Abstract
The circulation of carbapenemase-producing Escherichia coli (CPEC) in our society is a serious concern for vulnerable patients in nosocomial environments. However, the genomic epidemiology of the circulation of CPEC bacteria among companion animals remains largely unknown. In this study, epidemiological analysis was conducted using complete genome identification of CPEC ST410 isolates obtained from companion animals. To estimate the genomic distance and relatedness of the isolates, a total of 37 whole-genome datasets of E. coli ST410 strains were downloaded and comparatively analysed. As a result of the analysis, the genomic structure of the chromosomes and plasmids was identified, revealing the genomic positions of multiple resistance and virulence genes. The isolates in this study were grouped into the subclade H24/RxC, with fimH24, and substituted quinolone resistance-determining regions (QRDRs) and multiple beta-lactamases, including extended-spectrum β-lactamase (ESBL) and carbapenemase. In addition, the in silico comparison of the whole-genome datasets revealed unidentified ST410 H24/Rx subgroups, including either high pathogenicity islands (HPIs) or H21 serotypes. Considering the genetic variations and resistance gene dissemination of the isolates carried by companion animals, future approaches for preventive measurement must include the “One Health” perspective for public health in our society.
Similar content being viewed by others
Introduction
Nosocomial infections caused by carbapenemase-producing Escherichia coli (CPEC) are emerging as a major clinical threat. Dissemination of carbapenemase-producing bacteria among companion animals, which live close to humans in modern society, should be considered an urgent threat to public health [1, 2]. Carbapenem usage in animals is prohibited worldwide. However, the unauthorized usage of carbapenems is prevalent and not systematically monitored; thus, the dissemination of CPEC among companion animals is an unaddressed threat.
E. coli ST410, as an emerging opportunistic pathogen with both pathogenicity and resistance, is a successfully disseminating clone that spreads among humans, animals and the environment [3, 4]. It has been speculated that the E. coli ST410 strain first appeared in approximately the 1800s [5]. In the human clinical environment, carbapenemase-producing E. coli ST410 outbreaks have been reported in multiple countries, including Denmark [6, 7], Italy [8] and China [9]. In Germany, extended-spectrum β-lactamase (ESBL)-producing E. coli ST410 strains have been reported as circulating clones among humans, animals (wildlife and companion animals) and the environment [3, 10]. As emerging multidrug-resistant (MDR) pathogens, fluoroquinolone- and extended-spectrum cephalosporin (ESC)-resistant (i.e., B3/H24Rx) and carbapenem-resistant (i.e., B4/H24RxC) clades have been reported to be spreading in Europe and North America [5, 11].
In South Korea, a nationwide study investigated nosocomial strains isolated between 2011 and 2015 [12]. The investigation reported that Klebsiella pneumoniae carbapenemase-2 (KPC-2) and New Delhi metallo-β-lactamase-1 (NDM-1) were the dominant carbapenemase types in the Korean nosocomial environment. Thus far, ST410 strains have not been discovered in ongoing human investigations. To date, in South Korea, two separate studies have discovered a total of 7 strains of CPEC from companion animals [13, 14]. Unlike the results from the human investigation, all companion animal-derived CPEC strains harboured the IncX3 plasmid, which encodes blaNDM-5, and were identified as multilocus sequence type (MLST) ST410. The discrepancy between the findings in humans and animals could be attributed to insufficient data regarding companion animals or unidentified transmission events within our environment and animal community. However, given the growing importance of companion animals in human society, it is crucial to consider the possibility of human-animal transmission of CPEC pathogens.
MDR pathogenic bacteria, which have already become a major public health concern, are no longer confined to the realm of human health. As a natural phenomenon derived from ancient bacterial genomes [15], resistance genes can be shared and transferred through horizontal gene transfer among pathogenic bacteria in humans, animals, and the environment [16,17,18]. Therefore, it was necessary to conduct a resistance and virulence gene distribution analysis combining potential sources of shared yet undetected resistance genes while considering the comprehensive perspective of the “One Health” approach.
In this study, the genomic distance between companion animal-derived CPEC pathogens and previously identified strains was measured using whole-genome phylogenetic analysis. Since all the strains discovered in our country were identified solely as ST410, worldwide ST410 datasets were selected as the reference. To conduct the phylogenetic analysis, we performed whole-genome sequencing on four CPEC isolates obtained from companion dogs, including three isolates (DMCPEC2, DMCPEC3 and DMCPEC7) that were included in our previous study [13]. The whole-genome datasets were screened and analysed based on the public database to identify undiscovered genes that could be potential threats.
