Abstract
Diseases outbreaks in pond aquaculture have resulted in huge losses to the aquaculture industry. The emergence of non-antimicrobial and environment friendly agents (probiotics) is the potential consideration for the healthy shrimp aquaculture. The present study was aimed to compare the bacterial community compositions in shrimp ponds and surrounding seawater, as well as isolate probiotic bacteria from the shrimp ponds. Based on the high-throughput of 16S rRNA gene sequencing, all sequences were assigned to 3584 unique operational taxonomic units (OTUs) at 97% similarity levels, which were affiliated with 24 phyla, 54 classes, 235 families, and 367 genera. The 10 most abundant phyla were Bacteroidota, Proteobacteria, Actinobacteriota, Planctomycetota, Cyanobacteria, Chloroflexi, Firmicutes, Desulfobacterota, Patescibacteria and Verrucomicrobiota. Notably, the alpha diversity (Shannon diversity) of shrimp ponds was significantly differences (P < 0.05) with that of surrounding seawater. There were 2498 and 791 unique OTUs in shrimp ponds and surrounding seawater, respectively. A total of 15 isolates were obtained in the culturable bacterial diversity, and the antibacterial activities were recorded for potential probiotic bacterial isolates against different tested bacterial isolates including pathogenic bacteria. An isolate Hallobacillus marinus HMALI004 showed strong inhibitory effects against three pathogenic bacteria, Vibrio cholerae CECT 514, non AHPND V. parahaemolyticus BCRC12959 and AHPND V. parahaemolyticus PD-2. The isolates Algophigus sanaruensis AGALI005, Algoriphagus taiwanensis ATALI009 and Bacillus aequororis BAALI008 were also identified as potential probiotics strains.
Similar content being viewed by others
Introduction
Marine aquaculture is a fast-growing food industry, accounting about half of the total seafood supply (Garcia and Rosenberg 2010). In the production of mariculture between 1976 and 2016, 245% increase has been reported, which ultimately significantly enhance the exports from USD 8 billion (in 1976) to USD 143 billion in 2016, while in 2017 the demand values increased by 7% to peak at an estimated 152 billion USD (FAO 2018). On the mariculture, shrimp farming is one of the most important aquaculture industries, with a global record production of 512 kt worth USD 32 billion in 2016 (FAO 2018). Shrimps are usually cultivated in small seawater ponds (< 1 ha), which provide many advantages to farmers, including easy management, adjustable stocking density, and large production in a unit area (Li et al. 2010; Martınez-Cordova et al. 2015).
Nevertheless, emerging pathogens restrain the production of aquaculture affecting both economy and socio-economy of countries. There are various methods used so far to control the aquaculture diseases, such as, antibiotics and synthetic chemicals. Antimicrobial resistance (AMR) is considering the most serious problem to human health and animals. It is estimated that in 2050 around 10 million peoples will die from this resistant infection with associated economic cost of up to £60 trillion (O-Neill 2016). Around 70% of the antibiotics were used in livestock and this need of consumption is assumed to rise by 67% in 2030 (Van Boeckel et al. 2015). Till now this is a major concern since the overuse of antibiotics has been identified as the single most important factor pointing toward rise in resistant infections (CDC 2013).
Antibiotics are commonly used in larval cycle and in growth phases on aquaculture production. Antibiotic residues accumulated in the edible shrimp tissues that may alter intestinal flora of human and cause allergy and food poisoning problems (Thornber 2020). Most frequently used antibiotics to combat bacterial diseases include sarafloxacin, florfenicol, enrofloxacin and oxytetracycline (Roque et al. 2001; SotoRodríguez et al. 2006). Other antibiotics are also used globally such as ciprofloxacin, oxolinic acid, gentamicin, chlortetracycline, quinolones, norfloxacin, perfloxacin, tiamulinand and sulfamethazine (Holmstrom et al. 2003). Expensive chemotherapy for overcoming aquaculture diseases has been criticized for their more unsupportive effects. Such diseases controlling methods are ineffective against newly emerging pathogens in large aquaculture ponds. Therefore, substitutive methods are the demand of the present hour for the development of healthy microbial ecosystem in aquaculture for the maintenance of aquatic organism’s health (Liu et al. 2013). Disease outbreaks in shrimp aquaculture is the major challenge that causes huge economic losses (Hai 2015). The common infectious pathogenic bacteria in shrimp aquaculture are Gram-negative bacteria such as strains of Pseudomonas, Yersinia, Vibrio, Flavobacterium, and Aeromonas. These pathogenic bacteria cause furunculosis, enteric red mouth disease, vibriosis, septicemia, hemorrhage (Dawood et al. 2019; Hamid et al. 2017; Wanna et al. 2021; Wiklund 2016; Won et al. 2020; Zommiti et al. 2018). Bacteria that caused shrimp diseases have extracellular DNA encoding toxins to result in high mortality via peeling hepatopancreatic tissues, ultimately leading to the death of shrimp. This rate of mortality leads to considerable losses in economy and shrimp farming industry (Restrepo et al. 2021).
Probiotics are an emerging and promising alternative to prevent infectious diseases in shrimp aquaculture. Probiotic application is shrimp aquaculture has been reported to improve health (Mehrabi et al. 2018), growth (Gobi et al. 2018), and immunity of shrimp (Ramesh and Souissi 2018), as well as improve water quality and prevent diseases by modifying the microbial composition of water and sediment in the farm (Deng et al. 2018; Meidong et al. 2018). Although there are many studies on the beneficial effects of probiotics in shrimp aquaculture, however, the research about the native bacterial diversity as potential probiotics in the shrimp pond system is still under consideration. Therefore, the present study investigated the native bacterial diversity in shrimp aquaculture ponds through culture-dependent and culture-independent techniques and explored potential probiotic bacterial strains in shrimp ponds through cross-talk experiments.
