Abstract
Retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) are cytosolic pattern recognition receptors that initiate innate antiviral immunity. Recent reports found that duck RLRs significantly restrict duck plague virus (DPV) infection. However, the molecular mechanism by which DPV evades immune responses is unknown. In this study, we first found that the DPV UL41 protein inhibited duck interferon-β (IFN-β) production mediated by RIG-I and melanoma differentiation-associated gene 5 (MDA5) by broadly downregulating the mRNA levels of important adaptor molecules, such as RIG-I, MDA5, mitochondrial antiviral signalling protein (MAVS), stimulator of interferon gene (STING), TANK-binding kinase 1 (TBK1), and interferon regulatory factor (IRF) 7. The conserved sites of the UL41 protein, E229, D231, and D232, were responsible for this activity. Furthermore, the DPV CHv-BAC-ΔUL41 mutant virus induced more duck IFN-β and IFN-stimulated genes (Mx, OASL) production in duck embryo fibroblasts (DEFs) than DPV CHv-BAC parent virus. Our findings provide insights into the molecular mechanism underlying DPV immune evasion.
Similar content being viewed by others
Introduction
Retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), including RIG-I, MDA5 and laboratory of genetics and physiology 2 (LGP2), are critical cytosolic RNA sensors that trigger the innate immune response [1]. Both RIG-I and MDA5 consist of two N-terminal cysteine/aspartic protease (caspase) and activation and recruitment domains (CARDs), a helicase domain, and a C-terminal domain (CTD). Upon sensing diverse cytosolic double-stranded RNAs (dsRNAs), RIG-I undergoes conformational changes, oligomerization, and exposure of the N-terminal CARD domains to interact with the CARD domain of a signalling adaptor, mitochondrial antiviral signalling protein (MAVS) [2]. MAVS transmits signals to downstream signalling molecules, the IκB kinase (IKK)-related kinases TBK1 and IKKε, and TBK1 and IKKε then phosphorylate IRF3/7 and NF-κB and induce their nuclear translocation, resulting in inflammatory cytokine and interferon (IFN) production [3]. In addition, STING is a central adaptor molecule that links DNA- and RNA-sensing pathways to activate IFN-β production. STING also interacts with RIG-I and MAVS, but not with MDA5, to form a stabilized complex upon virus infection [4]. Therefore, STING plays a critical role in RIG-I-mediated antiviral signalling [5, 6]. IFNs are classified into three types (type I, type II, and type III), among which type I IFNs (including a multigene α subtype with 13 members in humans and single β, ε, κ, τ, δ, and ω genes) are quintessential antiviral cytokines because of their central roles in the antiviral immune response, and IFN-α/β in birds and mammals are not grouped together in the phylogenetic tree [7].
Duck plague is an acute and contagious fatal disease with high morbidity and mortality in domestic waterfowl which causes substantial economic losses in the commercial waterfowl industry [8,9,10,11,12,13,14,15,16]. DPV, the causative agent of infectious disease, belongs to the Alphaherpesvirinae subfamily [17]. The DPV virion is composed of an envelope, a tegument layer and a spherical nucleocapsid that contains double-stranded DNA [18, 19]. Alphaherpesviruses encode 23 tegument proteins that play roles in promoting viral replication and viral assembly, regulating viral and host protein synthesis, and immune evasion [20].
Host shutoff has emerged as a key process that facilitates the reallocation of cellular resources for viral replication and evasion of host antiviral immune responses [21]. The virion host shutoff (VHS) protein, encoded by the herpes simplex virus-1 (HSV-1) UL41 gene, is a late tegument protein and an endoribonuclease with a substrate specificity for ribonuclease (RNase) A [22]. The VHS protein specifically degrades a wide array of mRNAs and induces the rapid shutoff of host protein synthesis [23, 24]. On the one hand, the VHS protein facilitates the turnover of all kinetic classes of viral mRNAs [25]. On the other hand, the VHS protein plays crucial roles in escaping the host innate immune response. The VHS protein downregulates the expression of major histocompatibility complex (MHC) class I/II molecules, impairs antigen presentation [26,27,28], suppresses the production of proinflammatory chemokines and cytokines, inactivates human monocyte-derived dendritic cells [21, 29], and degrades the mRNAs of several IFN-stimulated genes (ISGs), such as ch25h [30], ZAP [31], tetherin [32], viperin [33], IFIT3 [34] and TNF-α [35].
The duck RIG-I, MDA5 and STING proteins play pivotal roles in the innate immune response of the host to DPV infection [36,37,38]. In addition, DPV is immunosuppressive and exhibits broad cell tropism [39, 40]. While knowledge on the molecular mechanism of DPV immune evasion is limited, the UL47 protein is known to interact with signal transducer and activator of transcription 1 (STAT1) to inhibit duck IFN-β production [41]. In this study, we found that the DPV UL41 protein abrogated RIG-I/MDA5-mediated duck IFN-β production by downregulating the mRNA levels of important adaptors, and the conserved sites of the UL41 protein, E229, D231, and D232, were responsible for this activity. Our findings provide new insights into host-virus interactions and contribute to the development of new antiviral drugs.
Materials and methods
Viruses, cells, and vectors
The DPV CHv-BAC [42] and DPV CHv-BAC-ΔUL41 [43] recombinant viruses were blindly passaged up to 10 passages in DEFs. Passage 10 viruses were used for all experiments described in the manuscript.
DEF cells were cultured in minimal essential medium (MEM; Gibco, Meridian Road Rockford, USA) supplemented with 10% (v/v) foetal bovine serum (FBS; Gibco, Meridian Road Rockford, USA) at 37 °C and 5% CO2. For viral infections, maintenance medium supplemented with 2% FBS was added. Commonly used reagents were prepared in our laboratory. HEK293T cells were cultured in RPMI-1640 medium supplemented with 10% FBS at 37 °C and 5% CO2 [43].
All primers were designed by Primer Premier 5 software (Table 1). The recombinant plasmids pcaggs-MDA5-Flag, pcaggs-MAVS-Flag, pcaggs-STING-Flag, pcaggs-TBK1-Flag, pcaggs-IRF7-Flag, pcaggs-UL41-HA, pcaggs-mUL41-HA and IFN-β-Luc express firefly luciferase under the control of the duck IFN-β promoter (−96 to +1) were prepared in our laboratory [38, 43, 44]. The entire RIG-I open reading frame (ORF) (accession no. KC869660.1) was inserted into the pcaggs vector to generate pcaggs-RIG-I-Flag. The pRL-TK internal control vector and pGL4.45 vector were purchased from Promega.