Materials and methods
Bacterial strain isolation and minimum inhibitory concentration
A total of 4 isolates of E. coli ST410 strains obtained from companion animals were included in this study. Three strains (DMCPEC2, DMCPEC3 and DMCPEC7), which were identified as carrying blaNDM-5-encoding IncX3 plasmids in a previous study [13], were included among the four isolates. NB7CPEC was isolated from a screening rectal swab at a local veterinary clinic in Seoul, South Korea. Meropenem-impregnated (1 μg/mL) MacConkey (MIM) agar was used to identify the carbapenem-resistant gram-negative phenotype from the rectal swab of a mixed Pomeranian dog. The host dog (6-year-old mixed Pomeranian canine, spayed female) had no specific clinical disease condition and was swabbed for carbapenemase screening with a normal rectal swab collected by professional veterinarians in accordance with the Guide for the Care and Use of Laboratory Animals and the Animal Welfare Act. The antimicrobial resistance minimum inhibitory concentration (MIC) profile was determined using the broth microdilution method. E. coli strain ATCC 25922 was used as a quality control strain for the MIC determination, following the Clinical and Laboratory Standards Institute (CLSI) recommendations for performance and interpretation [19].
Total DNA isolation followed by in vitro genotyping
The Wizard Genomic DNA purification kit (Promega, Madison, WI) was used for total DNA purification, and carbapenemase gene screening was performed using previously designed multiplex PCR primers and protocols [20]. To compare the sequences with available GenBank data, Sanger sequencing was performed using the Basic Local Alignment Search Tool (BLAST) network service [21].
Bacterial species were determined using a matrix-assisted laser desorption ionization–time of flight-mass spectrometry (MALDI–TOF–MS; Bruker Daltonik GmbH, Bremen, Germany) biotyper and 16S rRNA sequencing.
Classical MLST was performed using a previously described protocol [22] to evaluate seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA and recA), and the results were further confirmed on the online database [23].
Combined complete genome sequencing and de novo assembly
The whole-genome DNA samples of 4 isolates of CPEC ST410 were purified with a Wizard Genomic DNA purification kit (Promega, Madison, WI, USA) from overnight cultures. For high-quality sequencing and assembly, both long and short genomic DNA libraries were prepared. Short-read sequencing was performed using an Illumina NovaSeq 6000 (Illumina, San Diego, CA, USA) platform following a paired-end 2 × 150-bp protocol. The Oxford Nanopore platform (Oxford Nanopore Technologies, Oxford, UK) was employed for long-read sequencing. The ONT library was constructed and sequenced by using the Ligation Sequencing Kit (SQK-LSK109), the Flow Cell Priming Kit (EXP-FLP002) and Flowcell (FLO-MIN106).
Basecalling and demultiplexing of barcodes were conducted with Guppy basecaller and barcoder v6.0.7 [24], followed by combined trimming and filtering of the reads with FiltLong v0.2.0 [25]. De novo assembly was performed with Flye v2.8.3 [26], and the results were combined with short reads using the Unicycler v0.4.8 hybrid assembler [27]. Examinations for overlaps and circularization were performed with Circlator v1.5.5 [28]. Pilon v1.23 [29] was employed for data polishing. The genome completeness for the generated assembled sequences was confirmed through calculations using BUSCO v4.1.2 [30]. Genome structural annotation was conducted using Prokka v1.14.6 [31], and functional annotation was performed with DIAMOND v 0.9.30 [32] and Blast2GO v4.1.9 [33] to perform gene ontology [34] analysis.
In silico typing and identification for bioinformatic comparison
For comparative analysis, 37 genomic datasets of E. coli ST410 strains were downloaded from the National Center for Biotechnology Information [35] and compared with the 4 isolates sequenced in this study. The assembled genomes were screened for comparison of the resistance genes, virulence genes, serotypes, SPI sites, MLST, plasmid types and fimH types on the Center for Genomic Epidemiology (CGE) server [36] for in silico utilization of ResFinder 4.1, VirulenceFinder 2.0, SerotypeFinder 2.0, SPIFinder 2.0, MLST 2.0, PlasmidFinder 2.1 and FimTyper 1.0. The schematic complete genome structure maps of the chromosomes and plasmids were generated with the CGView server [37]. High-quality whole-chromosome SNPs were identified from the chromosomal reference sequence of YD786 (GenBank no. CP013112.1) for concatenated alignment using the standard settings of CSI Phylogeny [38]. A maximum likelihood (ML) tree was constructed with 1000 bootstrap replicates in MEGA 11 software [39]. The comparative gene distribution annotations and heatmaps were annotated, and a phylogenetic tree was constructed and visualized on iTOLs [40].