Materials and methods
Samples collection and environmental parameters measurement
Water samples were collected from the ponds of four local shrimp farms (Pond 1 = P1, Pond 2 = P2, Pond 3 = P3, Pond 4 = P4) and (Surrounding seawater = S) in Shantou, Guangdong province, Southern China (23.36°N, 116.66°E) on September 2019. All four ponds are adjacent to each other at a distance of 115–140 m, and separated by surrounding seawater at a distance of 970 m. The length, width, and depth of the ponds were about 200, 60, and 1.5 m, respectively (Additional file 1: Table S3). All water samples were collected three three different locations of each pond and surrounding seawater from a depth of about 28 cm and stored in sterile polycarbonate bottles (Additional file 1: Fig. S2). Samples were kept at 4 °C and transferred to the laboratory for further analysis within 12 h. Water samples were subsampled for further investigation. Physicochemical parameters such as pH, temperature, and oxidation-reduction potential (ORP), were measured through HQ 30d multi-parameter water quality analyzer (HACH, Loveland, co). Salinity was measured by portable refractometer (Suwei, Guangzhou, China). Chemical oxygen demand (COD) was measured by using the dichromate method (Dedkov et al. 2000), and ammonia and nitrite were measured as described by Lu et al. (2009). Subsamples for DNA extraction were filtered through a nucleopore filter (diameter of 0.45 μm) and the filters were kept at − 80 °C until DNA extraction.
Analysis of bacterial communities using high throughput sequencing
To study the bacterial community structure in shrimp ponds and surrounding seawater, environmental whole genome DNA was extracted from the filter membranes using a PowerSoil DNA Extraction Kit (MoBio Laboratories, Solana Beach, CA, USA) according to the manufacturer’s instruction. Bacterial communities were characterized at each time point of the incubation at 515 F and 907R primers (Raina et al. 2022), which mainly target the V4-V5 region of the bacterial 16S rRNA gene, which were used for PCR amplification of extracted DNA. PCR reaction included 6 µL of template, 2.5 µmol of each deoxyribonucleotide triphosphate (Bioline), 1 µL of UltraPure Bovine Serum Albumin (Thermo Fisher), 0.25 µL of Velocity DNA polymerase and 5 × PCR buffer (Bioline), 10 pmol of each primer (resuspended in UV sterilized water) with the following adaptors (5′/-GTGCCAGCMGCCGCGG-515 F-3/) and (5/-CCGTCAATTCMTTTRAGT-907R-3/). PCR programmed condition were 98 °C for 2 min, followed by 30 cycles of denaturation at 98 °C for 30 s, annealing for 30 Sect. (46 °C for 3 cycles, 48 °C for 3 cycles, and 50 °C for 24 cycles), 72 °C for 30 s, and final extension at 72 °C for 10 min. PCR clean-up, indexing and sequencing (llumina MiSeq (2 × 300 bp)) were performed at the BGI-Shenzhen (The Beijing Genomics Institute, Shenzhen, Guangdong, China) for sequencing.
After obtaining raw sequencing data, the barcodes were removed, and the primer and adapter sequence reading and metadata were imported into QIIME 2 and analyzed according to the tutorial (Caporaso et al. 2010). Briefly, the DADA2 algorithm (Callahan et al. 2016) was used for demultiplexing, denoising, truncating reads, and joining forward and reversed read pairs. Chimeras were identified using the VSEARCH ‘uchime_denovo’ method (Rognes et al. 2016) and subsequently removed. Respective reads were then summarized in a feature table. Features were then divided to the genus level, and a reduced table containing only highly abundant taxa (> 1% mean relative abundance) was used for generating bar charts.
Shannon-Wiener diversity index and phylogenetic diversity index in each sample were calculated using qiime2 (v2019.10.0) with diversity plugin, and their significance was measured by Kruskal-Wallis test. Microbial beta-diversity was determined by non-metric multidimensional scaling (NMDS) using a Bray–Curtis distance matrix between the samples based on the genus level, which was performed using an R module called ggvegan (v2.5-7).
Isolation and identification of culturable bacteria
Culturable bacteria were isolated from shrimp ponds and surrounding seawater via serial dilution and cultural technique using marine agar 2216E plates (peptone 5.0 g, magnesium chloride 8.8 g, ferric citrate 0.1 g, potassium chloride 0.55 g, yeast extract 1.0 g, calcium chloride 1.8 g, sodium sulfate 3.24 g, sodium bicarbonate 0.16 g, potassium bromide 0.08 g, agar 15.0 g, sodium chloride 19.54 g, strontium chloride 34.0 mg, boric acid 22.0 mg, sodium fluoride 2.4 mg, sodium silicate 4.0 mg, ammonium nitrate 1.6 mg, disodium phosphate 8.0 mg in one liter of purified water). The most dominant and morphologically distinct colonies (in terms of size, shape, colour, fast-growing) were isolated, sub-cultured, and purified on marine agar 2216E plates. Pure colonies were preserved in 60% glycerol and 40% marine broth (v/v) and preserved at − 80 °C.
Moreover, DNA was extracted from purified bacterial colonies by using Chelex and proteinase K as lyzing agents. Briefly, aliquots (a spot) of bacterial colonies were added into 50 µL DNAse free water with 5% Chelex and 2.5 µL of proteinase K (20 mg/ml), and further incubated at 37 °C for 30 min to release DNA. The 16 S rRNA gene was amplified using bacterial specific universal primers 27 F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TGACTGACTGAGGYTACCTTGTTACGACTT-3′) (Wang et al. 2012). The PCR mixture composed of 1.25 µL of 50 mM MgCl2, 2.5 µL of 10x PCR buffer, 0.2 µL of 25 mM dNTPs, 0.25 µL of Taq DNA polymerase (5 U/µL), 0.4 µL of each primer (25 pmol/µL), and 1 µL of template DNA. The final volume was implemented to 25 µL with DNAse free water. The amplification was carried out in a thermocycler (Mastercycler Personal 5332) under the following conditions, initial denaturation at 95 °C for 2 min followed by 30 cycles of denaturation at 95 °C for 90 s, annealing at 52 °C for 1 min and extension at 72 °C for 90 s; and a final extension at 72 °C for 10 min. The amplified products were then subjected to 1% agarose gel electrophoresis with ethidium bromide as a stain and visualized on a transilluminator under UV light.