RT-qPCR
Total RNA was extracted from DEF cells using TRIzol reagent l (Invitrogen, CA, USA) according to the manufacturer’s recommendations, and complementary DNA (cDNA) was generated using the PrimeScript® RT reagent kit with gDNA Eraser (Takara, Dalian, China). Target genes were detected using previously described primers (Table 1), and the threshold cycle (Ct) values were normalized to that of 18S rRNA. RT-qPCR was performed according to the manufacturer’s protocol, and the reaction mixture was comprised of the following components: 10 μL of SYBR Green I Mix, 2 μL of each primer, 2 μL of standard template, and autoclaved double-filtered nanopure water added to a final volume of 20 μL. Amplification reactions were performed with preliminary denaturation at 95 °C for 1 min, followed by 40 cycles of denaturation at 95 °C for 20 s, annealing at 60 °C for 30 s and extension at 72 °C for 30 s. All reactions were performed in triplicate with at least three independent experiments. The relative gene expression levels were determined with the 2−ΔΔCt method [9].
Western blotting analysis
Cells were harvested at 36 h post-transfection (hpt). The samples were separated by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto polyvinylidene fluoride (PVDF) membranes. The membranes were blocked with 5% skim milk for 2 h at 37 °C, incubated with a mouse anti-Flag antibody (MBL, Japan, 1:5000), mouse anti-HA antibody (MBL, Japan, 1:4000) or mouse anti-GAPDH antibody (Proteintech, Beijing, 1:20 000) overnight at 4 °C, and then probed with an HRP-conjugated secondary antibody (Bio-Rad, CA, USA) for 1 h at 37 °C. Proteins were detected with Western Blot Chemiluminescence HRP Substrate (Takara, Dalian, China) according to the manufacturer’s instructions [45].
Indirect immunofluorescence assay (IFA)
HEK293T cells were transfected with pcaggs-UL41-HA, pcaggs-mUL41-HA or empty vector, collected at 36 hpt, fixed with 4% paraformaldehyde overnight at 4 °C, and permeabilized with 0.25% Triton X-100 for 30 min at 4 °C. The cells were rinsed three times with PBST (containing 0.1% Tween-20), blocked with 5% BSA PBS for 2 h at 37 °C, and then incubated with a mouse anti-HA antibody (MBL, Japan, 1:100), goat anti-mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, and Alexa Fluor 488 (Thermo Fisher Scientific, Meridian Road Rockford, USA, 1:1000). All antibodies were diluted in 1% BSA PBS. Finally, cell nuclei were visualized with DAPI (Roche, Mannheim, Germany). Coverslips were sealed with glycerin buffer, and the cells were visualized using a fluorescence microscope (Nikon ECLIPSE 80i, Japan) [46].
Luciferase reporter assay
DEF cells were co-transfected with IFN-β-Luc (400 ng/well) and the internal control pRL-TK (4 ng/well) together with the specific expression plasmid (400 ng/well), pcaggs-UL41-HA (400 ng/well), pcaggs-mUL41-HA or empty vector using Lipofectamine 3000 (Invitrogen, CA, USA) according to the manufacturer’s instructions. The cells were harvested at 36 hpt, and firefly luciferase activity was measured by the dual-luciferase assay system (Promega) according to the manufacturer’s instructions [47].
Tissue culture infectious dose 50 (TCID50)
DEF cells were infected with DPV CHv-BAC or DPV CHv-BAC-ΔUL41 at an MOI of 5 and incubated at 37 °C for 2 h. The culture supernatant of virus-infected cells was discarded, and maintenance medium supplemented with 2% FBS was added. At 4 h post-infection (hpi), the cytoplasmic samples were washed twice with PBS and collected to determine the TCID50 using tenfold serial dilutions. All samples were tested in triplicate with at least three independent experiments.
Statistical analysis
Different groups were compared with one-way ANOVA using GraphPad Prism 7.0 software (La Jolla, CA, USA). All experiments were repeated at least three times independently. The data are expressed as the means and standard error of the mean (SEM). Asterisks indicate the level of statistical significance (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001).
Results
The DPV UL41 protein inhibited duck IFN-β signalling activation induced by poly(I:C) stimulation
First, we evaluated whether the UL41 protein affects duck IFN-β production. DEF cells were transfected with pcaggs-UL41-HA or an empty vector as a control and then stimulated with the dsRNA analogue poly(I:C). We found that the UL41 protein negatively regulated duck IFN-β and ISG (Mx, OASL) production in DEF cells (Figure 1A). We further co-transfected IFN-β-Luc, pRL-TK, empty vector, or pcaggs-UL41-HA into DEF cells and then stimulated by poly(I:C). As shown in Figure 1B, the UL41 protein significantly inhibited IFN-β-Luc luciferase activity in a dose-dependent manner. These results indicated that the UL41 protein inhibits duck IFN-β production in DEF cells.
The DPV UL41 protein inhibited duck IFN-β signalling activation induced by poly(I:C). All transfected samples were collected at 36 hpt. A The DPV UL41 protein inhibited duck IFN-β production induced by poly(I:C) in DEF cells. DEF cells were transfected with pcaggs-UL41-HA or empty vector and then stimulated with 50 µg/mL poly(I:C) after 12 hpt. The cells were harvested and detected by RT-qPCR after 24 h of stimulation. B The DPV UL41 protein significantly inhibited duck IFN-β-Luc luciferase activity in a dose-dependent manner. DEF cells were co-transfected with the duck IFN-β-Luc luciferase reporter plasmid, pRL-TK, 1 μg/well pcaggs-UL41-HA, 2 μg/well pcaggs-UL41-HA or empty vector and then stimulated with 50 µg/mL poly(I:C) at 12 hpt. The cells were harvested and detected by the dual-luciferase assay at 24 hpt. Protein expression was confirmed by Western blotting. The data were analysed by one-way ANOVA. *p < 0.05, **p < 0.01.
