotics. Today, a prompt empirical antibiotherapy remains strongly recommended in critical patients suspected of sepsis. The use of procalcitonin algorithms may expose to the risk to delay antibiotic treatment and therefore to increase mortality. Despite previous randomized trials [10,11,12,13,14,15,16,17,18] reporting that PCT-based algorithm appears safe and reduces antibiotic exposure in critically ill patients, trials specially designed to demonstrate the noninferiority of PCT-guided strategies with respect to mortality in the ICU are scarce [11, 12, 23] and inconsistent. Differences in the PCT-guided antibiotic algorithms may explain this heterogeneity. In procalcitonin algorithms providing rules for the initiation, continuation and discontinuation of antibiotic treatment [11, 23], one study [11] reported that 60-day mortality was non-inferior in the experimental group (non-inferiority margin of 10% almost reached) compared to the standard of care group, while our group [23] failed to demonstrate noninferiority with respect to 3-month mortality (non-inferiority margin of 12% exceeded). In one study [12] focusing only on the de-escalation of antibiotic therapy, 28-day and 1-year mortality were significantly lower in the PCT group than in the control group and the non-inferiority margin was 8%. These results suggest that PCT might be more useful for stopping than initiating antibiotics in critically ill patients.
Our large pooled data analysis supports the recommendation that PCT alone should not replace clinical decision for antibiotic initiation, in particular among severe AECOPD. In addition, any delay in antibiotic prescription in such situation could lead to poorer outcome. In this line, a Cochrane meta-analysis [32] concluded that antibiotics reduced the risk of treatment failure in patients with severe AECOPD hospitalized in the ICU and reduced mortality. One explanation could be that antibiotics prevent the risk of bacterial infection due to an impaired phagocytosis of alveolar macrophage in COPD patient. Interestingly, the impairment of phagocytosis could be both mediated by virus infections such as human rhinovirus and related to disease severity [33]. Nevertheless, further studies are needed to assess the usefulness and safety of PCT to withhold or stop antibiotics in critically ill AECOPD patients.
Several limitations of the study warrant discussion. First, this study was a retrospective analysis of two different prospective cohorts [23,24,25]. However, a separate analysis of each cohort showed similar results (data not shown) indicating homogenous data among the two studies. Second, we chose to include in the BAC + group all patients in whom a positive bacterial culture of respiratory tract samples was observed. However, there is no absolute “gold standard” for the diagnosis of bacterial infection in AECOPD since a bacterial colonization is frequently detected. Therefore, a risk of misclassification is not totally excluded. This limitation is not specific to our study. Third, despite a large microbiological investigation, we reported a high percentage of patients without documented pathogen. This finding might be explained by a relatively high proportion of patients with antibiotic at ICU inclusion. In contrast, the proportion of positive PCR results is consistent with recent large cohort of AECOPD requiring hospital admission [34]. Fourth, considering the number of documented bacterial and viral infection, we cannot exclude a lack of power of the study. Fifth, antibiotic treatment before the inclusion of some patients may have affected PCT levels. However, previous reports showed that PCT levels were similar among patients pretreated and not pretreated with antibiotics [3, 4, 23,24,25]. Moreover, higher PCT values were observed for the subgroup of patients with antibiotics in the documented bacterial group (see the Additional file 1: Appendix).
Conclusion
In this study, PCT predicted bacterial infection with poor accuracy in patients with severe AECOPDs admitted in ICU. Therefore, a PCT-based strategy to initiate antibiotic treatment should be considered with cautious in this setting. Further studies are needed to assess the usefulness and safety of PCT to withhold or stop antibiotics in critically ill AECOPD patients.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AECOPD:
-
Acute exacerbation of chronic obstructive pulmonary disease
- AUC:
-
Area under the curve
- ICU:
-
Intensive care unit
- IQR:
-
Interquartile range
- PCT:
-
Procalcitonin
- ROC:
-
Receiver operating characteristic
References
Christ-Crain M, Jaccard-Stolz D, Bingisser R, Gencay MM, Huber PR, Tamm M, et al. Effect of procalcitonin-guided treatment on antibiotic use and outcome in lower respiratory tract infections: cluster-randomised, single-blinded intervention trial. Lancet. 2004;363:600–7. https://doi.org/10.1016/s0140-6736(04)15591-8.
Schuetz P, Christ-Crain M, Thomann R, Falconnier C, Wolbers M, Widmer I, et al. Effect of procalcitonin-based guidelines vs standard guidelines on antibiotic use in lower respiratory tract infections: the ProHOSP randomized controlled trial. JAMA. 2009;302:1059–66. https://doi.org/10.1001/jama.2009.1297.