Furthermore, the amplified 16S rRNA gene was sequenced by BGI-Guangzhou (The Beijing Genomics Institute, Guangzhou, Guangdong, China). ContigExpress (http://www.contigexpress.com/index.html) was used to assembled the sequences of 16S rRNA gene. Sequences were identified by comparing with the closest relatives in the GenBank database using BLAST (Basic Local Alignment Search Tool) (Additional file 1: Table S2). 16S rRNA gene sequences of representative genus from the families Moraxellaceae, Aeromonadaceae, Bacillaceae, Cyclobacteriaceae, Vibrionaceae, Nocardioidaceae, Erythrobacteraceae, and Shewanellaceae were downloaded and used to construct Neighbor-joining phylogenetic tree through MEGA-X. Halobacterium salinarum JCM 8978 (AB663362) was used as an outgroup. Bootstrap support is shown for cases where the value was greater than 50% based on 1000 replications (Ali et al. 2021). The scale bar indicates 0.01 substitution per nucleotide position. And GeneBank accession numbers were listed after each sequence name (Fig. 3). The major strains used in this study were deposited in China Center of Industrial Culture Collection (CICC).
Cross-talk experiment to check the antimicrobial activity
To analyze the binary interaction between isolates, the stationary phase culture of each strain was tested for sensitivity against the other 15 strains. The 15 cryopreserved strains isolated in this study were re-cultured in marine broth for 24 h, with lawn as the target strain. The inhibitory activity of the strains was performed in Petri plates containing marine agar medium 2216E. About 50–100 µL of bacterial inoculum was spread uniformly on a sterile marine agar plate using a sterile cotton swab. A full loop of growing bacteria was placed on the surface of the lawn of the target bacteria and was incubated at 25 °C temperature for 24 to 72 h. The inhibition of the pathogenic bacterial growth was measured according to the size of the inhibition zone (Smania et al. 1999). Pathogenic Vibrio strains like non AHPND V. parahaemolyticus BCRC12959 and V. parahaemolyticus PD-2 were received from shrimp disease and immunity Lab, Collage of Science, Shantou University.
Statistical analysis
The differences between shrimp ponds and surrounding seawater were analyzed by the Kruskal-Wallis test (also called one-way ANOVA on ranks) to check the significance and the P value < 0.05 was considered significant.
Results
Physicochemical characteristics of water samples
The physicochemical parameters of water in shrimp ponds and surrounding seawater varied in chemical oxygen demand (COD), oxidation-reduction potential (ORP), salinity, and ammonia (NH4) concentrations, while slight differences were detected for pH value. The ORP of shrimp ponds varied from 39 mv to 86 mv, as compared to the surrounding seawater which was 71 mv. The salt concentration was low in all four ponds ranging from 0.5 to 0.9%, compared to seawater samples recorded 1%. The COD level in different shrimp ponds varied from 35.3 mg/L to 45.4 mg/L, while the value for the seawater sample was 55.5 mg/L. The ammonia (NH4) concentrations in different shrimp ponds ranged from 2.2 mg/L to 3.7 mg/L, and the value for seawater was 2.2 mg/L. The observed pH of different shrimp ponds varied from 7.7 to 8.5, while the value for the seawater sample was 7.2. The temperature of different shrimp ponds was in the range of 28.3 to 29.5 °C, while for seawater samples was 29.5 °C (Additional file 1: Table S1).
Composition of bacterial communities in shrimp ponds and surrounding seawater
Through Illumina high throughput sequencing analysis of bacterial diversity in the shrimp ponds and surrounding seawater, a total of 762,863 sequences were obtained. The range of effective reads in the water samples was from 50,568 to 51,275. All sequences were assigned to 3584 unique OTUs, belonging to 24 phyla, 54 classes, 235 families, and 367 genera. Significant differences (P < 0.05) were detected between alpha-bacterial communities (Shannon index) of seawater samples and shrimp ponds (Fig. 1a). Pond 3 (P3) showed substantial phylogenetic diversity compared to other samples (Fig. 1b). Interestingly, P1 and P2 showed similarities in bacterial diversity to P4 while distinct from surrounding seawater samples (S) (Fig. 1b). Data was also clustered to NMDS plot for beta-diversity (Fig. 1c), in which bacterial diversity of shrimp ponds was significantly different from those in the surrounding seawater.
Notably, few bacterial phyla (Bacteriodetes, Proteobacteria, Actinobacteria, Planctomycetota, Cyanobacteria, Chloroflexi, and some other phyla with low concentration) were detected in both shrimp ponds and the surrounding seawater. In shrimp ponds, the major bacterial phyla were Bacteriodetes (27.5–35.4%), Proteobacteria (12.5–61.4%), Actinobacteria (13.5–22.2%), Planctomycetota (12.8–23.1%), while Cyanobacteria (50.45%) were the dominant phyla in surrounding seawater (Additional file 1: Fig. S1).