The substitution of crucial sites on the DPV UL41 protein rescued IFN-β signalling
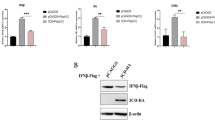
Based on previous reports [48], genetic and biochemical assays showed that the VHS protein and homologues of other alphaherpesviruses share sequence similarities with a family of nucleases and have some conserved residues that are responsible for RNase activity, such as three amino acid residues of the HSV-1 VHS protein, E192, D194, and D195 [49]. In addition, we compared the DPV UL41 protein with homologues of other alphaherpesviruses and found conserved motifs in the DPV UL41 protein (Figure 2). Based on these findings, we mutated the residues E229, D231, and D232 of the DPV UL41 protein to “alanine (A)”, and we found that the mRNA level of the UL41 gene was increased in the pcaggs-mUL41-HA group (Figure 3A). In addition, DEF cells were transfected with pcaggs-UL41-HA and then treated with DMSO, the proteasome inhibitor MG132 (10 μM), or the lysosomotrophic neutralizing agent NH4Cl (20 mM). The expression of UL41 protein was not significantly different among these groups, but was increased in the pcaggs-mUL41-HA group, as determined by Western blotting and IFA (Figures 3B and C). These results indicated that the UL41 protein is not subject to lysosomal or proteasomal degradation in transfected cells. We further determined that duck IFN-β production and IFN-β-Luc activity stimulated by poly(I:C) were increased in the mUL41 group (Figures 3D and E).
Mutation of crucial DPV UL41 residues rescued UL41 protein expression and IFN-β signalling. A HEK293T cells were transfected with the pcaggs-UL41-HA, pcaggs-mUL41-HA or empty vector. The cells were then harvested and subjected to RT-qPCR. B DEF cells were transfected with pcaggs-UL41-HA, pcaggs-mUL41-HA or empty vector and then treated with DMSO, 10 μM MG132, or 20 mM NH4Cl. C HEK293T cells were transfected with the pcaggs-UL41-HA, pcaggs-mUL41-HA or empty vector. Immunofluorescence analysis revealed that the fluorescence (green) was significantly stronger in the mUL41 group than in the UL41 group. D DEF cells were transfected with the UL41 or mUL41 expression plasmid or empty vector and then stimulated with 50 µg/mL poly(I:C) at 12 hpt. The cells were harvested and subjected to RT-qPCR after 24 h of stimulation. E DEF cells were co-transfected with IFN-β-Luc, pRL-TK, and 1 μg/well pcaggs-UL41-HA, 2 μg/well pcaggs-UL41-HA, 2 μg/well pcaggs-mUL41-HA or empty vector and then stimulated with 50 μg/mL poly(I:C) at 12 hpt. The cells were harvested and detected by the dual-luciferase assay at 24 hpt. Protein expression was confirmed by Western blotting. The data were analysed by one-way ANOVA. *p < 0.05, **p < 0.01.
The DPV UL41 protein inhibited RIG-I/MDA5-mediated IFN-β-Luc activation
DEF cells were co-transfected with empty vector, pcaggs-UL41-HA or pcaggs-mUL41-HA and IFN-β-Luc along with plasmids expressing important adaptor proteins, including, RIG-I, MDA5, MAVS, STING, TBK1 and IRF7, and IFN-β-Luc luciferase activity was detected [8]. As shown in Figure 4, all the expression constructs resulted in a 150- to 800-fold induction of IFN-β-Luc luciferase activity, and the UL41 protein dramatically reduced duck IFN-β promoter activation mediated by RIG-I, MDA5, MAVS, STING, TBK1 and IRF7. Moreover, the luciferase activity of IFN-β-Luc induced by all the expression constructs was significantly increased in the mUL41 group (Figure 4). Thus, these results showed that the UL41 protein significantly inhibits IFN-β-Luc activation induced by important adaptor molecules.
The DPV UL41 protein inhibited IFN-β-Luc activation by every important adaptor molecule in the RIG-I/MDA5 innate immune pathway. DEF cells were co-transfected with empty vector, pcaggs-UL41-HA or pcaggs-mUL41-HA together with IFN-β-Luc and plasmids expressing important adaptor proteins in the RIG-I/MDA5 innate immune pathway, namely, RIG-I, MDA5, MAVS, STING, TBK1 and IRF7, and then subjected to luciferase reporter assay to detect IFN-β-Luc promoter activity. Protein expression was confirmed by Western blotting. The data were analysed by one-way ANOVA. *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001.
The DPV UL41 protein decreased the mRNA levels of important adaptor molecules
To further clarify the underlying molecular mechanism, we investigated whether the DPV UL41 protein also affects the mRNA levels of these proteins. As shown in Figure 5A, the transcription of RIG-I, STING, and IRF7 was stimulated by poly(I:C), and the UL41 protein decreased the mRNA levels of various adaptor proteins, especially RIG-I, STING, and IRF7. In addition, DEF cells were co-transfected with the pcaggs-UL41-HA vector or empty vector along with the RIG-I, MDA5, MAVS, STING, TBK1 or IRF7 expression plasmids. We found that the UL41 protein significantly decreased the mRNA and protein levels of RIG-I, MDA5, MAVS, STING, TBK1 and IRF7, which were recovered in the mUL41 group (Figure 5B). These results showed that DPV UL41 protein inhibits duck IFN-β production by broadly degrading mRNAs.
The DPV UL41 protein broadly decreased the mRNA levels of important adaptor molecules in the RIG-I/MDA5 innate immune pathway. A The DPV UL41 protein inhibited the expression of important adaptor molecules induced by poly(I:C), including, RIG-I, MDA5, MAVS, STING, TBK1 and IRF7, in DEF cells. DEF cells were transfected with pcaggs-UL41-HA, pcaggs-mUL41-HA or empty vector and then stimulated with 50 µg/mL poly(I:C) at 12 hpt. The cells were harvested and detected by RT-qPCR after 24 h of stimulation. B DEF cells were co-transfected with pcaggs-UL41-HA, pcaggs-mUL41-HA or empty vector and the RIG-I, MDA5, MAVS, STING, TBK1 and IRF7 expression plasmids. Then, the cells were harvested and subjected to RT-qPCR and Western blot analysis. C HEK293T cells were co-transfected with pcaggs-UL41-HA, pcaggs-mUL41-HA or empty vector and the RIG-I, MDA5, MAVS, STING, TBK1 and IRF7 expression plasmids. Then, the cells were harvested and subjected to RT-qPCR and Western blot analysis. The data were analysed by one-way ANOVA. *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001.