Christ-Crain M, Stolz D, Bingisser R, Müller C, Miedinger D, Huber PR, et al. Procalcitonin guidance of antibiotic therapy in community-acquired pneumonia: a randomized trial. Am J Respir Crit Care Med. 2006;174:84–93. https://doi.org/10.1164/rccm.200512-1922OC.
Stolz D, Christ-Crain M, Bingisser R, Leuppi J, Miedinger D, Müller C, et al. Antibiotic treatment of exacerbations of COPD: a randomized, controlled trial comparing procalcitonin-guidance with standard therapy. Chest. 2007;131:9–19. https://doi.org/10.1378/chest.06-1500.
Kristoffersen KB, Søgaard OS, Wejse C, Black FT, Greve T, Tarp B. Antibiotic treatment interruption of suspected lower respiratory tract infections based on a single procalcitonin measurement at hospital admission–a randomized trial . Clin Microbiol Infect. 2009;15:481–7. https://doi.org/10.1111/j.1469-0691.2009.02709.x.
Long W, Deng X, Zhang Y, Lu G, Xie J, Tang J, et al. Procalcitonin guidance for reduction of antibiotic use in low-risk outpatients with community-acquired pneumonia. Respirology. 2011;16:819–24. https://doi.org/10.1111/j.1440-1843.2011.01978.x.
Corti C, Fally M, Fabricius-Bjerre A, Mortensen K, Jensen BN, Andreassen HF, et al. Point-of-care procalcitonin test to reduce antibiotic exposure in patients hospitalized with acute exacerbation of COPD. Int J Chron Obstruct Pulmon Dis. 2016;11:1381–9. https://doi.org/10.2147/copd.s104051.
Ni W, Bao J, Yang D, Xi W, Wang K, Xu Y, et al. Potential of serum procalcitonin in predicting bacterial exacerbation and guiding antibiotic administration in severe COPD exacerbations: a systematic review and meta-analysis. Infect Dis . 2019;51:639–50. https://doi.org/10.1080/23744235.2019.1644456.
Li Z, Yuan X, Yu L, Wang B, Gao F, Ma J. Procalcitonin-guided antibiotic therapy in acute exacerbation of chronic obstructive pulmonary disease: an updated meta-analysis. Medicine (Baltimore). 2019;98:e16775. https://doi.org/10.1097/md.0000000000016775.
Nobre V, Harbarth S, Graf JD, Rohner P, Pugin J. Use of procalcitonin to shorten antibiotic treatment duration in septic patients: a randomized trial. Am J Respir Crit Care Med. 2008;177:498–505. https://doi.org/10.1164/rccm.200708-1238OC.
Bouadma L, Luyt CE, Tubach F, Cracco C, Alvarez A, Schwebel C, et al. Use of procalcitonin to reduce patients’ exposure to antibiotics in intensive care units (PRORATA trial): a multicentre randomised controlled trial. Lancet. 2010;375:463–74. https://doi.org/10.1016/s0140-6736(09)61879-1.
De Jong E, Van Oers JA, Beishuizen A, Vos P, Vermeijden WJ, Haas LE, et al. Efficacy and safety of procalcitonin guidance in reducing the duration of antibiotic treatment in critically ill patients: a randomised, controlled, open-label trial. Lancet Infect Dis. 2016;16:819–27. https://doi.org/10.1016/s1473-3099(16)00053-0.
Stolz D, Smyrnios N, Eggimann P, Pargger H, Thakkar N, Siegemund M, et al. Procalcitonin for reduced antibiotic exposure in ventilator-associated pneumonia: a randomised study. Eur Respir J. 2009;34:1364–75. https://doi.org/10.1183/09031936.00053209.
Bloos F, Trips E, Nierhaus A, Briegel J, Heyland DK, Jaschinski U, et al. Effect of sodium selenite administration and procalcitonin-guided therapy on mortality in patients with severe sepsis or septic shock: a randomized clinical trial. JAMA Intern Med. 2016;176:1266–76. https://doi.org/10.1001/jamainternmed.2016.2514.
Layios N, Lambermont B, Canivet JL, Morimont P, Preiser JC, Garweg C, et al. Procalcitonin usefulness for the initiation of antibiotic treatment in intensive care unit patients. Crit Care Med. 2012;40:2304–9. https://doi.org/10.1097/CCM.0b013e318251517a.
Shehabi Y, Sterba M, Garrett PM, Rachakonda KS, Stephens D, Harrigan P, et al. Procalcitonin algorithm in critically ill adults with undifferentiated infection or suspected sepsis. A randomized controlled trial. Am J Respir Crit Care Med. 2014;190:1102–10. https://doi.org/10.1164/rccm.201408-1483OC.
