Abstract
Diarrhea caused by pathogenic Escherichia coli (E. coli) is one of the most serious infectious diseases in humans and animals. Due to antibiotics resistance and the lack of efficient vaccine, more attention should be paid to find potential versatile vaccine candidates to prevent diseases. In this study, the sequence homology analysis indicated that OmpF from E. coli CVCC 1515 shares a high identity (90−100%) with about half of the E. coli (46.7%) and Shigella (52.8%) strains. Then the recombinant OmpF was supposed to be developed as a versatile vaccine to prevent E. coli infection. OmpF was expressed in E. coli BL21 (DE3) using the auto-induction method. The recombinant OmpF (rOmpF) protein had an average molecular weight of 40 kDa with the purity of 90%. Immunological analysis indicated that the titers of anti-rOmpF sera against rOmpF and whole cells were 1:240,000 and 1:27,000, respectively. The opsonophagocytosis result showed that 72.21 ± 11.39 and 11.04 ± 3.90% of bacteria were killed in the rOmpF immunization and control groups, respectively. The survival ratio of mice immunized with rOmpF ranged between 40 and 60% as observed within 36 h after challenge, indicating mice were partially protected from E. coli CVCC 1515 infection. The expressed rOmpF protein induced an effective immune response, but only provide a weak protection against pathogenic E. coli CVCC 1515 and a small reduction in E. coli CICC 21530 (O157:H7) excretion in a mouse infection model. Native forms of the OmpF antigen may be studied for immunogenicity and potential protective efficacy.
Similar content being viewed by others
Introduction
Bacterial diarrhea caused by enterotoxigenic Escherichia coli is the main infectious disease in humans and animals worldwide (Johnson et al. 2010). Enterotoxigenic E. coli is transmitted by food or water contaminated with animal or human feces. The E. coli CVCC 1515 (O149:K91 and K88ac) strain, a predominant serotype, occurred more frequently in neonatal and postweaning pigs (Noamani et al. 2003; Maynard et al. 2003). Urease-positive E. coli CVCC 1515 was responsible for over 90% of cases of post-weaning diarrhea in recent outbreaks in Canada, which leading to substantial economic losses (Noamani et al. 2003). None of the attempted solutions to the problem of post-weaning diarrhea due to enterotoxigenic E. coli in pigs has been consistently effective. Another enteric pathogen E. coli CICC 21530 (O157:H7) strain is major cause of food-borne diarrheal disease, and can produce large quantities of one or more related potent toxins that cause severe damage to the lining of the intestine. Close to 75,000 cases of E. coli O157:H7 infection with 2–10% deaths are now estimated to occur annually in the United States (Perna et al. 2001; Vali et al. 2004). Although antibiotics and vaccines are currently available to prevent E. coli-induced diarrheas, antibiotics residues may pose severe health hazards in human and there are few available vaccines against homologous E. coli challenge. Therefore, more attention should be paid to find potential versatile vaccine candidates to prevent diseases induced by E. coli.
Outer membrane proteins (OMPs), exposed on the surface of gram-negative bacteria, are quickly recognized as extracellular foreign particles by the host immune system, thereby generating an immune response against bacterial pathogens (Osman and Marouf 2014). Highly immunogenic OMPs can thus be exploited as vaccine candidates against several bacterial species such as Chlamydia trachomatis, Neisseria meningitides, Aeromonas hydrophila and Edwardsiella tarda (Pal et al. 1997; Wright et al. 2002; Khushiramani et al. 2012; Yadav et al. 2014; Okamura et al. 2012). Among OMPs, the outer membrane protein F (OmpF) and OmpC are the two most common porins that make 2% of the total cellular protein, and OmpF is the best-characterized porin protein in terms of structural and functional characteristics (Williams et al. 2000). OmpF consists of 16 antiparallel β-strands forming a barrel embedded in the membrane and displays eight domains of the surface antigen at the N-terminal extracellular domain (http://www.uniprot.org/) (Williams et al. 2000). Several attempts have been made to evaluate the OmpF immunogenicity of gram-negative bacteria. Secundino et al. showed that OmpF of Salmonella typhi could induce a sustained, lifelong and specific bactericidal antibody response (Secundino et al. 2006). Synthetic peptides representing certain epitopes of the OmpF of Pseudomonas aeruginosa have been reported to confer protection against P. aeruginosa infections in a mouse model (Hughes and Gilleland 1995). Liu et al. demonstrated that the recombinant OmpC and OmpF proteins from E. coli stimulated strong immunoglobulin G (IgG) antibody responses, and provided 62.5 and 87.5% protection against E. coli PCN033, respectively (Liu et al. 2012). Immunization with OmpF of Yersinia pseudotuberculosis not only resulted in production of high-avidity antibodies, but also stimulated bactericidal activity of peritoneal macrophages (Sidorova et al. 2014). Sharma et al. suggested that the OmpF epitope (66–80) in fusion with a carrier protein is a promising vaccine candidate against A. hydrophila (Sharma and Dixit 2015).
In the present study, the OmpF protein clusters in E. coli, Shigella and Salmonella were obtained from UniProtKB database and the homology was analyzed. As a conservative protein, the ompF gene was cloned from the genomic DNA of E. coli CVCC 1515 and expressed in E. coli BL21 (DE3) by the auto-induction method. After purification by Ni2+-NTA affinity chromatography, the recombinant OmpF (rOmpF) was used as an antigen to immunize mice. The protection efficiency of rOmpF vaccine was evaluated against the pathogenic E. coli CVCC 1515 and CICC 21530 (O157:H7) strains in vitro and in vivo.
Materials and methods
Bacterial strains and plasmids
Strains of E. coli CVCC 1515, Salmonella enteritidis CVCC 3377 and Salmonella pullorum CVCC 503 were purchased from the China Veterinary Culture Collection Center (CVCC) (Beijing, China). Shigella dysenteriae CMCC 51252 and Shigella flexneri CMCC 51571 were purchased from the National Center for Medical Culture Collection (CMCC) (Beijing, China). E. coli CICC 21530 (O157:H7), Pseudomonas aeruginosa CICC 10419 and CICC 21630 strains were purchased from the China Center of Industrial Culture Collection (CICC) (Beijing, China). E. coli DH5α and BL21 (DE3) strains were purchased form TransGen Biotech Co., Ltd. (Beijing, China). The pMD™ 19-T Simple and pET-28a(+) vectors with His tags were obtained from TaKaRa Biotechology Co., Ltd. (Dalian China) and Novagen (America), respectively.