The DPV UL41 protein inhibits duck IFN-β activation in DEF cells
These findings indicated that the DPV UL41 protein inhibits RIG-I/MDA5-mediated IFN-β production to promote viral infection by broadly affecting mRNA levels in vitro. DEF cells were infected with DPV CHv-BAC or DPV CHv-BAC-ΔUL41 and then collected at 4 hpi. We detected duck IFN-β and ISGs (Mx, OASL) production under viral infection. DPV CHv-BAC-G-ΔUL41 mutant virus infection induced more duck IFN-β and ISGs (Mx, OASL) production than infection with the DPV CHv-BAC parent virus. The viral titers in the cytoplasm did not differ between the DPV CHv-BAC-ΔUL41 mutant virus and the DPV CHv-BAC parent virus (Figure 6). These results further showed that the UL41 protein effectively inhibits duck IFN-β production.
DPV downregulated duck IFN-β and ISG production via the UL41 protein. DEF cells were infected with the DPV CHv-BAC-G parent virus or the DPV CHv-BAC-G-ΔUL41 mutant virus at an MOI of 5 and then harvested and subjected to RT-PCR analysis at 4 hpi. A Quantification of IFN-β. B Quantification of Mx and OASL. C The viral titers in cytoplasmic samples were determined by TCID50. The data were analysed by one-way ANOVA. **p < 0.01, ***p < 0.001 and ****p < 0.0001.
Knockdown of IRF7 expression increased the replication of the DPV CHv-BAC-ΔUL41 mutant virus
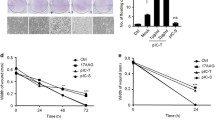
Because IRF3 is absent in the avian genome, IRF7 is an important adaptor and performs some of the IRF3 functions [50]. Previous studies reported that duck IRF7 activates IFN-I transcription and inhibits the in vitro replication of viruses, such as duck Tembusu virus (DTMUV) [51]. We knocked down the expression of IRF7 in DEF cells and examined the knockdown efficiency by RT-qPCR and Western blot. Compared with that in the shNC group, the expression of IRF7 was markedly downregulated in the shIRF7 group (Figure 7A). Then, we further detected the viral replication of the DPV CHv-BAC and DPV CHv-BAC-ΔUL41 recombinant viruses with IRF7 knockdown. As shown in Figure 7B, knockdown of IRF7 expression did not affect the replication of the DPV CHv-BAC parent virus but facilitated the replication of the DPV CHv-BAC-ΔUL41 mutant virus.
Knockdown of IRF7 expression increased the replication of the DPV CHv-BAC-ΔUL41 mutant virus. A DEF cells were transfected with pGPU6/GFP/Neo-shIRF7 or pGPU6/GFP/Neo-shNC, and the transcription and expression of endogenous IRF7 was detected by RT-qPCR and Western blotting, respectively, using a rabbit anti-IRF7 antibody (ABclonal, China, 1:1000). B DEF cells were transfected with pGPU6/GFP/Neo-shIRF7 or pGPU6/GFP/Neo-shNC and then infected with the DPV CHv-BAC parent virus or DPV CHv-BAC-ΔUL41 mutant virus at an MOI of 0.1 after 12 hpt. The viral titers were determined by the TCID50 at 24 hpi. The data were analysed by one-way ANOVA. *p < 0.05.
Discussion
In this study, we reached three major conclusions. First, the DPV UL41 protein significantly inhibited duck IFN-β production. Second, the DPV UL41 protein inhibited the RIG-I/MDA5 innate immune pathway by broadly decreasing mRNA levels. Third, the conserved residues E229, D231 and D232 of the DPV UL41 protein were responsible for this activity.
Pattern recognition receptors (PRRs) sense viral nucleic acids or other virus-specific components, activating a series of signalling cascades to induce IFN and proinflammatory defence mechanisms in response to pathogens [52, 53]. RLRs function as cytosolic PRRs that initiate innate antiviral immunity by detecting exogenous viral RNAs. Significant amounts of dsRNA can be detected for viruses with positive-strand RNA, dsRNA and DNA genomes [54]. Therefore, RLRs also play an important role in the antiviral response to DNA viruses, such as Kaposi’s sarcoma-associated herpesvirus (KSHV) [55], Epstein-Barr virus (EBV) [56], murine gammaherpesvirus 68 (MHV68) [57], HSV [58] and DPV [36]. DPV infection activates high levels of RIG-I and MDA5 expression both in vitro and in vivo. Overexpression of RIG-I inhibits DPV infection, while its knockdown promotes DPV infection [36]. In addition, DPV infection was significantly suppressed in MDA5-overexpressing DEF cells, while the siRNA-mediated knockdown of MDA5 markedly enhanced DPV growth. LGP2 is a concentration-dependent switch that plays a role in MDA5-mediated antiviral activity against DPV [37]. These results collectively suggest that RIG-I and MDA5 act as anti-DPV molecules, and further studies are required to explore the molecular mechanism underlying the antiviral activity of RIG-I and MDA5 in ducks. However, all herpesviruses establish latent infections, a state from which the virus can be reactivated, resulting in recurring disease [59], and manipulation of the host immune response is required to accomplish this feat. KSHV four viral interferon regulatory factors (vIRF4) specifically inhibit IRF7 dimerization [60]. HSV-1 ICP27 targets the TBK1-activated STING signalosome to prevent IFN-I production [61], and the US11 protein binds to RIG-I and MDA5 to inhibit their downstream signalling pathway [62]. In this study, we first found that the UL41 protein inhibited duck RIG-I/MDA5-mediated duck IFN-β production via mRNA degradation activity. Infection with the DPV CHv-BAC-ΔUL41 mutant virus induced more duck IFN-β and ISGs (Mx and OASL) production than DPV CHv-BAC parent virus infection in DEF cells. Collectively, these results showed that DPV infection might evade immune responses via the UL41 protein.
VHS protein has been identified as an IFN-α/β resistance factor. HSV-2 VHS-deficient mutants induce > 50-fold more IFN-α/β than wild-type and VHS-rescued viruses in primary murine embryonic fibroblast (MEF) cells. HSV-2 VHS-deficient mutants are greatly attenuated in vivo, and replication and virulence are largely restored to the levels of the wild-type virus in mice lacking the IFN-α/β receptor [21, 63,64,65]. In addition, the HSV-1 VHS protein acts as a critical determinant of viral pathogenesis, and VHS-deficient mutants induce IFN and ISG production and increase susceptibility to IFN in cells [64]. However, the MDV UL41 gene deletion mutant replicated in cell culture, and the degrees of tumour lesions and neurovirulence were equivalent to those of the lesions induced by the parental virus [66]. In this study, we found that the DPV UL41 protein induced increased, physiologically active levels of duck IFN-β and increased amounts of ISGs in DEF cells. Moreover, the DPV UL41 protein was shown to function as a duck IFN-β resistance factor to facilitate viral replication.