Hochreiter M, Köhler T, Schweiger AM, Keck FS, Bein B, Von Spiegel T, et al. Procalcitonin to guide duration of antibiotic therapy in intensive care patients: a randomized prospective controlled trial. Crit Care. 2009;13:R83. https://doi.org/10.1186/cc7903.
Schroeder S, Hochreiter M, Koehler T, Schweiger AM, Bein B, Keck FS, et al. Procalcitonin (PCT)-guided algorithm reduces length of antibiotic treatment in surgical intensive care patients with severe sepsis: results of a prospective randomized study. Langenbecks Arch Surg. 2009;394:221–6. https://doi.org/10.1007/s00423-008-0432-1.
Heyland DK, Johnson AP, Reynolds SC, Muscedere J. Procalcitonin for reduced antibiotic exposure in the critical care setting: a systematic review and an economic evaluation. Crit Care Med. 2011;39:1792–9. https://doi.org/10.1097/CCM.0b013e31821201a5.
Prkno A, Wacker C, Brunkhorst FM, Schlattmann P. Procalcitonin-guided therapy in intensive care unit patients with severe sepsis and septic shock–a systematic review and meta-analysis. Crit Care. 2013;17:R291. https://doi.org/10.1186/cc13157.
Schuetz P, Wirz Y, Sager R, Christ-Crain M, Stolz D, Tamm M, et al. Effect of procalcitonin-guided antibiotic treatment on mortality in acute respiratory infections: a patient level meta-analysis. Lancet Infect Dis. 2018;18:95–107. https://doi.org/10.1016/s1473-3099(17)30592-3.
Huang HB, Peng JM, Weng L, Wang CY, Jiang W, Du B. Procalcitonin-guided antibiotic therapy in intensive care unit patients: a systematic review and meta-analysis. Ann Intensive Care. 2017;7:114. https://doi.org/10.1186/s13613-017-0338-6.
Daubin C, Valette X, Thiollière F, Mira JP, Hazera P, Annane D, et al. Procalcitonin algorithm to guide initial antibiotic therapy in acute exacerbations of COPD admitted to the ICU: a randomized multicenter study. Intensive Care Med. 2018;44:428–37. https://doi.org/10.1007/s00134-018-5141-9.
Daubin C, Parienti JJ, Fradin S, Vabret A, Ramakers M, Terzi N, et al. Procalcitonin levels and bacterial aetiology among COPD patients admitted to the ICU with severe pneumonia: a prospective cohort study. BMC Infect Dis. 2009;9:157. https://doi.org/10.1186/1471-2334-9-157.
Daubin C, Parienti JJ, Vabret A, Ramakers M, Fradin S, Terzi N, et al. Procalcitonin levels in acute exacerbation of COPD admitted in ICU: a prospective cohort study. BMC Infect Dis. 2008. https://doi.org/10.1186/1471-2334-8-145.
Simon L, Gauvin F, Amre DK, Saint-Louis P, Lacroix J. Serum procalcitonin and C-reactive protein levels as markers of bacterial infection: a systematic review and meta-analysis. Clin Infect Dis. 2004;39:206–17. https://doi.org/10.1086/421997.
Wacker C, Prkno A, Brunkhorst FM, Schlattmann P. Procalcitonin as a diagnostic marker for sepsis: a systematic review and meta-analysis. Lancet Infect Dis. 2003;13:426–35. https://doi.org/10.1016/s1473-3099(12)70323-7.
Hoeboer SH, Van Der Geest PJ, Nieboer D, Groeneveld AB. The diagnostic accuracy of procalcitonin for bacteraemia: a systematic review and meta-analysis. Clin Microbiol Infect. 2015;21:474–81. https://doi.org/10.1016/j.cmi.2014.12.026.
Tang JH, Gao DP, Zou PF. Comparison of serum PCT and CRP levels in patients infected by different pathogenic microorganisms: a systematic review and meta-analysis. Braz J Med Biol Res. 2018;51:e6783. https://doi.org/10.1590/1414-431x20176783.
Pfister R, Kochanek M, Leygeber T, Brun-Buisson C, Cuquemelle E, Machado MB, et al. Procalcitonin for diagnosis of bacterial pneumonia in critically ill patients during 2009 H1N1 influenza pandemic: a prospective cohort study, systematic review and individual patient data meta-analysis. Crit Care. 2014;18:R44. https://doi.org/10.1186/cc13760.
Parlato M, Philippart F, Rouquette A, Moucadel V, Puchois V, Blein S, et al. Circulating biomarkers may be unable to detect infection at the early phase of sepsis in ICU patients: the CAPTAIN prospective multicenter cohort study. Intensive Care Med. 2018;44:1061–70. https://doi.org/10.1007/s00134-018-5228-3.