Results
Isolate profiles and minimum inhibitory concentrations
The NDM-5-harbouring E. coli ST410 isolate NB7CPEC was discovered from a rectal swab of a 6-year-old mixed Pomeranian canine (Table 1) hospitalized in a local veterinary clinic. The minimum inhibitory concentration value was measured for NB7CPEC and combined with the MIC results from a previous study (DMCPEC2, DMCPEC3 and DMCPEC7). All 4 isolates were identified as resistant strains against 3 tested carbapenems (Additional file 1), namely, ertapenem (MIC value ≥ 32 μg/mL, ≥ 2 μg/mL as the resistance breakpoint standard), imipenem (MIC value ≥ 16 μg/mL, ≥ 4 μg/mL as the resistance breakpoint standard) and meropenem (MIC value ≥ 32 μg/mL, ≥ 2 μg/mL as the resistance breakpoint standard) [19]. The isolates also showed high MIC values against ceftazidime, cefepime (3rd- and 4th-generation cephalosporins, respectively), gentamicin (aminoglycosides), ampicillin (aminopenicillins) and tetracycline (tetracyclines). The MIC of tobramycin (aminoglycosides), chloramphenicol (phenicols), and trimethoprim (folate pathway inhibitors) showed different values according to the isolates. The only susceptible option in common was colistin (MIC values lower than 0.5 μg/mL) for the CPEC isolates.
Qualification of the sequenced whole-genome datasets
The 4 E. coli ST410 isolates identified from 4 different companion dogs were subjected to whole-genome sequencing, and a high-quality nucleotide sequence was generated (Additional file 2). Whole-genome sequencing identified chromosomes and 2 plasmids from each strain. In NB7CPEC, 2 additional plasmids were identified (Table 2).
Identification of characteristic genes in the chromosomes and plasmids of the ST410 strains
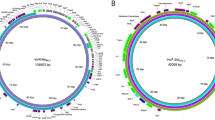
The identified chromosomal length of each strain was between 4.71 and 4.83 Mbp (Figure 1). The ESBL-encoding genes of CMY-2 and CMY-121 were identified. The chromosomes included variant sites of quinolone resistance-determining regions (QRDRs) of the gyrA (S83L and D87N), parC (S80I) and parE (S458A) genes. A total of 5 CRISPR region sites were also discovered, with a CAS-type IE cluster in the chromosome. The virulence genes (csgA, fimH, lpfA, hlyE and yehABCD) were also identified, and their positions were marked on the visualization map.
Circular map of comparative chromosomes of the 4 ST410 isolates identified in this study. The whole chromosomes of the E. coli ST410 strains identified in this study were comparatively mapped. The chromosomal map of DMCPEC2 (4 832 084 bp) was utilized as the backbone for visualization and is depicted as a black circle. The circular map was generated by using CGView.
Two heterogeneous plasmids were carried by all isolates, including the blaNDM-5-encoding IncX3 plasmid (Figure 2). The other plasmid (Figure 3) was discovered as an integrated form of three types of plasmids, namely, IncFIA (% identity; 99.74), IncFIB (AP001918) (% identity; 98.39) and IncFII (pAMA1167-NDM-5) (% identity; 100).
Schematic visualization map of IncX3 plasmids encoding blaNDM-5 carried by ST410 strains. The approximately 46 kbp-long IncX3-type plasmids encoding the carbapenemase gene blaNDM-5 were carried by all 4 strains. The IncX3 plasmids were consistent with the datasets reported in a previous study [13]. The data for pNB7-NDM5 were included in the visual map. The plasmid map of pEC3-NDM5 (46 749 bp) was used as the backbone for visualization and is depicted as a black circle. The circular map was generated by using CGView.