The VHS protein is an mRNA-specific RNase that evades the host innate immune response. The HSV-1 VHS protein directly degrades cGAS mRNA to downregulate IFN-β activation [67]. The HSV-2 VHS protein inhibits TLR2/3- and RIG-I/MDA5-mediated antiviral pathways [68]. The bovine herpesvirus 1 (BoHV-1) VHS protein does not affect TBK1- or IRF3-induced IFN-β production but suppresses the antiviral innate immune response by directly targeting the STAT1 transcript [69]. In this study, we first found that the DPV UL41 protein broadly abrogated RIG-I-, MDA5-, MAVS-, STING-, TBK1- and IRF7-mediated IFN-β-Luc activation and significantly inhibited the mRNA levels of these important adaptor molecules. Therefore, consistent with the UL41 proteins of other alphaherpesviruses, the DPV UL41 protein evades the host innate immune response by broadly regulating mRNA levels.
On the one hand, the VHS protein cleaves RNA on the 3’ sides of U and C residues in vitro [22]. The VHS protein degrades ribosome-associated mRNA by interacting with the cap-binding initiation factor in vivo [23]. Compared with the HSV-1 VHS protein, the DPV UL41 protein might cleave different mRNA sites to broadly degrade the molecules, but more experiments are required to explore this hypothesis. On the other hand, the VHS protein specifically degrades ARE-containing RNAs [70]. The HSV-1 VHS protein degrades cGAS mRNA, which contains three ARE core motifs (ATTTA) in the 3’ untranslated region (UTR) [67]. The BoHV-1 VHS protein binds the second ARE motif of STAT1 mRNA [69]. We also predicted the ARE motifs in the duck RIG-I, MDA5, MAVS, STING, TBK1 and IRF7 proteins and found they existed in the 3′UTRs of MAVS, STING, and TBK1 but not in that of RIG-I or IRF7. Therefore, we speculated that the mechanism by which the DPV UL41 protein degrades mRNA differs from that of the HSV-1 VHS protein. We also speculated that the DPV UL41 protein inhibits duck IFN-β production through other mechanisms. In addition, the HSV-1 VHS protein limits the accumulation of dsRNA [71]. We also speculated that the DPV UL41 protein directly destabilize dsRNA to downregulate the activation of important adaptor proteins in the RIG-I/MDA5 immune pathway.
The VHS protein and its homologues are present in only the Alphaherpesvirinae subfamily, and the VHS polypeptides of alphaherpesviruses are highly conserved [48]. A smaller but significant number of conserved residues, E192, D194, and D195, in the VHS protein were identified as crucial RNase active sites [50, 72]. We also identified and mutated the three conserved residues of E229, D231, and D232 in the DPV UL41 protein to alanine, and the mUL41 protein rescued duck IFN-β production and the mRNA levels of the important adaptor proteins. These results further showed that the active sites were highly conserved. In summary, this study is the first time to confirm that the DPV UL41 protein suppresses RIG-I/MDA5-mediated duck IFN-β production.
Availability of data and materials
The datasets analysed in this study are available from the corresponding author upon reasonable request.
Abbreviations
- DPV:
-
duck plague virus
- RIG-I:
-
retinoic acid-inducible gene I
- IFN-β:
-
interferon-β
- MDA5:
-
melanoma differentiation-associated gene 5
- MAVS:
-
mitochondrial antiviral signalling protein
- STING:
-
stimulator of interferon gene
- TBK1:
-
TANK-binding kinase 1
- IRF7:
-
interferon regulatory factors 7
- dsRNA:
-
double-stranded RNA
- VHS:
-
virion host shutoff
- HSV-1:
-
herpes simplex virus-1
- MHC:
-
major histocompatibility complex
- ISGs:
-
IFN-stimulated genes
- STAT1:
-
signal transducer and activator of transcription 1
- DEFs:
-
duck embryo fibroblasts
- FBS:
-
foetal bovine serum
- IFA:
-
indirect immunofluorescence assay
- KSHV:
-
Kaposi’s sarcoma-associated herpesvirus
- EBV:
-
Epstein–Barr virus
- MHV68:
-
murine gammaherpesvirus 68
References
Jahan AS, Biquand E, Munoz-Moreno R, Le Quang A, Mok CK, Wong HH, Teo QW, Valkenburg SA, Chin AWH, Man Poon LL, Te Velthuis A, García-Sastre A, Demeret C, Sanyal S (2020) OTUB1 is a key regulator of RIG-I-dependent immune signaling and is targeted for proteasomal degradation by influenza A NS1. Cell Rep 30:1570–1584. https://doi.org/10.1016/j.celrep.2020.01.015
Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S (2011) Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 147:423–435. https://doi.org/10.1016/j.cell.2011.09.039
Belgnaoui SM, Paz S, Hiscott J (2011) Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr Opin Immunol 23:564–572. https://doi.org/10.1016/j.coi.2011.08.001
Ishikawa H, Barber GN (2008) STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455:674–678. https://doi.org/10.1038/nature07317
Nazmi A, Mukhopadhyay R, Dutta K, Basu A (2012) STING mediates neuronal innate immune response following Japanese encephalitis virus infection. Sci Rep 2:347. https://doi.org/10.1038/srep00347
Zevini A, Olagnier D, Hiscott J (2017) Crosstalk between cytoplasmic RIG-I and STING sensing pathways. Trends Immunol 38:194–205. https://doi.org/10.1016/j.it.2016.12.004
Gan Z, Yang YC, Chen SN, Hou J, Laghari ZA, Huang B, Li N, Nie P (2018) Unique composition of intronless and intron-containing type I IFNs in the Tibetan Frog Nanorana parkeri provides new evidence to support independent retroposition hypothesis for Type I IFN genes in amphibians. J Immunol 201:3329–3342. https://doi.org/10.4049/jimmunol.1800553