The genus-level bacterial community structure of shrimp ponds and surrounding seawater is summarized in Fig. 2. Bacterial community in shrimp ponds was diverse than the surrounding seawater community in terms of composition and their relative abundance. The most abundant bacterial genera in shrimp ponds were Pirellula, while Cyanobium was the dominant bacterial community in surrounding seawater. The relative abundance of Pirellula in shrimp ponds ranged from 18.1 to 58%, but it was not detected in surrounding seawater. In shrimp ponds, the relative abundance of hgcl-clade bacteria (8.4–16.5%) was significantly higher (P < 0.05) than that in surrounding seawater (7.6%). The relative abundance of genus Cyanobium in surrounding seawater (52.6%) was significantly higher (P < 0.05) than that in shrimp ponds (0.3–4.5%). The relative abundance of Rhodopirellula in shrimp ponds was 1.4–9.4%, while it was not found in surrounding seawater. In shrimp ponds, the relative abundance of unidentified Kapabacteria from phylum Bacteroidetes was 1.8–10.6%, but it was not detected in surrounding seawater. The relative abundance of unidentified Roseiflexaceae, belonging to phylum Chloroflexi, in shrimp ponds (5.4–23%) was significantly higher (P < 0.05) than surrounding seawater (1.3%).
Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences showing the phylogenetic position of all isolated strains. Halobacterium salinarum JCM 8978 (AB663362) was used as the outgroup. Bootstrap support is shown for cases where the value was greater than 50% based on 1000 replications. The scale bar indicates 0.01 substitution per nucleotide position. Strains from the shrimp ponds were highlighted as green shade, and the strains from surrounding seawater were highlighted as orange shade, while the reference strains were marked with red dots, and GeneBank accession numbers listed after each sequence name
The genus Algoriphagus in shrimp ponds was found (1.4–6.9%) abundant than surrounding seawater (2.5%). Rheinheimera from the family Alteromonadaceae in shrimp ponds ranged from 1.3 to 14.4%, however, the relative abundance of this genus in the surrounding seawater sample was 1.9%. The relative abundance of unidentified Saprospiraceae in shrimp ponds (1.7–3.3%) was significantly higher (P < 0.05) than that in surrounding seawater (0.2%). In addition, the abundance of Bacillus in shrimp ponds ranged from 0.03 to 0.3%, but it was not detected in surrounding seawater. The relative abundance of unidentified Comamonadaceae in shrimp ponds (0.9–1.6%) was significantly lower (P < 0.05) than that in surrounding water (3.7%). Furthermore, the pathogenic bacteria V. cholera was identified in shrimp pond 1 (P1).
Isolation of bacterial strains
Bacterial strains were isolated and 15 strains were selected based on their distinct morphology. The findings of 16S rRNA gene sequence indicated that the bacterial isolates from shrimp ponds were Bacillus (3 strains), Algoriphagus (3 strains), Vibrio (1 strain), Aeromonas (1 strain), Acinetobacter (1 strain), Exiguobacterium (1 strain), and Halobacillus (2 strains). In contrast, Nocardioides (1 strain), Erthrobacter (1 strain), and Shewanella (1 strain) were found to be the dominant genera in surrounding seawater (Fig. 3). In the phylogenetic tree, strains from the shrimp ponds were highlighted as green shade, and the strains from surrounding sea water were highlighted as orange shade, while the reference strains were marked with red dots (Fig. 3). Detailed information of the BLAST results of individual strains were provided in (Additional file 1: Table S2).
Candidate probiotics strain through cross-talk
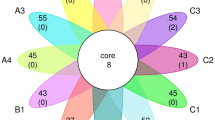
To test the inhibitory interaction among 15 bacterial isolates, each isolate was tested for potential growth inhibition against all other isolates. The results indicated that more than 225 pairings of the 15 isolates were monitored for pathogenicity in shrimp ponds. Notably, 17 (15 were isolated via culturable method and 2 were collected from shrimp disease and immunity Lab, Collage of Science, Shantou University, Shantou Guangdong China) strains showed binary inhibition (Fig. 4a). Four stains showed inhibitory binary interaction (Fig. 4b). Most of the observed inhibition was due to the activity of strains in two bacterial phyla, Bacteriodetes and Firmicutes. Strain ATALI009 (CICC accession number 25,154) showed the highest 16S rRNA gene similarity with Algoriphagus taiwanensis CC-PR82 (99.4%), and inhibited the growth of strains BVALI010 (Bacillus vietnamensis), NAALI012 (Nocardioides aestuarii) (pathogenic), SDALI015 (Shewanella decolorationis) (pathogenic), and EAALI014 (Erthrobacter alti) (pathogenic), with an inhibition zone of 0.3 mm, 0.2 mm, 0.3 mm, and 0.2 mm, respectively. Strain AGALI005 (CICC accession number 25,153) which is also grouped into the genus of Algoriphagus (99.1% similarity with Algoriphagus sanaruensis M8-2), could inhibit the growth of strains HDALI003 (Halobacillus dabanensis), NAALI012 (N. aestuarii), BVALI010 (B. vietnamensis), and SDALI015 (S. decolorationis) by 0.4 mm, 0.4 mm, 0.5 mm, and 0.3 mm, respectively. Strain BAALI008 (CICC accession number 25,152) belongs to the genus Bacillus (alternative of Sutcliffiella showed 99.7% similarity with Bacillus horikoshii M-8), which could inhibit the growth of strain NAALI012 (N. aestuarii), with an inhibition zone of 0.3 mm. HMALI004 (H. marinus) (CICC accession number 25,155) inhibited the growth of strains AVALI006 (Aeromonas veronii), NAALI012 (N. aestuarii), EAALI014 (E. alti), and SDALI015 (S. decolorationis), with inhibition zones of 0.5 mm, 0.4 mm, 0.8 mm, and 0.4 mm, respectively. Strain HMALI004 with the highest similarity to H. marinus KGW1 (99.7%) was further applied to assess the inhibitory potential against well-studied bacterial marine pathogens, including V. cholerae CECT 514T, V. parahaemolyticus PD-2, and non AHPND V. parahaemolyticus BCRC12959. Strain HMALI004 showed dramatic inhibitory activity against all tested model pathogens, and the inhibiting zones of CECT 514, PD-2, and BCRC12959 were 0.5 mm, 0.7 mm, and 0.4 mm against strains, respectively (Fig. 4a).