Cloning of the ompF gene
The primer pairs of ompF-fw-BamHI: 5′-GGATCCGCAGAAATATATAACAAAGATGGC-3′ and ompF-rev-XhoI: 5′-CTCGAGTTAGAACTGATAAACGATACCCACA-3′ were designed according to the sequence of the ompF gene in E. coli UMNK88 (GenBank Accession No. CP002729.1) using Primer Premier 5.0. Genomic DNA was extracted from E. coli CVCC 1515 using a TIANamp Bacteria DNA Kit (Tiangen Biotech, Beijing, China) following the manufacturer’s instructions and used as a PCR template. The ompF gene was amplified and cloned into the pMD19-T Simple vector. The resultant positive pMDompF plasmid was isolated using the TIANprep Mini Plasmid Kit (Tiangen Biotech, Beijing, China), and digested with BamHI and XhoI (NEB, Beijing, China). The digested fragment was inserted into the pET-28a(+) vector digested with same enzymes, and transformed into E. coli BL21 (DE3). The positive transformants were confirmed by colony PCR and DNA sequencing, respectively.
Homological analysis of OmpF
After obtaining the ompF gene and amino acid sequence of E. coli CVCC 1515, the Basic Local Alignment Search Tool (BLAST) was used to find the local similarity between sequences of OmpF in typical E. coli strains (CICC 21530 (O157:H7), K12 and BL21). Then the similar protein clusters of E. coli, Shigella and Salmonella in the data base of uniprot uniref100 (http://www.uniprot.org/blast/) were also obtained for homology searches. MEGA5.1 software was used for the construction of a phylogenetic tree and the clusters whose size were less than 4 (E. coli) or 3 (Shigella and Salmonella) were omitted.
Expression and purification of the rOmpF protein
The positive transformants were cultured for 24 h at 30 °C in ZYM-5052 auto-inducing media (300 mL in the 1 L shaking flask, 100 μg/mL kanamycin) (Studier 2005; Guan et al. 2015). The positive transformant was cultured in LB medium on a platform shaker (37 °C, 250 rpm) to an optical density at 600 nm (OD600 nm) of 0.4–0.6. Cells were inoculated to auto-induction media (1% inoculum density) and cultured for 24 h on a platform shaker (37 °C, 300 rpm). 1 mL of cultured cells were collected at 4, 6, 8, 10, 12, 14, and 24 h respectively by centrifugation (8000×g, 2 min), and analyzed by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). After 24 h of auto-induction, the cultured cells were harvested by centrifugation (5000×g, 30 min), resuspended in 50 mM Tris–HCl buffer (pH7.9, containing 5 mg of lysozyme and 5 μL of DNaseI type IV stock/g cell paste), and sonicated for 5−6 min with an Ultrasonic Crasher Noise Isolating Chamber (SCIENTZ, Ningbo Science Biotechnol Co., Ltd., China) on ice. The insoluble fractions of the cells were collected by centrifugation (14,000×g, 20 min), washed twice in 50 mM Tris–HCl buffer [pH 7.9, containing 1.5% (v/v) lauryldimethylamine oxide (LDAO)], and suspended in 10 mM Tris–HCl buffer [pH 7.5, containing 1 mM ethylenediamine tetraacetic acid (EDTA) and 8 M urea]. After centrifugation (14,000×g, 20 min), the supernatant was added into 20 mM Tris–HCl buffer [pH 7.9, containing 1 M NaCl and 5% (v/v) LDAO]. The solution was dialyzed in 20 mM Tris–HCl buffer [pH 7.9, containing 0.5 M NaCl and 0.1% (v/v) LDAO].
The rOmpF protein was purified by Ni2+-NTA affinity chromatography and refolded according to the previous methods (Guan et al. 2015; Saleem et al. 2012). Briefly, the cell lysis solution was loaded onto a Ni2+-nitriloacetate (NTA) resin column (QIAGEN, Germany), which was pre-equilibrated with 20 mM Tris–HCl buffer [pH 7.9, containing 0.5 M NaCl, 0.1% (v/v) LDAO and 40 mM imidazole]. The column was washed with 20 mM Tris–HCl buffer [pH 7.4, containing 0.5 M NaCl, 0.1% (v/v) LDAO and 500 mM imidazole]. rOmpF was then desalted with 20 mM Tris–HCl buffer [pH 7.4, containing 150 mM NaCl and 0.1% (v/v) LDAO] using a HiPrep 26/10 desalting column. All protein elutions were analyzed by 12% SDS-PAGE. The purity and yield of rOmpF protein was calculated by the Gel-Pro Analyzer™ version 6.3 (Media Cybernetics). The purified rOmpF protein was lyophilized in a freeze dryer (ALPHA 1-2 LD plus, Christ, Germany).
Mouse immunization and challenge
Forty female SPF BALB/c mice, 6–8 weeks old, were purchased from Vital River, Beijing, China. The mice were immunized with rOmpF (20 mice) or PBS (control, 20 mice) according to the previous method reported by Reddy et al. (2010). The first injection solution consisted of 25 μg of rOmpF in sterile PBS (75 μL) and complete Freund’s adjuvant (25 μL) (Sigma-Aldrich, Inc.). Mice were hypodermically injected with antigen mixture (100 μL/mouse). The mice in the control group were immunized with PBS instead of rOmpF. Subsequent two injections containing 25 μg of rOmpF in sterile PBS (75 μL) and incomplete Freund’s adjuvant (25 μL) (Sigma-Aldrich, Inc.) were given every 2 weeks. Five days after each immunization, 10 mice were bled from the tail vein, and the serum was isolated and stored at −20 °C until use.
Two weeks after the second immunization, all the mice (40) were randomly divided into four groups as follows: (i) 10 rOmpF-immunized mice (group 1) and 10 PBS-immunized mice (group 2, control) were challenged with 109 colony forming unit (CFU) E. coli CVCC 1515 (1 mL) by intraperitoneal injection; (ii) 10 rOmpF-immunized mice (group 3) and 10 PBS-immunized mice (group 4, control) were challenged with 1010 CFU E. coli CICC 21530 (O157:H7) (0.2 mL) by gastric tube. The mortality of mice and E. coli in fecal shedding of control and rOmpF immunized mice was recorded daily for 7 days.