Integrated map of the 3 plasmid types carried by the ST410 strains. Three distinct plasmid types (namely, FII (pAMA1167-NDM-5), FIA and FIB (AP001918)) were integrated into 77–82 kbp-long plasmids. The plasmid map of pEC3-FIIFIAFIB (82 232 bp) was used as the backbone for visualization and is depicted as a black circle. The circular map was generated by using CGView.
The bacterial strain NB7CPEC, which was isolated in 2021 from a local veterinary clinic, carried an additional 2 different plasmids (Figures 4 and 5). The IncFII (pHN7A8)-type plasmid (Figure 4) was identified as a 73 618 bp-long plasmid encoding the ESBL-harbouring genes blaTEM-1B and blaCTX-M-65. The structural positions of the resistance genes and the mobile gene elements were mapped on the schematic map of the whole plasmid. The p0111-type (% identity, 98.08) plasmid was also identified as a 96,439 bp-long plasmid (Figure 5), although it lacked mobile gene cassettes and resistance genes.
Whole-genome phylogeny and bioinformatic comparison of the characteristic genes
The complete genome datasets of 37 E. coli ST410 strains isolated between 2010 and 2020 from 17 different countries were downloaded and included in a complete genome comparison along with the data from this study (Additional file 3). The strains were isolated from China (n = 8), South Korea (n = 5, including 4 isolates from this study), the USA (n = 4), Switzerland (n = 4), the United Kingdom (n = 3), Brazil (n = 2), Germany (n = 2), Ghana (n = 2), India (n = 2), Malaysia (n = 2), and Canada, Cuba, Denmark, France, Norway, Portugal and Spain (n = 1 each).
The measured number of valid SNP positions of the whole chromosomes identified by the CSI Phylogeny pipeline was 5271. The pairwise SNP difference, adjusted according to YD786 (GenBank no. CP013112.1) as the reference genome, ranged from 0 (between CP024801 and CP026473) to 2,204 (between CP031231 and CP027205) (Additional file 4). Among the isolates from this study, the pairwise SNP differences were from 28 (between DMCPEC3 and DMCPEC7) to 533 (between DMCPEC2 and NB7CPEC). The input parameters, analysis quality and identity of each strain against the reference are displayed in Additional file 5. The resistance and virulence genes and plasmid types are denoted in Additional files 6, 7, 8.
A whole-chromosome SNP-based phylogenetic tree was then constructed and displayed along with the epidemiological datasets (Figure 6). All 41 isolates included amino acid substitutions in the QRDR and fimH24 genes, which could be classified as the B/H24R lineage.
Epidemiological comparison of the complete genome datasets of 41 ST410 strains. A maximum likelihood (ML) tree was constructed and visualized by iTOLs based on whole-chromosome SNPs. The isolates originating from this study are highlighted with a black background. The coloured ranges covering the strain labels show the identified subgroups: Group A (red); Group B (blue); Group C (green). The coloured and labelled columns indicate the epidemiological information of the ST410 strains. From left to right, geographic locations; O antigen types; H antigen types; quinolone resistance-determining regions (QRDRs) variation sites of gyrA; QRDR variation sites of parC/parE; levels of identity of salmonella pathogenicity islands (%). The geographic locations of the strains are denoted in distinguishable colours according to their continents of origin: Africa (grey), Asia (yellow), Europe (cyan), North America (red) and South America (green).
The analysis of the subclade ST410 B/H24R identified the following distinguishable groups: (i) Group A, including the H9 antigen and the type 1 high-pathogenicity island (HPI) of the Salmonella pathogenicity islands, (ii) Group B, identified with the H21 antigen, and (iii) Group C (B4/H24RxC), carrying the ESBLs and carbapenemase genes. Three of the strains from this study (DMCPEC3, DMCPEC7 and NB7CPEC) encoding blaCMY-6 and blaNDM-5 could be included in Group C.
The isolates included in phylogenetic group A included HPIs. The discovered HPI gene dataset was identical to the type 1 HPI (identity > 98%) identified from Salmonella enterica group VI [41]. The SNP differences ranged from 37 (between CP018965 and CP035325) to 662 (between CP035123 and CP073926). Several virulence genes were identified from Group A with high priority, namely, iucC (aerobactin synthetase), ituA (ferric aerobactin receptor) and sitA (iron transport protein). The tetracycline resistance gene tet(A) was carried by the isolates of Group A with higher priority than the other groups.