Inhibitory activity (a) and Chord-diagram (b), Inhibitory zone in a is labeled with 1 for strong, 2 for weak, and 3 for no inhibition based on halo size. In Chord-diagram the targeted strains (inhabited) marked in red color, whereas strains with probiotic activity was highlighted in black color. The findings show that probiotics bacterial isolates such as H. marinus HMALI004, B. aequororis BAALI008, A. sanaruensis AGALI005 and A. taiwanensis ATALI009 inhabited the growth of N. aestuarii NAALI012, E. alti EAALI014, S. decolorationis SDALI015, B. vietnamensi BVALI010, H. dabanensis HDALI003, V. parahaemolyticus PD-2, V. cholerae CECT 514T, and non AHPND V. parahaemolyticus BCRC12959. The number at the end of nodes represents the inhibitory values in milimeter
Discussion
Microorganisms are essential in aquatic environments due to their potential to degrade pollutants, improve water quality and prevent the growth of pathogens (Martinez-Cordova et al. 2015). In addition, they play a pivotal role in the health of aquatic life and the sustainability of aquaculture ecosystems (Ninawe and Selvin 2009), the microbial diversity of the shrimp ponds system remains to be studied (Alfiansah et al. 2018; Hastutia et al. 2021; Martinez-Porchas and Vargas-Albores 2017; Zeng et al. 2020; Zhang et al. 2019).
In the present study, the relative abundance of Bacteriodetes, Proteobacteria, Actinobacteria, Planctomycetota, and Chloroflexi in shrimp ponds were significantly higher than in surrounding seawater, while the relative abundance of Cyanobacteria in surrounding seawater was significantly higher than that in shrimp ponds. These findings are consistent with a report that Proteobacteria, Bacteroidetes, and Actinobacteria were the leading phyla of pond aquaculture (Zheng et al. 2017, 2021). However, it contradicted with the results of Dabade et al. (2016), in which Proteobacteria, Flavobacteria, and Actinobacteria were the major phyla in shrimp aquaculture ponds. This abundance of microbial communities in our study might be due to similar condition of water in all shrimp ponds.
Genus hgcl-clade belongs to Actinobacteria, and its relative abundance in shrimp ponds was very high. This bacterium has been reported to be a potential probiotic in aquaculture ponds (Das et al. 2008). Actinobacteria possess the potential of probiotics to improve the digestibility and immune system of shrimp to counteract the pathogenicity of pathogens (Costantini et al. 2017). Genus Rhodopirellula belonging to Actinobacteria was observed in ponds water, while it was not found in surrounding seawater. Genomic analysis of Rhodopirellula sp. revealed the stigmatellin, myxothiazol, and myxamid antibiotics encoding genes (Huntley et al. 2012). This group of bacteria could be a potential probiotic in shrimp ponds. Another dominant bacteria in shrimp ponds was Rheinheimera sp., which was previously isolated from the rhizosphere of Echinacea purpurea (Carolina et al. 2019). Comamonadaceae is an aerobic organotrophs, Fe3+ reducing and anaerobic denitrifiers, hydrogen oxidizers, photoheterotrophic and photoautotrophic, and fermentative bacterial family. Most of the members of this family are environmental bacteria reported from water and soil habitats. Some members of the Comamonadaceae family are also probiotics (Willems 2014; Qin et al. 2016) studied the abundance and diversity of bacteria in pond water augmented with various feeds and found that the ponds supplied with sudangrass had better water conditions. Moreover, in the ponds supplied with sudangrass, pathogenic bacteria such as Acinetobacter and Aeromonas decreased significantly, while probiotic unclassified Comamonadaceae increased significantly. In this study, the relative abundance of Saprospiraceae sp. in shrimp ponds water was significantly higher than that in surrounding seawater. This species was mostly reported from marine water as predators of other bacteria and microalgae (Mcllroy and Nielsen 2014). Saprospiraceae sp. showed activity against the marine pathogen V. shiloi (Choi et al. 2015). The bacterial population in shrimp ponds is the key to protect the shrimp pond ecosystem from pathogens (Alfiansah et al. 2018).
The bacterial diversity in shrimp ponds was significantly higher than that in surrounding water. Since a high microbial community is important for the stability of ponds and the digestion and immunity of shrimp, the diverse microbial community in shrimp ponds could improve the quality of water and shrimp health (Fan et al. 2019; Zhang et al. 2016). In the current study, four bacterial strains with antimicrobial activities were isolated through culturable method, and two strains were affiliated to Firmicutes (H. marinus HMALI004 and B. aequororis BAALI008), and the other two strains were affiliated to Bacteroidetes phylum (A. sanaruensis AGALI005 and A. taiwanensis ATALI009). Except for H. marinus HMALI004, the other three were found in 16S rRNA high-throughput sequencing. These bacterial strains act as probiotics in shrimp ponds. Jang et al. (2019) used Bacillus sp. as a dietary probiotic supplement in the feed of olive flounder to fight against the pathogenic challenge and found a significant abundance of Bacillus sp. in the intestine after 14 days, which enhanced the richness of the bacterial community, immune response, digestive enzymes activities, and protection from pathogens. Thus, the use of probiotic bacteria Bacillus sp. in aquaculture has become the best strategy to prevent pathogens and increase nutrient assimilation.
Bacteria affiliated with the genus of H. marinus HMALI004 were isolated from shrimp ponds. Through cross-talk experiments, this bacterial strain could inhibit the growth of Nocardioides, Erthrobacter, and Shewanella, which have been proved to be pathogens in aquaculture ponds (Mehta and Shamoo 2020; Tseng et al. 2018). Moreover, H. marinus HMALI004 also inhibited the growth of well-studied marine pathogens, including V. parahaemolyticus PD-2, V. cholerae CECT 514T, and non AHPND V. parahaemolyticus BCRC12959. Similar results were recorded by Mayavu et al. (2014), in which Hallobacillus isolated from parangipettai salt pan environment inhibited the growth of V. angullaram, V. paraheamolyticus, and V. alginolyticus. Likewise, based on the antagonistic activity against shrimp pathogens, AshokKumar and Mayavu (2014) isolated 25 halobacterial strains from the Marakanam salt pan environment, and among all 25 bacteria only one bacterial strain, Bacillus Mk22 showed high antagonistic activities. Recently, the use of halophilic bacterial applications has increased in industries. It secretes a wide range of various bioactive metabolites, such as enzymes (amylase, protease, cellulases, etc.), extra-cellular polysaccharides (EPS), proteins, etc. (Enache et al. 2010).