The animal protocol for the present study was approved by the Animal Care and Use Committee of the Feed Research Institute, Chinese Academy of Agricultural Sciences (Beijing, China), and all mice involved were cared for in accordance with the institutional guidelines from the above Committee.
Western blotting analysis of rOmpF
The SDS-PAGE was performed by loading purified rOmpF protein (about 0.1 μg) in gel for 120 min at 80 V. Subsequently, the protein was transferred to the PVDF membrane. Followed by blocking overnight with 5% BSA in TBST (25 mM Tris, 150 mM NaCl, and 0.05% (v/v) Tween-20, pH 7.4) at 4 °C, the PVDF membrane was washed three times with TBST and then incubated with the rOmpF sera (1:5000) for 2 h at room temperature. After another washing step, the membrane was incubated with secondary antibodies (Beijing CWBIO Co., Ltd.) at a dilution of 1:5000 for 2 h at room temperature. Finally, the bands were stained using BCIP/NBT solution (Beijing CWBIO Co., Ltd.) as substrate.
Detection of specific antibodies by the indirect enzyme-linked immunosorbent assay (iELISA)
The titers or capacities of antisera against rOmpF and bacteria (E. coli CVCC 1515, E. coli CICC 21530, S. dysenteriae CMCC 51252, S. flexneri CMCC 51571, S. enteritidis CVCC 3377, S. pullorum CVCC 503, P. aeruginosa CICC 21630, and P. aeruginosa CICC 10419) were measured by iELISA (Guan et al. 2015; Hu et al. 2010). rOmpF was dissolved in coating buffer (pH 9.6, 0.015 M sodium carbonate, 0.035 M sodium bicarbonate). The 96-well plates were coated with 2 μg/mL of the rOmpF solution or 106 CFU/mL bacteria solution (each well 100 μL), incubated overnight at 4 °C, and washed four times with 0.01 M PBS (containing 0.05% Tween 20). The plates were blocked for 2 h at 37 °C by adding 0.01 M PBS (containing 5% BSA), washed three times, and then incubated with serial dilutions of mice serum at 37 °C for 1.5 h. After washing as above, 100 μL of horseradish peroxidase (HRP) conjugated goat anti-mouse IgG (1:5000) was added into each well and incubated for 30 min at 37 °C. The plates were washed three times again. 100 μL of 3, 3′, 5, 5′-tetramethylbenzidine (TMB) was added to each well and incubated for 20 min in the dark at room temperature. Finally, the color reaction was stopped by adding 2 M H2SO4 (50 μL/well). The absorbance of each well at 450 nm was determined by an automatic ELISA plate reader (Perlong Medical, Beijing). The result was considered as positive when the ratio of the test group and negative control group was greater than 2.1 (Lunin et al. 2009; Xu et al. 2011).
Opsonophagocytosis assay
Murine peritoneal macrophages cells were isolated as previously described and adjusted to 4 × 106 CFU/mL (Guan et al. 2015; Zhang et al. 2008). Briefly, after incubation with 100 μL of anti-rOmpF sera or anti-PBS sera at 37 °C for 30 min, 400 μL of E. coli CVCC 1515 cells (4 × 106 CFU/mL) were incubated with 500 μL of macrophage suspension and 100 μL of baby rabbit complement (Cedarlane, Hornby, ON, Canada) at 30 °C for 1 h. Macrophages were then lysed by adding sterile water into the mixture (Rennermalm et al. 2001; Xu et al. 2011; Gressler et al. 2016). The mixture was then serially diluted for the plate count. The bacterial killing rate was calculated as the formula: [1− (number of bacteria recovered in the presence of phagocytes/number of bacteria recovered in the absence of phagocytes)] × 100% (Liu et al. 2012). Data from three independent experiments are expressed as percentage (mean ± standard deviation) of killed bacteria.
Serum bactericidal assay
The serum bactericidal test was carried out according to pervious protocols with some modification (Maslanka et al. 1997; Marzoa et al. 2012). 12.5 μL of E. coli CVCC 1515 cells (5–6 × 103 CFU/mL), 50 μL of serial twofold mouse serum, and 25 μL of baby rabbit complement were added into each well of a 96-well cell culture plate. Each group contained (i) bacteria (12.5 μL) + complement (25 μL) + immunized serum or unimmunized serum (50 μL, eightfold serially diluted in PBS) (complement-dependent manner) and (ii) bacteria + heat-inactivated complement + immunized serum or unimmunized serum (complement-independent manner). The plates were incubated at 37 °C for 60 min, and 20 μL of samples from each well were plated onto LB agar. After overnight incubation, the plates were counted.
All statistical analyses were performed using SPSS version 22.0. Differences were considered significant at p < 0.05.
Results
Homology analysis of OmpF
The BLAST result showed that proportion of the OmpF protein sequences in E. coli was up to 100% (Fig. 1A). The amino acid sequences of OmpF from E. coli CVCC 1515 shared 99, 100 and 100% identity among E. coli CICC 21530 (O157:H7), K12 and BL21, respectively. In order to predict the potential of OmpF porin as a universal vaccine against gram-negative bacteria, the homology analysis of OmpF protein clusters was carried out by MEGA5.1 software. The OmpF from E. coli CVCC 1515 was identical to that of C9E725 (E. coli) and P02931 (Shigella). It also shares a high identity (90–100%) with about half of the E. coli (46.7%) (Fig. 1B) and Shigella (52.8%) strains (Fig. 2). However, the identity with Salmonella strains was lower (59.5–85.1%), 67.2% of Salmonella protein clusters only share an identity of 62.4% with E. coli CVCC 1515 OmpF protein (Fig. 2). The results showed that the OmpF protein is highly conserved among E. coli and Shigella, and with a certain degree of homology with Salmonella.