Group B featured the H21 antigen type. The isolates of Group B carried the blaCMY-42 gene with relatively higher priority than the other groups. The differences in SNPs ranged from 5 (between CP035944 and CP042934) to 403 (between CP031653 and CP029369).
The isolates in Group C mainly originated from samples from Asia and Europe. The serotypes of the isolates in Group C were identified as O8:H9. The pairwise SNP difference matrix ranged between 15 (CP034958 and CP033401) and 103 (CP048344 and CP027205). Group C isolates were found to have carbapenemase genes (Figure 7), encoding at least one carbapenemase gene of NDM-5 or OXA-1. The ESBL genes of CMY-2, CMY-6 and TEM-1B were discovered in this group with higher priority, along with the IncFIA-, IncFII- and IncX3-type plasmids.
Whole-genome dataset comparison of the antimicrobial resistance genes and plasmid types identified from the ST410 strains. Different types of antimicrobial resistance genes were categorized and are shown in distinguishable colours and rectangular boxes: carbapenemases (black), ESBLs (red), aminoglycosides (magenta), dihydrofolate reductases (green), macrolides (brown), quinolones (purple), sulfonamides (blue) and tetracyclines (grey). A heatmap indicating the identified plasmid types is shown along with the antimicrobial resistance gene distribution map.
Discussion
The extraintestinal pathogenic E. coli ExPEC ST410 strain is known to circulate not only in humans but also in animals (wildlife and companion animals) and the environment [3, 10]. Through whole-genome phylogenetic analysis, it was discovered that the companion animal-derived isolates were closely grouped with strains identified from humans, animals and the environment (in subgroup C, Figures 6–8). The total number of whole-genome SNP differences of isolate DMCPEC7, carried by a companion dog, from the human blood isolate KBN10P04869 (GenBank no. CP026473) was 36. Previous investigations on Korean isolates have indicated differences in genetic characteristics, such as MLST types, between human and animal strains. The MLST types of the human-obtained CPEC isolates investigated by the national laboratory surveillance system were ST131, ST1642 and ST101, whereas ST410 was not reported [12]. In contrast, all CPEC isolates identified from companion animals in South Korea thus far have been typed as ST410. Despite the phylogenetic relatedness, the genetic studies conducted in this work are not suitable to provide direct evidence of the transmission between the isolative sources of those strains, including human-animal transmission. However, when considering the genetic evidence revealed by the whole-genome phylogeny, it is crucial to seriously consider the possibility of circulation among humans, animals and the environment. Therefore, an integrative “One Health” approach should be applied for control measures considering the E. coli ST410 strains.
Virulence gene distribution comparison identified from the whole genome datasets. Virulence genes were screened from the whole genome datasets adjusting VirulenceFinder 2.0 via the CGE server and visualized using iTOLs. Distinguished phylogenetic subgroups are indicated with coloured ranges: Group A (red); Group B (blue); Group C (green).
The E. coli ST410 strains, along with ST131, have been proposed as globally circulating strains of extraintestinal pathogenic E. coli (ExPEC) [10]. ExPEC bacteria are known to encode various extraintestinal virulence factors and have been attributed to various infectious diseases, including neonatal meningitis, urinary tract infections, bloodstream infections and pneumonia [42,43,44]. In these clinical situations, the administration of β-lactam antibiotics, including carbapenems, is an important treatment option. However, ExPEC bacteria may be resistant not only to β-lactam antibiotic agents due to the production of various β-lactamases, such as ESBL and carbapenemase but also to other antibiotics, including fluoroquinolones, tetracyclines and aminoglycosides [45,46,47]. In this study, companion animal-derived ST410 strains were found to carry various virulence genes and antimicrobial resistance genes. The MIC results (Additional file 1) revealed that colistin is currently the only viable option for treating these isolates. Therefore, the circulation of these strains in companion animals within our community should be addressed seriously, considering that these strains have the capacity to not only cause extraintestinal infections as ExPEC bacteria but also resist various antimicrobial agents as CPEC pathogens.