Cheng A (2015) Duck plague, 1st edn. China agriculture press, Beijing
Wu Y, Cheng A, Wang M, Zhang S, Zhu D, Jia R, Luo Q, Chen Z, Chen X (2011) Establishment of real-time quantitative reverse transcription polymerase chain reaction assay for transcriptional analysis of duck enteritis virus UL55 gene. Virol J 8:266. https://doi.org/10.1186/1743-422X-8-266
Zhao C, Wang M, Cheng A, Yang Q, Wu Y, Zhu D, Chen S, Liu M, Zhao X, Jia R, Sun K, Chen X (2018) Programmed cell death: the battlefield between the host and alpha-herpesviruses and a potential avenue for cancer treatment. Oncotarget 9:30704–30719. https://doi.org/10.18632/oncotarget.25694
You Y, Cheng AC, Wang MS, Jia RY, Sun KF, Yang Q, Wu Y, Zhu D, Chen S, Liu MF, Zhao XX, Chen XY (2017) The suppression of apoptosis by alpha-herpesvirus. Cell Death Dis 8:e2749. https://doi.org/10.1038/cddis.2017.139
Zhao C, Wang M, Cheng A, Yang Q, Wu Y, Jia R, Zhu D, Chen S, Liu M, Zhao X, Zhang S, Liu Y, Yu Y, Zhang L, Tian B, Rehman MU, Pan L, Chen X (2019) Duck plague virus promotes DEF cell apoptosis by activating caspases, increasing intracellular ROS levels and inducing cell cycle S-phase arrest. Viruses 11:196. https://doi.org/10.3390/v11020196
Huang J, Jia R, Wang M, Shu B, Yu X, Zhu D, Chen S, Yin Z, Chen X, Cheng A (2014) An attenuated duck plague virus (DPV) vaccine induces both systemic and mucosal immune responses to protect ducks against virulent DPV infection. Clin Vaccine Immunol 21:457–462. https://doi.org/10.1128/CVI.00605-13
Yang X, Qi X, Cheng A, Wang M, Zhu D, Jia R, Chen X (2010) Intestinal mucosal immune response in ducklings following oral immunisation with an attenuated duck enteritis virus vaccine. Vet J 185:199–203. https://doi.org/10.1016/j.tvjl.2009.04.011
Shen FX, Ma GP, Cheng AC, Wang MS, Li CF, Sun KF, Chang H, Zhu DK, Jia RY, Chen XY, Sun T (2010) Development and application of an indirect immunohistochemical method for the detection of duck plague virus vaccine antigens in paraffin sections and localization in the vaccinated duckling tissues. Poult Sci 89:1915–1923. https://doi.org/10.3382/ps.2010-00848
Yu X, Jia R, Huang J, Shu B, Zhu D, Liu Q, Gao X, Lin M, Yin Z, Wang M, Chen S, Wang Y, Chen X, Cheng A (2012) Attenuated Salmonella typhimurium delivering DNA vaccine encoding duck enteritis virus UL24 induced systemic and mucosal immune responses and conferred good protection against challenge. Vet Res 43:56. https://doi.org/10.1186/1297-9716-43-56
Wen Y, Cheng A, Wang M, Ge H, Shen C, Liu S, Xiang J, Jia R, Zhu D, Chen X, Lian B, Chang H, Zhou Y (2010) A thymidine kinase recombinant protein-based ELISA for detecting antibodies to duck plague virus. Virol J 7:77. https://doi.org/10.1186/1743-422X-7-77
Lian B, Cheng A, Wang M, Zhu D, Luo Q, Jia R, Liu F, Han X, Chen X (2011) Induction of immune responses in ducks with a DNA vaccine encoding duck plague virus glycoprotein C. Virol J 8:214. https://doi.org/10.1186/1743-422X-8-214
Chang H, Cheng A, Wang M, Jia R, Zhu D, Luo Q, Chen Z, Zhou Y, Liu F, Chen X (2011) Immunofluorescence analysis of duck plague virus gE protein on DPV-infected ducks. Virology J 8:19. https://doi.org/10.1186/1743-422X-8-19
Yang L, Wang M, Cheng A, Yang Q, Wu Y, Jia R, Liu M, Zhu D, Chen S, Zhang S, Zhao X, Huang J, Wang Y, Xu Z, Chen Z, Zhu L, Luo Q, Liu Y, Yu Y, Zhang L, Tian B, Pan L, Rehman MU, Chen X (2019) Innate immune evasion of Alphaherpesvirus tegument proteins. Front Immunol 10:2196. https://doi.org/10.3389/fimmu.2019.02196
He T, Wang M, Cheng A, Yang Q, Wu Y, Jia R, Liu M, Zhu D, Chen S, Zhang S, Zhao XX, Huang J, Sun D, Mao S, Ou X, Wang Y, Xu Z, Chen Z, Zhu L, Luo Q, Liu Y, Yu Y, Zhang L, Tian B, Pan L, Rehman MU, Chen X (2020) Host shutoff activity of VHS and SOX-like proteins: role in viral survival and immune evasion. Virol J 17:68. https://doi.org/10.1186/s12985-020-01336-8
Taddeo B, Roizman B (2006) The virion host shutoff protein (UL41) of herpes simplex virus 1 is an endoribonuclease with a substrate specificity similar to that of RNase A. J Virol 80:9341–9345. https://doi.org/10.1128/JVI.01008-06
Page HG, Read GS (2010) The virion host shutoff endonuclease (UL41) of herpes simplex virus interacts with the cellular cap-binding complex eIF4F. J Virol 84:6886–6890. https://doi.org/10.1128/JVI.00166-10
Smiley JR (2004) Herpes simplex virus virion host shutoff protein: immune evasion mediated by a viral RNase? J Virol 78:1063–1068. https://doi.org/10.1128/jvi.78.3.1063-1068.2004
Taddeo B, Zhang W, Roizman B (2013) The herpes simplex virus host shutoff RNase degrades cellular and viral mRNAs made before infection but not viral mRNA made after infection. J Virol 87:4516–4522. https://doi.org/10.1128/JVI.00005-13
Tigges MA, Leng S, Johnson DC, Burke RL (1996) Human herpes simplex virus (HSV)-specific CD8+ CTL clones recognize HSV-2-infected fibroblasts after treatment with IFN-gamma or when virion host shutoff functions are disabled. J Immunol 156:3901–3910. https://doi.org/10.1089/aid.1996.12.741
Gopinath RS, Ambagala AP, Hinkley S, Srikumaran S (2002) Effects of virion host shut-off activity of bovine herpesvirus 1 on MHC class I expression. Viral Immunol 15:595–608. https://doi.org/10.1089/088282402320914539