Homology and phylogenetic analysis of the OmpF protein. A Proportion of the OmpF protein sequences in E. coli (%); B phylogenetic analysis of the OmpF protein in E. coli (omit the clusters less than 4 in size). Black symbol indicates the protein clusters which were identical to that of E. coli CVCC 1515 OmpF
Expression and purification of the rOmpF protein
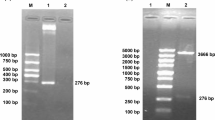
The 1023 bp ompF gene was successfully amplified from the genomic DNA of E. coli CVCC 1515 (Fig. 3A), and cloned into pMD19-T Simple vector. After being digested with BamHI and XhoI, the ompF fragment was inserted into a pET-28a(+) vector, and the resultant plasmid was named as pET-28a(+)-ompF. DNA sequencing result showed that the open reading frame of the ompF gene was composed of 1023 nucleotides and encoded a protein (341 amino acids) with a predicted molecular weight of 37.51 kDa. ompF shares 100% nucleotide sequence identity with that of the published ompF gene (GenBank Accession No. JQ886179.1), and the pET-28a(+)-ompF plasmid was successfully constructed.
Cloning, expression, purification and Western blotting analysis of OmpF. aA PCR products of the ompF gene. Lane M1 Trans DNA Marker II (1500, 900, 700, 500, 400, 200 and 100 bp); lane 1 ompF PCR products. Lane 2 the recombinant pET-28a(+)-ompF plasmid. Lane 3 the recombinant pET-28a(+)-ompF plasmid digested with BamHI and XhoI; lane M2 Trans5K DNA Marker (5000, 3000, 2000, 1500, 1000, 800, 500 and 300 bp); B schematic representation of the pET-28a(+)-ompF plasmid; C SDS-PAGE analysis of the expression of rOmpF in auto-inducing media at different time. 1 mL of cultured cells were collected at 4, 6, 8, 10, 12, 14, and 24 h, respectively. Lane M protein marker (94.4–14.4 kDa); lane 1 4 h; lane 2 6 h; lane 3 8 h; lane 4 10 h; lane 5 12 h; lane 6 14 h; lane 7 24 h; D SDS-PAGE analysis of the purified rOmpF protein. Lane M 5 µL of protein marker (94.4–14.4 kDa); lane 1 total proteins after auto-induction of E. coli BL21 (DE3) containing pET-28a(+)-ompF (15 µL of cell lysis); lane 2 precipitation containing inclusion body after sonication and centrifugation (15 µL of renaturation solution); Lane 3 flowthrough (15 µL of elution solution); Lane 4 the eluent washed with 70% elution buffer (15 µL of the purified OmpF protein); E Western blotting analysis of the OmpF protein. Lane M, protein marker; lane 1, the OmpF protein
As shown in Fig. 3B, the OmpF protein fused with the 6× His tag (353 amino acids) was successfully expressed in E. coli BL21 (DE3) by auto-inducing for 24 h at 30 °C. The molecular weight of rOmpF with an N-terminal 6× His tag was approximately 40 kDa, which matches with the expected size of 38.83 kDa. After purification by His-tag Ni affinity chromatography, the purity of the rOmpF protein was about 90%, and approximate 3 mg of the purified protein was obtained from 1 L of cultures (Fig. 3D).
Immune response to the rOmpF vaccination
The results of Western blotting showed that the purified OmpF proteins had a main band with the size of 40 kDa (Fig. 3E). It indicated that the anti-OmpF serum mainly bound with OmpF and suggest that OmpF is an immunogenic protein.
The anti-rOmpF sera titer was also tested by an indirect enzyme-linked immunosorbent assay (iELISA) using rOmpF as antigen. After the first immunization, the titers of anti-rOmpF against rOmpF were 1:100–1:300, and rised sharply to 1:27,000–1:240,000 after the second and third immunization (Fig. 4A). Antisera were used for following experiments after the third immunization. The antibody titers of the control mice immunized with PBS-adjuvant were only 1:30–1:100.
Serum responses in mice immunized with rOmpF. Mice were immunized with rOmpF at day 0 and boosted at the 3rd-week and 5th-week respectively, and sera were collected at 5-day intervals after each immunization. iELISA was used to detect the antibody titers. Results were shown as mean ± SD for 10 mice. A Titers of the anti-rOmpF sera against rOmpF. Asterisk indicates a significant difference between the OmpF immunized group and the control group (p < 0.05). Different lower case letters indicate a significant difference between each immunized (p < 0.05); B cross-reaction properties of the anti-rOmpF sera against E. coli, Shigella, Salmonella and Pseudomonas. Y axis indicates the optical density (OD 450) values of antibody response with different bacteria in iELISA
Additionally, the capacities for the anti-rOmpF sera binding to bacterial cells were performed among the strains of E. coli CVCC 1515, E. coli CICC 21530 (O157:H7), S. dysenteriae CMCC 51252, S. flexneri CMCC 51571, S. enteritidis CVCC 3377, S. pullorum CVCC 503, P. aeruginosa CICC 21630, and P. aeruginosa CICC 10419 in vitro. The antibody titers against E. coli, Shigella and Salmonella were 1:27,000, 1:27,000 and 1:9000, respectively. The OD values of 9000-fold diluted antiserum response in iELISA were shown in Fig. 4B. Antibody response of anti-rOmpF sera against E. coli, Shigella and Salmonella were significantly higher than the anti-PBS group. However, there was no significant discrepancy between the anti-rOmpF sera and the PBS immunized sera against P. aeruginosa (Fig. 4B). This indicated that the rOmpF sera strongly reacted with E. coli, Shigella and Salmonella strains, but not with P. aeruginosa strains.
Opsonophagocytosis and serum bactericidal test in vitro
Sera obtained from mice immunized with rOmpF plus adjuvant or PBS plus adjuvant were analyzed for their ability to promote opsonophagocytic killing of E. coli CVCC 1515 by murine peritoneal macrophages. The opsonophagocytosis result showed that only 11.04 ± 3.90% of E. coli CVCC 1515 could be killed in the control group (the anti-PBS serum), but 72.21 ± 11.39% of the bacteria were killed in the anti-rOmpF serum group, which suggested that the antibodies against rOmpF were effective for mediating opsonophagocytosis of E. coli (Fig. 5A).