E. coli strains are largely classified into phylogroups A, B1, B2, C, D, E or F, followed by further categorization of sequence types, clades and subclades [48]. The carbapenemase gene is mainly carried by E. coli strains in phylogroups A and B1 [49], and E. coli ST410 strains are grouped into phylogroup A. ST410 has been reported as a high-risk clone in previous studies [48]. The subclade of ST410 carrying fimH24 has been classified by B2/H24R (gyrA and parC mutations), B3/H24Rx (additional carriage of the ESBL gene) and B4/H24RxC (carbapenemase introduction) [5]. In this study, further specific groups were identified in the B2/H24R subclade by bioinformatic gene identification based on whole-genome data. In particular, the distribution of virulence factor genes (Figure 8) was greater in Group A strains relative to the other strains, such as HPIs, iucC, ituA and sitA. HPI and ExPEC bacteria are strongly correlated and have been described as the causative agents of various extraintestinal infections [50, 51]. Conversely, Group C isolates were identified as having a relatively larger dissemination of antimicrobial resistance genes. To continue to define the genetic characteristics of the ST410 strains identified in this study, further investigations are needed, such as additional isolate collection and antimicrobial and virulence phenotyping.
Whole-genome analyses revealed the genomic potential of CPEC isolates identified from companion animals, indicating that these strains should be considered a potential threat to public health. Therefore, it is crucial to consider new measures for controlling the dissemination of CPEC ST410 strains in our society, adopting a combined public health approach with a “One Health” perspective.
Availability of data and materials
All referred sequences in this study are available from the NCBI BioProject number PRJNA858561. All data generated or analysed during this study have been submitted with this manuscript. All genetic information of the plasmids was deposited in GenBank. Therefore, all data from this study are available to the public.
Abbreviations
- BLAST:
-
Basic Local Alignment Search Tool
- CPEC:
-
carbapenemase-producing Escherichia coli
- CGE:
-
Center for Genomic Epidemiology
- CLSI:
-
Clinical and Laboratory Standards Institute
- CRE:
-
carbapenem-resistant Enterobacterales
- ESBL:
-
extended-spectrum β-lactamase
- ESC:
-
extended-spectrum cephalosporin
- ExPEC:
-
extraintestinal pathogenic E. coli
- GO:
-
Gene Ontology
- Inc:
-
incompatibility type
- MALDI-TOF:
-
matrix-assisted laser desorption ionization–time of flight-mass spectrometry
- MDR:
-
multidrug resistant
- MIC:
-
minimum inhibitory concentration
- MIM:
-
meropenem-impregnated MacConkey
- ML:
-
maximum likelihood
- MLST:
-
multilocus sequence type
- ORF:
-
open reading frame
- QRDR:
-
quinolone resistance-determining region
- SNP:
-
single nucleotide polymorphism
- SPIs:
-
Salmonella Pathogenicity islands
References
Shaheen BW, Nayak R, Boothe DM (2013) Emergence of a New Delhi metallo-β-lactamase (NDM-1)-encoding gene in clinical Escherichia coli isolates recovered from companion animals in the United States. Antimicrob Agents Chemother 57:2902–2903
Stolle I, Prenger-Berninghoff E, Stamm I, Scheufen S, Hassdenteufel E, Guenther S, Bethe A, Pfeifer Y, Ewers C (2013) Emergence of OXA-48 carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in dogs. J Antimicrob Chemother 68:2802–2808
Falgenhauer L, Imirzalioglu C, Ghosh H, Gwozdzinski K, Schmiedel J, Gentil K, Bauerfeind R, Kämpfer P, Seifert H, Michael GB (2016) Circulation of clonal populations of fluoroquinolone-resistant CTX-M-15-producing Escherichia coli ST410 in humans and animals in Germany. Int J Antimicrob Agents 47:457–465
Guenther S, Ewers C, Wieler LH (2011) Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front Microbiol. https://doi.org/10.3389/fmicb.2011.00246/full
Roer L, Overballe-Petersen S, Hansen F, Schønning K, Wang M, Røder BL, Hansen DS, Justesen US, Andersen LP, Fulgsang-Damgaard D (2018) Escherichia coli sequence type 410 is causing new international high-risk clones. Msphere 3:e00337-e418
Roer L, Hansen F, Thomsen MCF, Knudsen JD, Hansen DS, Wang M, Samulioniené J, Justesen US, Røder BL, Schumacher H (2017) WGS-based surveillance of third-generation cephalosporin-resistant Escherichia coli from bloodstream infections in Denmark. J Antimicrob Chemother 72:1922–1929