Trgovcich J, Johnson D, Roizman B (2002) Cell surface major histocompatibility complex class II proteins are regulated by the products of the gamma(1)34.5 and U(L)41 genes of herpes simplex virus 1. J Virol 76:6974–6986. https://doi.org/10.1128/jvi.76.14.6974-6986.2002
Samady L, Costigliola E, MacCormac L, McGrath Y, Cleverley S, Lilley CE, Smith J, Latchman DS, Chain B, Coffin RS (2003) Deletion of the virion host shutoff protein (VHS) from herpes simplex virus (HSV) relieves the viral block to dendritic cell activation: potential of vhs− HSV vectors for dendritic cell-mediated immunotherapy. J Virol 77:3768–3776. https://doi.org/10.1128/jvi.77.6.3768-3776.2003
You H, Yuan H, Fu W, Su C, Wang W, Cheng T, Zheng C (2017) Herpes simplex virus type 1 abrogates the antiviral activity of Ch25h via its virion host shutoff protein. Antiviral Res 143:69–73. https://doi.org/10.1016/j.antiviral.2017.04.004
Su C, Zhang J, Zheng C (2015) Herpes simplex virus 1 UL41 protein abrogates the antiviral activity of hZAP by degrading its mRNA. Virol J 12:203. https://doi.org/10.1186/s12985-015-0433-y
Zenner HL, Mauricio R, Banting G, Crump CM (2013) Herpes simplex virus 1 counteracts tetherin restriction via its virion host shutoff activity. J Virol 87:13115–13123. https://doi.org/10.1128/JVI.02167-13
Shen G, Wang K, Wang S, Cai M, Li ML, Zheng C (2014) Herpes simplex virus 1 counteracts viperin via its virion host shutoff protein UL41. J Virol 88:12163–12166. https://doi.org/10.1128/JVI.01380-14
Jiang Z, Su C, Zheng C (2016) Herpes simplex virus 1 tegument protein UL41 counteracts IFIT3 antiviral innate immunity. J Virol 90:11056–11061. https://doi.org/10.1128/JVI.01672-16
Lin HW, Hsu WL, Chang YY, Jan MS, Wong ML, Chang TJ (2010) Role of the UL41 protein of pseudorabies virus in host shutoff, pathogenesis and induction of TNF-α expression. J Vet Med Sci 72:1179–1187. https://doi.org/10.1292/jvms.10-0059
Huo H, Wang Y, Wang D, Wang Y, Chen X, Zhao L, Chen H (2019) Duck RIG-I restricts duck enteritis virus infection. Vet Microbiol 230:78–85. https://doi.org/10.1016/j.vetmic.2019.01.014
Huo H, Zhao L, Wang D, Chen X, Chen H (2019) LGP2 plays a critical role in MDA5-mediated antiviral activity against duck enteritis virus. Mol Immunol 116:160–166. https://doi.org/10.1016/j.molimm.2019.10.006
Chen S, Wu Z, Zhang J, Wang M, Jia R, Zhu D, Liu M, Sun K, Yang Q, Wu Y, Zhao X, Cheng A (2018) Duck stimulator of interferon genes plays an important role in host anti-duck plague virus infection through an IFN-dependent signalling pathway. Cytokine 102:191–199. https://doi.org/10.1016/j.cyto.2017.09.008
Tian B, Cai D, He T, Deng L, Wu L, Wang M, Jia R, Zhu D, Liu M, Yang Q, Wu Y, Zhao X, Chen S, Zhang S, Huang J, Ou X, Mao S, Yu Y, Zhang L, Liu Y, Cheng A (2020) Isolation and selection of duck primary cells as pathogenic and innate immunologic cell models for duck plague virus. Front Immunol 10:3131. https://doi.org/10.3389/fimmu.2019.03131
Liu T, Cheng A, Wang M, Jia R, Yang Q, Wu Y, Sun K, Zhu D, Chen S, Liu M, Zhao X, Chen X (2017) RNA-seq comparative analysis of Peking ducks spleen gene expression 24 h post-infected with duck plague virulent or attenuated virus. Vet Res 48:47. https://doi.org/10.1186/s13567-017-0456-z
He T, Wang M, Cheng A, Yang Q, Jia R, Wu Y, Huang J, Chen S, Zhao XX, Liu M, Zhu D, Zhang S, Ou X, Mao S, Gao Q, Sun D, Wen X, Tian B, Liu Y, Yu Y, Zhang L, Pan L, Chen X (2020) Duck enteritis virus pUL47, as a late structural protein localized in the nucleus, mainly depends on residues 40 to 50 and 768 to 777 and inhibits IFN-beta signalling by interacting with STAT1. Vet Res 51:135. https://doi.org/10.1186/s13567-020-00859-w
Wu Y, Cheng A, Wang M, Zhu D, Jia R, Chen S, Zhou Y, Chen X (2012) Comparative genomic analysis of duck enteritis virus strains. J Virol 86:13841–13842. https://doi.org/10.1128/JVI.01517-12
He T, Wang M, Cheng A, Yang Q, Jia R, Wu Y, Huang J, Tian B, Liu M, Chen S, Zhao XX, Zhu D, Zhang S, Ou X, Mao S, Gao Q, Sun D (2021) DPV UL41 gene encoding protein induces host shutoff activity and affects viral replication. Vet Microbiol 255:108979. https://doi.org/10.1016/j.vetmic.2021.108979
Wu Z, Zhang W, Wu Y, Wang T, Wu S, Wang M, Jia R, Zhu D, Liu M, Zhao X, Yang Q, Wu Y, Zhang S, Liu Y, Zhang L, Yu Y, Pan L, Merits A, Chen S, Cheng A (2019) Binding of the duck Tembusu virus protease to STING is mediated by NS2B and is crucial for STING cleavage and for impaired induction of IFN-β. J Immunol 203:3374–3385. https://doi.org/10.4049/jimmunol.1900956
Liu C, Cheng A, Wang M, Chen S, Jia R, Zhu D, Liu M, Sun K, Yang Q, Wu Y, Zhao X, Chen X (2017) Regulation of viral gene expression by duck enteritis virus UL54. Sci Rep 7:1076. https://doi.org/10.1038/s41598-017-01161-0
Chang H, Cheng A, Wang M, Jia R, Zhu D, Luo Q, Chen Z, Zhou Y, Liu F, Chen X (2011) Immunofluorescence analysis of duck plague virus gE protein on DPV-infected ducks. Virol J 8:19. https://doi.org/10.1186/1743-422X-8-19
Zhang W, Jiang B, Zeng M, Duan Y, Wu Z, Wu Y, Wang T, Wang M, Jia R, Zhu D, Liu M, Zhao X, Yang Q, Wu Y, Zhang S, Liu Y, Zhang L, Yu Y, Pan L, Chen S, Cheng A (2020) Binding of duck Tembusu virus nonstructural protein 2A to duSTING disrupts the induction of its signal transduction cascade to inhibit IFN-β induction. J Virol 94:e01850-e1919. https://doi.org/10.1128/JVI.01850-19