Phagocytosis and bactericidal activity of the macrophages modulated by the anti-rOmpF sera in vitro. A Effect of the anti-rOmpF sera on opsonophagocylic killing of E. coli by murine macrophages. Data are expressed as percentage (mean ± SD) of killed bacteria; B bactericidal activity of serum complement on E. coli. Escherichia coli CVCC 1515 was incubated for 1 h with the anti-rOmpF sera and complement (or heat-inactivated complement) (the rOmpF+ adjuvant group), the anti-PBS sera and complement (or heat-inactivated complement) (the PBS+ adjuvant group), and PBS and complement (or heat-inactivated complement) (the negative control group), respectively. Viable bacteria were counted, and data are expressed as mean ± SD. Lower case letters indicate a significant difference between two groups (p < 0.05)
The bactericidal efficiency of the complement pathway was further determined. As shown in Fig. 5B, the viable count of E. coli was lower in the anti-rOmpF sera group (rOmpF + adjuvant) than in the anti-PBS sera (PBS + adjuvant) and negative control groups. In the presence of both the anti-rOmpF sera and complement, the minimum number of bacterial cells were observed, indicating complement-mediated opsonic activity of the anti-rOmpF sera for E. coli in vitro.
Protection efficacy after immunization with rOmpF in vivo
The immune protection efficiency of rOmpF was investigated in a murine model. After being challenged with E. coli CVCC 1515 within 12–36 h, the survival ratio of the rOmpF-immunized mice was decreased from 60 to 40%, but kept to 30% at 48 h postchallenge, which was higher than that of the PBS-immunized mice (control group) (Fig. 6A).
Protection efficacy after immunization with rOmpF against E. coli CVCC 1515 and CICC 21530 (O157:H7) in vivo. After 2 weeks of the final immunization, the mice were challenged with E. coli CVCC 1515 by intraperitoneal injection (A) or E. coli CICC 21530 (O157:H7) by gastric tube (B), and observed for 7 days after challenge. Data are expressed as mean ± SD. A The survival ratio of the mice immunized with rOmpF or PBS; rOmpF+ adjuvant: group 1; PBS+ adjuvant: group 2, control; B fecal shedding of the mice immunized with rOmpF or PBS; rOmpF+ adjuvant: group 3; PBS+ adjuvant: group 4, control. Asterisk indicates a significant difference between two groups (p < 0.05)
In our study, mice were administered orally with E. coli CICC 21530 (O157:H7), and E. coli in fecal shedding was examined by plate counting. As shown in Fig. 6B, a significant decrease in fecal shedding of E. coli was observed at 2 days postchallenge in the rOmpF-immunized mice and the PBS-immunized mice, but there was no significant difference in the bacterial count in fecal shedding between the immunized and control group at the subsequent time point. The duration of the E. coli shedding in mice lasted no longer than 1 day.
Discussion
Considering the diversity of pathogenic E. coli serotype, we took more attention on developing a versatile vaccine that provides heterologous protection for E. coli, even other gram-negative pathogens such as Salmonella or Shigella. Lots of diarrheal illness were co-infected by these strains (O’Ryan et al. 2015). As we all known, porins exist most frequently as trimers and the sequence homology among porins of several genera such as Escherichia and Neisseria has shown a highly conserved nature (Yadav et al. 2014). Therefore, as a porin of E. coli, the OmpF protein was selected to be a candidate vaccine in our study.
Previously, we have studied the OmpA and OmpC of E. coli, the result showed that they all shared a high identity with some typical E. coli, Salmonella and Shigella strains, and possessed satisfied immunogenicity (Guan et al. 2015; Wang et al. 2015). Therefore, in this study, the homology of OmpF protein clusters in these strains were further analyzed, thus leading to a better manifestation of the sequence conservation. The result implies that OmpF may be a shared antigen among E. coli and Shigella strains and these consensus regions should be helpful in designing universal vaccines against a broad range of gram-negative pathogens.
The OmpF protein from E. coli has antigenic epitopes located on several extracellular loops, indicating that it may have some immune properties (Liu et al. 2012; Klebba et al. 1990; Fourel et al. 1993). In our study, the protein was expressed in the formation of inclusion bodies (IBs). In order to fully expose extracellular epitopes and then obtain better immunogenic response for the whole cell, the rOmpF was further resolubilized by denaturant (urea), and refolded by the assistant of detergent (LDAO) which could simulate the natural environment (Saleem et al. 2012).
Previous studies demonstrated that antigen proteins with higher purity, such as M2 (>90%), the culture filtrate proteins (CFP) (>96%), PfEBA-175II F2 (>95%) and Mtb72F (>98%) displayed good immunogenicity (Frace et al. 1999; Roberts et al. 1995; Zhang and Pan 2005; Skeiky et al. 2004). However, some recombinant proteins such as Bm95, E6/E7, OprF-OprI and EspA-Stx2A1 with low purity of 80–90% also display the effective immunogenicity (García-García et al. 2000; De Bruijn et al. 1998; von Specht et al. 1995; Cheng et al. 2009). In comparison, the purity of rOmpF (90%) obtained in this study falls within the scope of the above antigen proteins and meets the purity requirement of vaccine preparation.
As expected, BALB/c mice vaccinated with the renatured and purified rOmpF protein elicited a significant immunogenic response, which was consistent with the previous report (Sidorova et al. 2014). The iELISA results showed that the antiserum not only had high affinity against rOmpF (1: 240,000 dilution) but also against the whole cell (1:27,000 dilution) (Fig. 4). It was demonstrated that the rOmpF extracellular epitopes especially the conformations can restore after renaturation, some of the antibody against them can specifically recognize the bacteria. Therefore, the protein got the prerequisite for being used as a subunit vaccine. Moreover, significant antigenic cross-reactivity responses to Shigella and Salmonella strains were observed which is in accordance with that of homology analysis, indicating that rOmpF may be a potential candidate for a universal vaccine.
It was well known that antibody could mediate phagocytosis of organisms when the antibody constant region was recognized by the Fc receptors of phagocytes such as neutrophils and macrophages. Additionally, the Fc regions of antibodies maybe bind and activate complement proteins that can directly cause bacterial death (Clemens et al. 2003). All the bactericidal function implementation depends on the affinity of the Fab region with the antigenic components of bacterial pathogens, which has been proved by the iELISA results. In present study, a significant bactericidal effect was achieved in the opsonophagocytosis assay and the serum bactericidal assay (Fig. 5), indicating a phagocytosis and classical complement pathway killing mediated by the antiserum. It was consistent with the results of the previous studies (Liu et al. 2012; Wang et al. 2015; Marzoa et al. 2012; Seder and Mascola 2003). However, the role of OmpF in macrophage adherence and cytokine production needs to be further evaluated.