Overballe-Petersen S, Roer L, Ng K, Hansen F, Justesen US, Andersen LP, Stegger M, Hammerum AM, Hasman H (2018) Complete nucleotide sequence of an Escherichia coli sequence type 410 strain carrying blaNDM-5 on an IncF multidrug resistance plasmid and bla OXA-181 on an IncX3 plasmid. Genome Announc 6:e01542-e1617
Piazza A, Comandatore F, Romeri F, Pagani C, Floriano AM, Ridolfo A, Antona C, Brilli M, Mattioni Marchetti V, Bandi C (2018) First report of an ST410 OXA-181 and CTX-M-15 coproducing Escherichia coli clone in Italy: a whole-genome sequence characterization. Microb Drug Resist 24:1207–1209
Liu Y, Feng Y, Wu W, Xie Y, Wang X, Zhang X, Chen X, Zong Z (2015) First report of OXA-181-producing Escherichia coli in China and characterization of the isolate using whole-genome sequencing. Antimicrob Agents Chemother 59:5022–5025
Schaufler K, Semmler T, Wieler LH, Wöhrmann M, Baddam R, Ahmed N, Müller K, Kola A, Fruth A, Ewers C (2016) Clonal spread and interspecies transmission of clinically relevant ESBL-producing Escherichia coli of ST410—another successful pandemic clone? FEMS Microbiol. https://doi.org/10.1093/femsec/fiv155
Patiño-Navarrete R, Rosinski-Chupin I, Cabanel N, Gauthier L, Takissian J, Madec J-Y, Hamze M, Bonnin RA, Naas T, Glaser P (2020) Stepwise evolution and convergent recombination underlie the global dissemination of carbapenemase-producing Escherichia coli. Genome Med 12:10
Yoon E-J, Yang JW, Kim JO, Lee H, Lee KJ, Jeong SH (2018) Carbapenemase-producing Enterobacteriaceae in South Korea: a report from the national laboratory surveillance system. Future Microbiol 13:771–783
Kyung SM, Choi S-W, Lim J, Shim S, Kim S, Im YB, Lee N-E, Hwang C-Y, Kim D, Yoo HS (2022) Comparative genomic analysis of plasmids encoding metallo-β-lactamase NDM-5 in Enterobacterales Korean isolates from companion dogs. Sci Rep 12:1569
Hong JS, Song W, Park H-M, Oh J-Y, Chae J-C, Han J-I, Jeong SH (2019) First detection of New Delhi metallo-β-lactamase-5-producing Escherichia coli from companion animals in Korea. Microb Drug Resist 25:344–349
D’Costa VM, King CE, Kalan L, Morar M, Sung WW, Schwarz C, Froese D, Zazula G, Calmels F, Debruyne R (2011) Antibiotic resistance is ancient. Nature 477:457–461
Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MO, Dantas G (2012) The shared antibiotic resistome of soil bacteria and human pathogens. Science 337:1107–1111
Bengtsson-Palme J, Larsson D (2015) Antibiotic resistance genes in the environment: prioritizing risks. Nat Rev Microbiol 13:396–396
Finley RL, Collignon P, Larsson DJ, McEwen SA, Li X-Z, Gaze WH, Reid-Smith R, Timinouni M, Graham DW, Topp E (2013) The scourge of antibiotic resistance: the important role of the environment. Clin Infect Dis 57:704–710
CLSI (2017) M100–S23 Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement An informational supplement for global application developed through the Clinical and Laboratory Standards Institute consensus process, 27th edn. Clinical and Laboratory Standards Institute, Wayne
Poirel L, Walsh TR, Cuvillier V, Nordmann P (2011) Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H (2006) Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151
Zhou Z, Alikhan N-F, Mohamed K, Fan Y, Achtman M, Brown D, Chattaway M, Dallman T, Delahay R, Kornschober C (2020) The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res 30:138–152
Wick RR, Judd LM, Holt KE (2019) Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol 20:129
Kolmogorov M, Yuan J, Lin Y, Pevzner PA (2019) Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37:540–546
Wick RR, Judd LM, Gorrie CL, Holt KE (2017) Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595
Hunt M, Silva ND, Otto TD, Parkhill J, Keane JA, Harris SR (2015) Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol 16:294
Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK (2014) Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069
Buchfink B, Reuter K, Drost H-G (2021) Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods 18:366–368
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Dimmer EC, Huntley RP, Alam-Faruque Y, Sawford T, O’Donovan C, Martin MJ, Bely B, Browne P, Mun Chan W, Eberhardt R (2012) The UniProt-GO annotation database in 2011. Nucleic Acids Res 40:D565–D570
O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D (2016) Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44:D733–D745