Bacteria affiliated with the genus of A. taiwanensis ATALI009 were isolated from shrimp ponds, and OTUs in this genus were also detected in 16S rRNA high throughput sequencing. Through cross-talk experiments, these bacterial strains could inhibit the growth of Nocardioides, Shewanella, and Erthrobacter, which have been proved to be pathogens in aquaculture ponds (Mehta and Shamoo 2020; Tseng et al. 2018). Algoriphagus sp. has a carotenoid gene cluster (ispH, crtYcd), which plays a vital role in photosynthesis, protection against oxidative damage, and nutrition (Tao et al. 2006). Carotenoids are pigments that may convert to vitamin A, and it can also act as an antioxidant (Sajjad et al. 2020) to protect shrimps from infection by strengthening the immune system (Wilding et al. 2000). The present study provides the first insight in the role of Algoriphagus as a probiotic in pond culture. In conclusion, the bacterial community in Shantou shrimp ponds was dominated by Proteobacteria, Firmicutes, Bacteriodetes, Actinobacteria, and Planctomycetes. Through unculture method, 16S rRNA high throughput sequencing, probiotic bacteria isolated from shrimp ponds were genus hgcl-clade, Rhodopirellula, Rheinheimera sp, Comamonadaceae, Saprospiraceae sp. Four probiotic bacterial strains such as H. marinus HMALI004, B. aequororis BAALI008, A. sanaruensis AGALI005 and A. taiwanensis ATALI009 were successfully isolated through culture dependent method luckily the OTUs for these strains were also present in unculturable method, some of which were first case reports of the species. Our study revealed that a novel source of antimicrobial metabolites from these probiotic strains could manage and reduce the risk of pathogen that causes shrimp diseases. Continuing the current research work in the future, the metabolites will be extracted and studied further for antimicrobial activities against various pathogenic bacterial strains. These findings provide insights to better understand the role of probiotic bacteria in shrimp ponds.
Availability of data and materials
All raw reads of 16S rRNA gene based High throughput sequencing were deposited in the sequencing read archive (SRA) of NCBI for diversity analysis with accession numbers from SAMN20970002 to SAMN20970016 under the Bioproject number-PRJNA757637. 16S rRNA gene sequences of all bacterial isolates were deposited to NCBI GenBank (accession numbers: OK001807- OK001820).
References
Alfiansah YR, Hassenruck C, Kunzmann A, Taslihan A, Harder J, Gardes A (2018) Bacterial abundance and community composition in pond water from shrimp aquaculture systems with different stocking densities. Front Microbiol 9:2457
Ali S, Xie J, Chen Y, Cai R, Juventus AJ, Hu Z, Zhang Y, Wang H (2021) Penaeicola halotolerans gen. nov., sp. nov., a novel bacterium of the family Cyclobacteriaceae isolated from a shrimp pond. Int J Syst Evol Micr 71:005047
Ashokkumar S, Mayavu P (2014) Screening, identification and antagonistic activity of halo stable Bacillus sp. Mk22 used as probiotic in Penaeus monodon Fabricius, 1798. Afr J Food Sci 8:48–53
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Carolina C, Pasqualetti C, Lanzoni O, Fagorzi C (2019) Harmful effect of Rheinheimera sp. against the protist Euplotes Aediculatus (Ciliophora, Spirotrichea): Insights into the ecological role of antimicrobial compounds from environmental bacterial strains. Front Microb 10:1–15
CDC (2013) Antibiotic resistance threats in the United States, 2013.Current,114pp. https://doi.org/CS239559-B
Choi EJ, Nam SJ, Paul L, Beatty D, Kauffman CA, Jensen PR, Fenical W (2015) Previously uncultured marine bacteria linked to novel alkaloid production. Chem Biol 22:1270–1279
Costantini L, Molinari R, Farinon B, Merendino N (2017) Impact of omega-3 fatty acids on the gut microbiota. Int J Mol Sci 18:2645
Dabade DS, Wolkers-Rooijackers JC, Azokpota P, Hounhouigan DJ, Zwietering MH (2016) Bacterial concentration and diversity in fresh tropical shrimps (Penaeus notialis) and the surrounding brackish waters and sediment. Int J Food Microbiol 218:96–104
Das S, Ward LR, Burke C (2008) Prospects of using marine actinobacteria as probiotics in aquaculture. App Microbiol Biotechnol 81:419–429
Dawood MAO, Koshio S, Abdel-Daim MM, Van Doan H (2019) Probiotic application for sustainable aquaculture. Rev Aquac 11:907–924
Dedkov YM, Elizarova OV, Kel’ina SY (2000) Dichromate method for the determination of chemical oxygen demand. J Anal Chem 55:77–781
Deng M, Chen J, Gou J, Hou J, Li D, He X (2018) The effect of different carbon sources on water quality, microbial community and structure of biofloc systems. Aquaculture 482:103–110
Enache M, Kamekura M (2010) Hydrolytic enzymes of halophilic microorganisms and their economic values. Rom J Biochem 47:47–59
Fan L, Wang Z, Chen M, Qu Y, Li J, Zhou A, Xie S, Zeng F, Zou J (2019) Microbiota comparison of Pacific white shrimp intestine and sediment at freshwater and marine cultured environment. Sci Total Environ 657:1194–1204