The porin protein not only has the ability to elicit a host immune response, but also protect the host against infection (Secundino et al. 2006). Above results demonstrated that rOmpF induced a strong immune response, but conferred no significant protection against E. coli CVCC 1515 in BALB/c mice, which was consistent with previous results reported by Toobak et al. (2013). Meenakshi et al. referred to an irrelevance of the increased antibody level as an indicator of a protective effect against Salmonella (Meenakshi et al. 1999). There are several possible reasons for lower protection in this study: (i) the characteristic of a prokaryotic expressed system cannot provide the glycosylation, which may affect the antigenicity; (ii) the native active configuration of rOmpF after denaturation and renaturation could be altered, formation of a partially folded or misfolded conformation, which may affect the immunogenicity (Dowling et al. 2007); and (iii) the anti-rOmpF sera did not reach or recognize the OmpF protein on the outer membrane of live E. coli due to the presence of lipopolysaccharide (LPS), pili, flagella, and other porin proteins, which can mask the OmpF protein (Okamura et al. 2012). It was reported that OmpF and OmpC are tightly bound to the LPS (Fourel et al. 1993). This viewpoint was supported by other researchers, who suggested that the potential limitation of the interaction between an antibody and OMPs of live bacteria by O-chain of LPS (Singh et al. 2003). Tarkka et al. also pointed out that it is difficult for the anti-OMP sera to reach OMP on live N. meningitides (Tarkka et al. 1989). Further increasing the yields of correctly folded rOmpF by addition of hydrophobic agents such as ethanol, DMSO (dimethylsulfoxide), and acetonitrile to the renaturation buffer and further study using different vaccination methods, including antigen dose, times, adjuvant and more mice need to be elucidated.
OMPs in gram-negative bacteria have been reported to induce the host humoral responses, and in turn inhibit postchallenge bacterial colonization (Okamura et al. 2012). Therefore, a preliminary evaluation of the wide spectrum of the rOmpF vaccine was carried out. Though the high bacterial count in fecal shedding was only last 1 day, which is shorter than that of the previous study (Fan et al. 2012), this difference may be due to the variation of strains that causes the antigenic variation. However, the microbe amount of the immunized group was significantly decreased before excreting from the bowel (Fig. 6B). It is suggested that by induction of a strong immune response, rOmpF conferred a cross protection against E. coli in BALB/c mice to a certain degree.
In conclusion, the expressed OmpF protein induced an effective immune response, including the high antibody titer and high affinity of the antibody that binds to the bacterial surface. Additionally, rOmpF increased the capacity of macrophages to clear bacteria, and it was found for the first time that the anti-rOmpF sera had a significant cross-reaction capacity against Escherichia, Shigella, and Salmonella strains in vitro. Although rOmpF only provide a weak protection against E. coli and reduce E. coli in fecal shedding in vivo, we believe OmpF in native forms might be suitable as a vaccine candidate.
Abbreviations
- OmpF:
-
outer membrane protein F
- rOmpF:
-
recombinant OmpF
- OMPs:
-
outer membrane proteins
- IgG:
-
immunoglobulin G
- iELISA:
-
indirect enzyme-linked immunosorbent assay
- LPS:
-
lipopolysaccharide
- DMSO:
-
dimethylsulfoxide
- CVCC:
-
China Veterinary Culture Collection Center
- CMCC:
-
National Center for Medical Culture Collection
- CICC:
-
China Center of Industrial Culture Collection
- EDTA:
-
ethylenediamine tetraacetic acid
- HRP:
-
horseradish peroxidase
- TMB:
-
3, 3′, 5, 5′-tetramethylbenzidine
- CAAS:
-
Chinese Academy of Agricultural Sciences
- IBs:
-
inclusion bodies
- SDS-PAGE:
-
sodium dodecyl sulfate polyacrylamide gel electrophoresis
References
Cheng Y, Feng Y, Luo P, Gu J, Yu S, Zhang WJ, Liu YQ, Wang QX, Zou QM, Mao XH (2009) Fusion expression and immunogenicity of EHEC EspA-Stx2Al protein: implications for the vaccine development. J Microbiol 47:498–505
Clemens JD, Koo H-W (2003) Trial design for vaccines. Part B. Phase 3 studies of vaccines. In: Bloom BR, Lambert P-H (eds) The Vaccine Book. New York: Elsevier, pp 95–117
De Bruijn ML, Schuurhuis DH, Vierboom MP, Vermeulen H, de Cock KA, Ooms ME, Ressing ME, Toebes M, Franken KL, Drijfhout JW, Ottenhoff TH, Offringa R, Melief CJ (1998) Immunization with human papillomavirus type 16 (HPV16) oncoprotein-loaded dendritic cells as well as protein in adjuvant induces MHC class I-restricted protection to HPV16-induced tumor cells. Cancer Res 58:724–731
Dowling W, Thompson E, Badger C, Mellquist JL, Garrison AR, Smith JM, Paragas J, Hogan RJ, Schmaljohn C (2007) Influences of glycosylation on antigenicity, immunogenicity, and protective efficacy of ebola virus GP DNA vaccines. J Virol 81:1821–1837
Fan HY, Wang L, Luo J, Long BG (2012) Protection against Escherichia coli O157: H7 challenge by immunization of mice with purified Tir proteins. Mol Biol Rep 39:989–997
Fourel D, Mizushima S, Bernadac A, Pagès JM (1993) Specific regions of Escherichia coli OmpF protein involved in antigenic and colicin receptor sites and in stable trimerization. J Bacteriol 175:2754–2757
Frace AM, Klimov AI, Rowe T, Black RA, Katz JM (1999) Modified M2 proteins produce heterotypic immunity against influenza A virus. Vaccine 17:2237–2244
García-García JC, Montero C, Redondo M, Vargas M, Canales M, Boue O, Rodríguez M, Joglar M, Machado H, González IL, Valdés M, Méndez L, de la Fuente J (2000) Control of ticks resistant to immunization with Bm86 in cattle vaccinated with the recombinant antigen Bm95 isolated from the cattle tick, Boophilus microplus. Vaccine 18:2275–2287