Center for Genomic Epidemiology. http://cge.cbs.dtu.dk/services/all.php. Accessed 21 Aug 2022
Stothard P, Wishart DS (2005) Circular genome visualization and exploration using CGView. Bioinformatics 21:537–539
Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O (2014) Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE 9:e104984
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027
Letunic I, Bork P (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245
Oelschlaeger T, Zhang D, Schubert S, Carniel E, Rabsch W, Karch H, Hacker J (2003) The high-pathogenicity island is absent in human pathogens of Salmonella enterica subspecies I but present in isolates of subspecies III and VI. J Bacteriol 185:1107–1111
Feng Y, Liu L, Lin J, Ma K, Long H, Wei L, Xie Y, McNally A, Zong Z (2019) Key evolutionary events in the emergence of a globally disseminated, carbapenem resistant clone in the Escherichia coli ST410 lineage. Commun Biol 2:322
Smith JL, Fratamico PM, Gunther NW (2007) Extraintestinal pathogenic Escherichia coli. Foodborne Pathog Dis 4:134–163
Russo TA, Johnson JR (2000) Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J Infect Dis 181:1753–1754
Knothe H, Shah P, Krcmery V, Antal M, Mitsuhashi S (1983) Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 11:315–317
Pitout JD (2010) Infections with extended-spectrum β-lactamase-producing Enterobacteriaceae. Drugs 70:313–333
Pitout JD (2012) Extraintestinal pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front Microbiol 3:9
Cummins EA, Snaith AE, McNally A, Hall RJ (2021) The role of potentiating mutations in the evolution of pandemic Escherichia coli clones. Eur J Clin Microbiol Infect Dis. https://doi.org/10.1007/s10096-021-04359-3
Gauthier L, Dortet L, Cotellon G, Creton E, Cuzon G, Ponties V, Bonnin RA, Naas T (2018) Diversity of carbapenemase-producing Escherichia coli isolates in France in 2012–2013. Antimicrob Agents Chemother 62:e00266-e318
Schubert S, Picard B, Gouriou S, Heesemann J, Denamur E (2002) Yersinia high-pathogenicity island contributes to virulence in Escherichia coli causing extraintestinal infections. Infect Immun 70:5335–5337
Schubert S, Cuenca S, Fischer D, Heesemann J (2000) High-pathogenicity island of Yersinia pestis in enterobacteriaceae isolated from blood cultures and urine samples: prevalence and functional expression. J Infect Dis 182:1268–1271
Funding
This work was supported by the National Research Foundation (NRF- 2020R1A2C200879414), BK21 FOUR Future Veterinary Medicine Leading Education and Research Center and Research Institute for Veterinary Science, Seoul National University, Seoul, Republic of Korea.
Author information
Authors and Affiliations
Contributions
SMK performed the experiments and analysis and wrote the paper. JL and ESL performed the experiments and corrected the analysis. CYH collected and stored the samples. HSY reviewed and edited the paper. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Handling editor: Freddy Haesebrouck
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
MIC profiles of the ST410 strains.
Additional file 2:
Whole-genome profiles of sequenced E. coli strains.
Additional file 3:
Epidemiological profiles of analysed whole-genome datasets in this study.
Additional file 4:
The pairwise SNP difference matrix of whole-chromosome datasets extracted from the reference genome.
Additional file 5:
Quality of SNP extraction analysis, including input parameters and identity of each strain with the reference.
Additional file 6:
The dissemination metadata of antimicrobial resistance gene carriage in the whole-genome datasets, searched by in silico screening by the ResFinder 4.1 database.
Additional file 7:
Genomic metadata of virulence genes (presence or absence), serotypes, Salmonella pathogenicity islands (SPIs) and FimH types searched on the Center for Genomic Epidemiology (CGE) server.
Additional file 8:
Plasmid-type metadata identified by PlasmidFinder 2.1 and presented as gene identity (%).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kyung, S.M., Lee, J., Lee, ES. et al. Genomic molecular epidemiology of carbapenemase-producing Escherichia coli ST410 isolates by complete genome analysis. Vet Res 54, 72 (2023). https://doi.org/10.1186/s13567-023-01205-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13567-023-01205-6