Vollenweider DJ, Jarrett H, Steurer-Stey CA, GarciaAymerich J, Puhan MA. Antibiotics for exacerbations of chronic obstructive pulmonary disease. Cochrane Database Syst Rev. 2012. https://doi.org/10.1002/14651858.CD010257.
Finney LJ, Belchamber KBR, Fenwick PS, Kemp SV, Edwards MR, Mallia P, et al. Human Rhinovirus Impairs the Innate Immune Response to Bacteria in Alveolar Macrophages in Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med. 2019;199:1496–507. https://doi.org/10.1164/rccm.201806-1095OC.
Gunasekaran K, Ahmad M, Rehman S, Thilagar B, Gopalratnam K, Ramalingam S, et al. Impact of a positive viral polymerase chain reaction on outcomes of chronic obstructive pulmonary disease (COPD) exacerbations. Int J Environ Res Public Health. 2020;17:8072. https://doi.org/10.3390/ijerph17218072.
Acknowledgments
None. Members listed in the Additional file 1.
Funding
The BPCTrea study was supported by an unrestricted academic grant (PHRC inter régional 2010, IDRCB 2010-A00630-39) from the Hospital Program for Clinical Research, French Ministry of Health. The funder of the study had no role in the study design, data collection, data analysis, or data interpretation. The funder also had no role in the writing of the report. The corresponding author had full access to all the study data and assumed final responsibility for the decision to submit the paper for publication. The PROCALCIVIR study was funded by an academic unrestricted grant from the Caen Côte de Nacre University Hospital. BRAHMS (the manufacturer of procalcitonin assay) provided a portion of the assay material.
Author information
Authors and Affiliations
Consortia
Contributions
CD, FF, SA and JJP initiated and designed the study. CD and FF were responsible for data collection. SA, FF and JJP performed the statistical analysis and were involved in the interpretation of the results. CD wrote the manuscript, and JJP, SA, FF, NT, FT and MF helped to draft the manuscript. FD, MR AP, BF, XV and DDC contributed to the revision of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was conducted according to the principles of the Declaration of Helsinki and pooled two data sets of prospective studies: BPCTrea (ClinicalTrials.gov, NCT02521636) and PROCALCIVIR studies. For BPCTrea, the study protocol was approved by the local research ethics committee (Comité de Protection des Personnes Nord Ouest III) for all participating centers. A written informed consent was obtained from the patients or their surrogates upon their enrollment in the study. For PROCALCIVIR, an ethical board decided that approval was not necessary given the observational nature of the study in accordance with French legislation about observational studies at the time of the study. According to French legislation about observational studies at the time of the study, no informed consent was obtained from the patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Online Resource 1.
Detailed information regarding inclusion, no inclusion and exclusion criteria and definitions. Online Resource 2. PCT levels at inclusion (PCT-H0), at six hours (PCT-H6) and day 1 (PCT-H24) after inclusion, in subgroups of patients with (ATB+) (Panel a-c) and without (ATB-) (Panel d-f) antibiotics at inclusion. Online Resource 3. PCT levels at inclusion (PCT-H0), at six hours (PCT-H6) and day 1 (PCT-H24) after inclusion, in subgroups of patients with (PNP+) (Panel a-c) or without (PNP-) (Panel d-f) pneumonia at inclusion. Online Resource 4. Receiver operating characteristic (ROC) curves at any time (i.e., H0, H6 and H24 after inclusion) for the prediction of documented bacterial infection (including bacterial and viral coinfection) vs nondocumented bacterial infection (i.e., documented viral infection alone or absence of documented pathogen) for the PCT levels in subgroups of patients with (ATB+) (Panel a-c) and without (ATB-) (Panel d-f) antibiotics at inclusion. Online Resource 5. Receiver operating characteristic (ROC) curves at any time (i.e., H0, H6 and H24 after inclusion) for the prediction of documented bacterial infection (including bacterial and viral coinfection) vs nondocumented bacterial infection (i.e., documented viral infection alone or absence of documented pathogen) for the PCT levels in subgroups of patients with (PNP+) (Panel a-c) or without (PNP-) (Panel d-f) pneumonia at inclusion.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Daubin, C., Fournel, F., Thiollière, F. et al. Ability of procalcitonin to distinguish between bacterial and nonbacterial infection in severe acute exacerbation of chronic obstructive pulmonary syndrome in the ICU. Ann. Intensive Care 11, 39 (2021). https://doi.org/10.1186/s13613-021-00816-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13613-021-00816-6