Gressler LT, Bordin AI, McQueen CM, Cohen ND, de Vargas AC (2016) Chloroquine inhibits Rhodococcus equi replication in murine and foal alveolar macrophages by iron-starvation. Vet Microbiol 188:16–24
Guan Q, Wang X, Wang X, Teng D, Mao R, Zhang Y, Wang J (2015) Recombinant outer membrane protein A induces a protective immune response against Escherichia coli infection in mice. Appl Microbiol Biotechnol 99:5451–5460
Hu C, Gong R, Guo A, Chen H (2010) Protective effect of ligand-binding domain of fibronectin-binding protein on mastitis induced by Staphylococcus aureus in mice. Vaccine 28:4038–4044
Hughes EE, Gilleland HE Jr (1995) Ability of synthetic peptides representing epitopes of outer membrane protein F of Pseudomonas aeruginosa to afford protection against P. aeruginosa infection in a murine acute pneumonia model. Vaccine 13:1750–1753
Johnson AM, Kaushik RS, Hardwidge PR (2010) Disruption of transepithelial resistance by enterotoxigenic Escherichia coli. Vet Microbiol 141:115–119
Khushiramani RM, Maiti B, Shekar M, Girisha SK, Akash N, Deepanjali A, Karunasagar I, Karunasagar I (2012) Recombinant Aeromonas hydrophila outer membrane protein 48 (Omp48) induces a protective immune response against Aeromonas hydrophila and Edwardsiella tarda. Res Microbiol 163:286–291
Klebba PE, Benson SA, Bala S, Abdullah T, Reid J, Singh SP, Nikaido H (1990) Determinants of OmpF porin antigenicity and structure. J Biol Chem 265:6800–6810
Liu C, Chen Z, Tan C, Liu W, Xu Z, Zhou R, Chen H (2012) Immunogenic characterization of outer membrane porins OmpC and OmpF of porcine extraintestinal pathogenic Escherichia coli. FEMS Microbiol Lett 337:104–111
Lunin VG, Sharapova NE, Tikhonova TV, Poletaeva NN, Galushkina ZM, Aksenova EI, Grabko VI, Velikodvorskaya GA, Lavrova NV, Anan’ina YV (2009) In vivo study of the immunogenic properties of the recombinant cellulose-binding domain of Anaerocellum thermophilum. Mol Genet Microbiol Virol 24:24–31
Marzoa J, Sánchez S, Costoya L, Diéguez-Casal E, Freixeiro P, Brookes C, Allen L, Taylor S, Gorringe AR, Ferreirós CM, Criado MT (2012) Induction of immune responses by purified outer membrane protein complexes from Neisseria meningitidis. Vaccine 30:2387–2395
Maslanka SE, Gheesling LL, Libutti DE, Donaldson KB, Harakeh HS, Dykes JK, Arhin FF, Devi SJ, Frasch CE, Huang JC, Kriz-Kuzemenska P, Lemmon RD, Lorange M, Peeters CC, Quataert S, Tai JY, Carlone GM (1997) Standardization and a multilaboratory comparison of Neisseria meningitidis serogroup A and C serum bactericidal assays. The Multilaboratory Study Group. Clin Diagn Lab Immunol 4:156–167
Maynard C, Fairbrother JM, Bekal S, Sanschagrin F, Levesque RC, Brousseau R, Masson L, Larivière S, Harel J (2003) Antimicrobial resistance genes in enterotoxigenic Escherichia coli O149: K91 isolates obtained over a 23-year period from pigs. Antimicrob Agents Chemother 47:3214–3221
Meenakshi M, Bakshi CS, Butchaiah G, Bansal MP, Siddiqui MZ, Singh VP (1999) Adjuvanted outer membrane protein vaccine protects poultry against infection with Salmonella enteritidis. Vet Res Commun 23:81–90
Noamani BN, Fairbrother JM, Gyles CL (2003) Virulence genes of O149 enterotoxigenic Escherichia coli from outbreaks of postweaning diarrhea in pigs. Vet Microbiol 97:87–101
O’Ryan M, Vidal R, del Canto F, Salazar JC, Montero D (2015) Vaccines for viral and bacterial pathogens causing acute gastroenteritis: part I: overview, vaccines for enteric viruses and Vibrio cholerae. Hum Vaccines Immunother 11:584–600
Okamura M, Ueda M, Noda Y, Kuno Y, Kashimoto T, Takehara K, Nakamura M (2012) Immunization with outer membrane protein A from Salmonella enterica serovar Enteritidis induces humoral immune response but no protection against homologous challenge in chickens. Poult Sci 91:2444–2449
Osman KM, Marouf SH (2014) Comparative dendrogram analysis of OMPs of Salmonella enteric serotype Enteritidis with Typhimurium, Braendurup and Lomita isolated from pigeons. Int J Adv Biotechnol Res 2:952–960
Pal S, Theodor I, Peterson EM, de la Maza LM (1997) Immunization with an acellular vaccine consisting of the outer membrane complex of Chlamydia trachomatis induces protection against a genital challenge. Infect Immun 65:3361–3369
Perna NT, Plunkett G 3rd, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Pósfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR (2001) Genome sequence of enterohaemorrhagic Escherichia coli O157: H7. Nature 409:529–533
Reddy ST, Ge X, Miklos AE, Hughes RA, Kang SH, Hoi KH, Chrysostomou C, Hunicke-Smith SP, Iverson BL, Tucker PW, Ellington AD, Georgiou G (2010) Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat Biotechnol 28:965–969
Rennermalm A, Li YH, Bohaufs L, Jarstrand C, Brauner A, Brennan FR, Flock JI (2001) Antibodies against a truncated Staphylococcus aureus fibronectin-binding protein protect against dissemination of infection in the rat. Vaccine 19:3376–3383
Roberts AD, Sonnenberg MG, Ordway DJ, Furney SK, Brennan PJ, Belisle JT, Orme IM (1995) Characteristics of protective immunity engendered by vaccination of mice with purified culture filtrate protein antigens of Mycobacterium tuberculosis. Immunology 85:502–508
Saleem M, Moore J, Derrick JP (2012) Expression, purification, and crystallization of neisserial outer membrane proteins. Methods Mol Biol 799:91–106
Secundino I, López-Macías C, Cervantes-Barragán L, Gil-Cruz C, Ríos-Sarabia N, Pastelin-Palacios R, Villasis-Keever MA, Becker I, Puente JL, Calva E, Isibasi A (2006) Salmonella porins induce a sustained, lifelong specific bactericidal antibody memory response. Immunology 117:59–70