FAO (2018) The State of World Fisheries and Aquaculture 2018-Meeting the sustainable development goals. Licence: CC BY-NC-SA 3.0 IGO; FAO: Rome, Italy, 2018; ISBN 978-92-5-130562-1
Garcia SM, Rosenberg AA (2010) Food security and marine capture fisheries: characteristics, trends, drivers and future perspectives. Philos Trans R Soc Biol Sci 365:2869–2880
Gobi N, Vaseeharan B, Chen JC, Rekha R, Vijayakumar S, Anjugam M, Iswarya A (2018) Dietary supplementation of probiotic Bacillus licheniformis Dahb1 improves growth performance, mucus and serum immune parameters, antioxidant enzyme activity as well as resistance against Aeromonas hydrophila in tilapia Oreochromis mossambicus. Fish Shellfish Immunol 74:501–508
Hai NV (2015) The use of probiotics in aquaculture. J Appl Microbiol 119:917–935
Hamid NH, Hassan MD, Sabri MM, Hasliza AH, Hamdan RH, Afifah MN, Raina MS, Nadia AB, Fuad MM (2017) Studies on pathogenicity effect of Aeromonas hydrophila infection in juvenile red hybrid tilapia Oreochromis sp. In: Proceedings of International Seminar on Livestock Production and Veterinary Technology, pp. 532–539
Hastutia YP, Fatmab YS, Pitoyoc H, Nurussalama W, Ruhyana J (2021) Assessment of total bacterial diversity in white leg shrimps and its aquaculture environment in Pangkajene and Banyuwangi, Indonesia. AsPac J Mol Biol Biotechnol 29:26–37
Holmstrom K, Gräslund S, Wahlstrom A, Poungshompoo S, Bengtsson BE, Kautsky N (2003) Antibiotic use in shrimp farming and implications for environmental impacts and human health. Int J Food Sci 38:255–266
Huntley S, Zhang Y, Treuner-Lange A, Kneip S, Sensen CW, Søgaard-Andersen L (2012) Complete genome sequence of the fruiting myxobacterium Corallococcus coralloides DSM 2259. J Bacteriol 194:3012–3013
Jang WJ, Lee JM, Hasan MT, Lee BJ, Lim SG, Kong IS (2019) Effects of probiotic supplementation of a plant-based protein diet on intestinal microbial diversity, digestive enzyme activity, intestinal structure, and immunity in olive flounder (Paralichthys olivaceus). Fish shellfish Immunol 92:719–727
Liu YC, Zhou ZG, He SX, Yao B, Ringo E (2013) Microbial diversity in the sediment of a crab pond in Nanjing, China. Aquac Res 44:321–325
Lu G, Ocola LE, Chen J (2009) Reduced graphene oxide for room-temperature gas sensors. Nanotechnology 20:445502
Martinez-Cordova LR, Emerenciano M, Miranda‐Baeza A, Martinez‐Porchas M (2015) Microbial‐based systems for aquaculture of fish and shrimp: an updated review. Rev Aquac 7:131–148
Martinez-Porchas M, Vargas‐Albores F (2017) Microbial metagenomics in aquaculture: a potential tool for a deeper insight into the activity. Rev Aquac 9:42–56
Mayavu P, Sugesh S, Suriya M, Sundaram S (2014) Enumeration of halophilic forms in parangipettai saltpan and its antagonistic activities against Vibrio sp. J Appl Biol Biotechnol 2:019–021
Mehrabi F, Khalesi M, Hazaie K (2018) Effects of pre-and probiotics on growth, survival, body composition, and hematology of common carp (Cyprinus carpio L.) fry from the Caspian Sea. Turk J Fish Aquat Sci 18:597–602
Mehta HH, Shamoo Y (2020) Pathogenic Nocardia: a diverse genus of emerging pathogens or just poorly recognized? PLoS Pathog 16:e1008280
Meidong R, Khotchanalekha K, Doolgindachbaporn S, Nagasawa T, Nakao M, Sakai K, Tongpim S (2018) Evaluation of probiotic Bacillus aerius B81e isolated from healthy hybrid catfish on growth, disease resistance and innate immunity of Pla-mong Pangasius bocourti. Fish Shellfish Immunol 73:1–10
Ninawe S, Selvin J (2009) Probiotics in shrimp aquaculture: avenues and challenges. Crit Rev Microbiol 35:43–66
O’Neill J (2016) Tackling drug-resistant infections globally: final report and recommendations. Rev Antimicrob Resist 84
Qin Y, Hou J, Deng M, Liu Q, Wu C, Ji Y, He X (2016) Bacterial abundance and diversity in pond water supplied with different feeds. Sci Rep 6:1–13
Raina JB, Lambert BS, Parks DH, Rinke C, Siboni N, Bramucci A, Ostrowski M, Signal B, Lutz A, Mendis H, Rubino F (2022) Chemotaxis shapes the microscale organization of the ocean’s microbiome. Nature 605:132–138
Ramesh D, Souissi S (2018) Effects of potential probiotic Bacillus subtilis KADR1 and its subcellular components on immune responses and disease resistance in Labeo rohita. Aquac Res 49:367–377
Restrepo L, Domínguez-Borbor C, Bajaña L, Betancourt I, Rodríguez J, Bayot B, Reyes A (2021) Microbial community characterization of shrimp survivors to AHPND challenge test treated with an effective shrimp probiotic (V. diabolicus). Microbiome 9:1–20
Rognes T, Flouri T, Nichols B, Quince C, Mahé F (2016) VSEARCH: a versatile open-source tool for metagenomics. PeerJ 4:e2584
Roque A, Molina AA, Bolan MC, Gomez GB (2001) In vitro susceptibility to 15 antibiotics of vibrios isolated from penaeid shrimps in Northwestern Mexico. Int J Antimicrob Agents 17:383–387
Sajjad W, Din G, Rafiq M, Iqbal A, Khan S, Zada S, Ali S, Kang S (2020) Pigment production by cold-adapted bacteria and fungi: colorful tale of cryosphere with wide range applications. Extremophiles 24:447–473
Smania A, Monache FD, Smânia EFA, Cuneo RS (1999) Antibacterial activity of steroidal compounds isolated from Ganoderma applanatum (Pers.) Pat. (Aphyllophoro-mycetideae) Fruit body. Int J Med Mushrooms 1:325–330