Seder RA, Mascola JR (2003) Basic immunology of vaccine development. In: Bloom BR, Lambert PH (eds) The vaccine book. Academic Press, Boston, pp 51–72
Sharma M, Dixit A (2015) Identification and immunogenic potential of B cell epitopes of outer membrane protein OmpF of Aeromonas hydrophila in translational fusion with a carrier protein. Appl Microbiol Biotechnol 99:6277–6291
Sidorova OV, Khomenko VA, Portnyagina OY, Likhatskaya GN, Vakorina TI, Kim NY, Chistyulin DK, Solov’eva TF, Novikova OD (2014) Mutant OmpF porins of Yersinia pseudotuberculosis with deletions of external loops: structure–functional and immunochemical properties. Biochem Biophys Res Commun 445:428–432
Singh SP, Williams YU, Miller S, Nikaido H (2003) The C-terminal domain of Salmonella enterica serovar typhimurium OmpA is an immunodominant antigen in mice but appears to be only partially exposed on the bacterial cell surface. Infect Immun 71:3937–3946
Skeiky YA, Alderson MR, Ovendale PJ, Guderian JA, Brandt L, Dillon DC, Campos-Neto A, Lobet Y, Dalemans W, Orme IM, Reed SG (2004) Differential immune responses and protective efficacy induced by components of a tuberculosis polyprotein vaccine, Mtb72F, delivered as naked DNA or recombinant protein. J Immunol 172:7618–7628
Studier FW (2005) Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif 41:207–234
Tarkka E, Muotiala A, Karvonen M, Saukkonen-Laitinen K, Sarvas M (1989) Antibody production to a meningococcal outer membrane protein cloned into live Salmonella typhimurium aroA vaccine strain. Microb Pathog 6:327–335
Toobak H, Rasooli I, Talei D, Jahangiri A, Owlia P, Darvish Alipour Astaneh S (2013) Immune response variations to Salmonella enterica serovar Typhi recombinant porin proteins in mice. Biologicals 41:224–230
Vali L, Wisely KA, Pearce MC, Turner EJ, Knight HI, Smith AW, Amyes SG (2004) High-level genotypic variation and antibiotic sensitivity among Escherichia coli O157 strains isolated from two Scottish beef cattle farms. Appl Environ Microbiol 70:5947–5954
von Specht BU, Knapp B, Muth G, Bröker M, Hungerer KD, Diehl KD, Massarrat K, Seemann A, Domdey H (1995) Protection of immunocompromised mice against lethal infection with Pseudomonas aeruginosa by active or passive immunization with recombinant P. aeruginosa outer membrane protein F and outer membrane protein I fusion proteins. Infect Immun 63:1855–1862
Wang X, Guan Q, Wang X, Teng D, Mao R, Yao J, Wang J (2015) Paving the way to construct a new vaccine against Escherichia coli from its recombinant outer membrane protein C via a murine model. Process Biochem 50:1194–1201
Williams KM, Bigley EC 3rd, Raybourne RB (2000) Identification of murine B-cell and T-cell epitopes of Escherichia coli outer membrane protein F with synthetic polypeptides. Infect Immun 68:2535–2545
Wright JC, Williams JN, Christodoulides M, Heckels JE (2002) Immunization with the recombinant PorB outer membrane protein induces a bactericidal immune response against Neisseria meningitidis. Infect Immun 70:4028–4034
Xu H, Hu C, Gong R, Chen Y, Ren N, Xiao G, Xie Q, Zhang M, Liu Q, Guo A, Chen H (2011) Evaluation of a novel chimeric B cell epitope-based vaccine against mastitis induced by either Streptococcus agalactiae or Staphylococcus aureus in mice. Clin Vaccine Immunol 18:893–900
Yadav SK, Sahoo PK, Dixit A (2014) Characterization of immune response elicited by the recombinant outer membrane protein OmpF of Aeromonas hydrophila, a potential vaccine candidate in murine model. Mol Biol Rep 41:1837–1848
Zhang D, Pan W (2005) Evaluation of three Pichia pastoris-expressed Plasmodium falciparum merozoite proteins as a combination vaccine against infection with blood-stage parasites. Infect Immun 73:6530–6536
Zhang X, Gonçalves R, Mosser DM (2008) The isolation and characterization of murine macrophages. Curr Protoc Immunol. doi:10.1002/0471142735.im1401s83
Authors’ contributions
XW performed the immunizations and the animal experiments. QFG participated in the sequence alignment and preparing figures. XW and QFG drafted the manuscript, and XMW jointly wrote the manuscript. DT and RYM assisted with the mouse challenge experiments and “Materials and methods” section. YH served and participated in this work as professional technician. JHW and JHY participated in the design of the study and supervised overall experiments. All authors read and approved the final manuscript.
Acknowledgements
We thank all the colleagues at Gene Engineering Laboratory, Feed Research Institute, Chinese Academy of Agricultural Sciences for their technical assistance throughout this study.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Ethical approval and consent to participate
The animal protocol for the present study was approved by the Animal Care and Use Committee of the Feed Research Institute, Chinese Academy of Agricultural Sciences (Beijing, China), and all mice involved were cared for in accordance with the institutional guidelines from the above Committee.
Funding
This study was supported by the National Natural Science Foundation of China (Grant Nos. 31672456, 31572444, 31572445 and 31372346), the Project of the National Support Program for Science and Technology in China (Grant No. 2013BAD10B02), the Special Fund for Agro-scientific Research in the Public Interest in China (Grant No. 201403047), and the AMP Direction of the National Innovation Program of Agricultural Science and Technology in CAAS (CAAS-ASTIP-2013-FRI-02).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Wang, X., Teng, D., Guan, Q. et al. Escherichia coli outer membrane protein F (OmpF): an immunogenic protein induces cross-reactive antibodies against Escherichia coli and Shigella . AMB Expr 7, 155 (2017). https://doi.org/10.1186/s13568-017-0452-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13568-017-0452-8