Soto-Rodriguez S, Armenta M, Gomez-Gil B (2006) Effects of enrofloxacin and florfenicol on survival and bacterial population in an experimental infection with luminescent Vibrio campbellii in shrimp larvae of Litopenaeus vannamei. Aquaculture 255:48–54
Tao L, Yao H, Kasai H, Misawa N, Cheng Q (2006) A carotenoid synthesis gene cluster from Algoriphagus sp. KK10202C with a novel fusion-type lycopene β-cyclase gene. Mol Genet Genom 276:79–86
Thornber K, Verner-Jeffreys D, Hinchliffe S, Rahman MM, Bass D, Tyler CR (2020) Evaluating antimicrobial resistance in the global shrimp industry. Rev Aquacult 12:966–986
Tseng SY, Liu PY, Lee YH, Wu ZY, Huang CC, Cheng CC, Tung KC (2018) The pathogenicity of Shewanella algae and ability to tolerate a wide range of temperatures and salinities. Can J Infect Dis Med Microbiol
Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, Teillant A, Laxminarayan R (2015) Global trends in antimicrobial use in food animals. Proc Nat Acad Sci USA 112:5649–5654
Wang H, Laughinghouse IV, Anderson MA, Chen F, Willliams E, Place AR, Zmora O, Zohar Y, Zheng T, Hill RT (2012) Novel bacterial isolate from Permian groundwater, capable of aggregating potential biofuel-producing microalga Nannochloropsis oceanica IMET1. Appl Env Microbiol 78:1445–1453
Wanna W, Surachat K, Kaitimonchai P, Phongdara A (2021) Evaluation of probiotic characteristics and whole genome analysis of Pediococcus pentosaceus MR001 for use as probiotic bacteria in shrimp aquaculture. Sci Rep 11:1–17
Wiklund T (2016) Pseudomonas anguilliseptica infection as a threat to wild and farmed fish in the Baltic Sea. Microbiol Aust 37:135–136
Wilding EI (2000) Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci. J Bacteriol 182:4319–4327
Willems A (2014) The family comamonadaceae. The prokaryotes, Alphaproteobacteria and betaproteobacteria. Springer, Berlin, pp 777–851
Won S, Hamidoghli A, Choi W, Bae J, Jang WJ, Lee S, Bai SC (2020) Evaluation of potential probiotics Bacillus subtilis WB60, Pediococcus pentosaceus, and Lactococcus lactis on growth performance, immune response, gut histology and immune-related genes in whiteleg shrimp, Litopenaeus vannamei. Microorganisms 8:281
Zeng S, Khoruamkid S, Kongpakdee W, Wei D, Yu L, Wang H, Deng Z, Weng S, Huang Z, He J, Satapornvanit K (2020) Dissimilarity of microbial diversity of pond water, shrimp intestine and sediment in Aquamimicry system. AMB Express 10:180
Zhang M, Sun Y, Liu Y, Qiao F, Chen L, Liu WT, Du Z, Li E (2016) Response of gut microbiota to salinity change in two euryhaline aquatic animals with reverse salinity preference. Aquaculture 454:72–80
Zhang M, Pan L, Huang F, Gao S, Su C, Zhang M, He Z (2019) Metagenomic analysis of composition, function and cycling processes of microbial community in water, sediment and effluent of Litopenaeus vannamei farming environments under different culture modes. Aquaculture 506:280–293
Zheng X, Tang J, Zhang C, Qin J, Wang Y (2017) Bacterial composition, abundance and diversity in fish polyculture and mussel–fish integrated cultured ponds in China. Aquac Res 48:3950–3961
Zheng Z, Aweya JJ, Bao S, Yao D, Li S, Tran NT, Ma H, Zhang Y (2021) The microbial composition of penaeid shrimps’ hepatopancreas is modulated by hemocyanin. J Immunol 207:2733–2743
Zommiti M, Bouffartigues E, Maillot O, Barreau M, Szunerits S, Sebei K, Feuilloley M, Connil N, Ferchichi M (2018) In vitro assessment of the probiotic properties and bacteriocinogenic potential of Pediococcus pentosaceus MZF16 isolated from artisanal Tunisian meat “Dried Ossban.” Front Microbiol 9:2607
Acknowledgements
Not applicable.
Funding
Funding for this study was provided by the 2020 Li Ka Shing Foundation (LKSF) Cross-Disciplinary Research Grant (2020LKSFG07A), and Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (GML2019ZD0606).
Author information
Authors and Affiliations
Contributions
SA, RC and HW designed the project. SA and JX conducted the experiments and analyzed the metagenomics data. SA and SZ wrote the draft. ZH, YZ, RC and HW reviewed and revised the final draft. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: FigureS1:
Phylum level relative abundance and communitycompositions of shrimp ponds and surrounding sea water samples obtained by 16S rRNA High through put-sequencing in 15 samples. The phylum level distributionis based on the 97% similarity clusters OTUs. Sequences whose relativeabundance was lower than 1% were assigned as “Minority”. Figure S2: Actual figures of sampling locations. TableS1. Psychochemical parameters of shrimp ponds and surrounding sea water.Table S2. BLAST results of the isolated stains. Table S3. Details ofselected sampling area.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ali, S., Xie, J., Zada, S. et al. Bacterial community structure and bacterial isolates having antimicrobial potential in shrimp pond aquaculture. AMB Expr 12, 82 (2022). https://doi.org/10.1186/s13568-022-01423-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13568-022-01423-9