Everly DN, Feng P, Mian IS, Read GS (2002) mRNA degradation by the virion host shutoff (Vhs) protein of herpes simplex virus: genetic and biochemical evidence that VHS is a nuclease. J Virol 76:8560–8571. https://doi.org/10.1128/jvi.76.17.8560-8571.2002
Taddeo B, Zhang W, Roizman B (2006) The U(L)41 protein of herpes simplex virus 1 degrades RNA by endonucleolytic cleavage in absence of other cellular or viral proteins. Proc Natl Acad Sci USA 103:2827–2832. https://doi.org/10.1073/pnas.0510712103
Cormican P, Lloyd AT, Downing T, Connell SJ, Bradley D, O’Farrelly C (2009) The avian Toll-Like receptor pathway-Subtle differences amidst general conformity. Dev Comp Immunol 33:967–973. https://doi.org/10.1016/j.dci.2009.04.001
Chen S, Wang T, Liu P, Yang C, Wang M, Jia R, Zhu D, Liu M, Yang Q, Wu Y, Zhao X, Cheng A (2019) Duck interferon regulatory factor 7 (IRF7) can control duck Tembusu virus (DTMUV) infection by triggering type I interferon production and its signal transduction pathway. Cytokine 113:31–38. https://doi.org/10.1016/j.cyto.2018.06.001
Takeuchi O, Akira S (2010) Pattern recognition receptors and inflammation. Cell 140:805–820. https://doi.org/10.1016/j.cell.2010.01.022
Magor KE, Miranzo Navarro D, Barber MR, Petkau K, Fleming-Canepa X, Blyth GA, Blaine AH (2013) Defense genes missing from the flight division. Dev Comp Immunol 41:377–388. https://doi.org/10.1016/j.dci.2013.04.010
Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR (2006) Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol 80:5059–5064. https://doi.org/10.1128/JVI.80.10.5059-5064.2006
West JA, Wicks M, Gregory SM, Chugh P, Jacobs SR, Zhang Z, Host KM, Dittmer DP, Damania B (2014) An important role for mitochondrial antiviral signaling protein in the Kaposi’s sarcoma-associated herpesvirus life cycle. J Virol 88:5778–5787. https://doi.org/10.1128/JVI.03226-13
Jangra S, Yuen KS, Botelho MG, Jin DY (2019) Epstein–Barr virus and innate immunity: friends or foes? Microorganisms 7:183. https://doi.org/10.3390/microorganisms7060183
Zhao Y, Karijolich J (2019) Know thyself: RIG-I-like receptor sensing of DNA virus infection. J Virol 93:e01085-e1119. https://doi.org/10.1128/JVI.01085-19
Rasmussen SB, Jensen SB, Nielsen C, Quartin E, Kato H, Chen ZJ, Silverman RH, Akira S, Paludan SR (2009) Herpes simplex virus infection is sensed by both Toll-like receptors and retinoic acid-inducible gene-like receptors, which synergize to induce type I interferon production. J Gen Virol 90:74–78. https://doi.org/10.1099/vir.0.005389-0
Osterrieder K (2017) Cell biology of herpes viruses. Springer International Publishing, Cham, pp 1–18
Hwang SW, Kim D, Jung JU, Lee HR (2017) KSHV-encoded viral interferon regulatory factor 4 (vIRF4) interacts with IRF7 and inhibits interferon alpha production. Biochem Biophys Res Commun 486:700–705. https://doi.org/10.1016/j.bbrc.2017.03.101
Christensen MH, Jensen SB, Miettinen JJ, Luecke S, Prabakaran T, Reinert LS, Mettenleiter T, Chen ZJ, Knipe DM, Sandri-Goldin RM, Enquist LW, Hartmann R, Mogensen TH, Rice SA, Nyman TA, Matikainen S, Paludan SR (2016) HSV-1 ICP27 targets the TBK1-activated STING signalsome to inhibit virus-induced type I IFN expression. EMBO J 35:1385–1399. https://doi.org/10.15252/embj.201593458
Xing J, Wang S, Lin R, Mossman KL, Zheng C (2012) Herpes simplex virus 1 tegument protein US11 downmodulates the RLR signaling pathway via direct interaction with RIG-I and MDA-5. J Virol 86:3528–3540. https://doi.org/10.1128/JVI.06713-11
Duerst RJ, Morrison LA (2004) Herpes simplex virus 2 virion host shutoff protein interferes with type I interferon production and responsiveness. Virology 322:158–167. https://doi.org/10.1016/j.virol.2004.01.019
Murphy JA, Duerst RJ, Smith TJ, Morrison LA (2003) Herpes simplex virus type 2 virion host shutoff protein regulates alpha/beta interferon but not adaptive immune responses during primary infection in vivo. J Virol 77:9337–9345. https://doi.org/10.1128/jvi.77.17.9337-9345.2003
Pasieka TJ, Lu B, Crosby SD, Wylie KM, Morrison LA, Alexander DE, Menachery VD, Leib DA (2008) Herpes simplex virus virion host shutoff attenuates establishment of the antiviral state. J Virol 82:5527–5535. https://doi.org/10.1128/JVI.02047-07
Gimeno I, Silva RF (2008) Deletion of the Marek’s disease virus UL41 gene (VHS) has no measurable effect on latency or pathogenesis. Virus Genes 36:499–507. https://doi.org/10.1007/s11262-008-0215-3
Su C, Zheng C (2017) Herpes simplex virus 1 abrogates the cGAS/STING-mediated cytosolic DNA-sensing pathway via its virion host shutoff protein, UL41. J Virol 91:e02414-e2416. https://doi.org/10.1128/JVI.02414-16
Yao XD, Rosenthal KL (2011) Herpes simplex virus type 2 virion host shutoff protein suppresses innate dsRNA antiviral pathways in human vaginal epithelial cells. J Gen Virol 92:1981–1993. https://doi.org/10.1099/vir.0.030296-0
Ma W, Wang H, He H (2019) Bovine herpesvirus 1 tegument protein UL41 suppresses antiviral innate immune response via directly targeting STAT1. Vet Microbiol 239:108494. https://doi.org/10.1016/j.vetmic.2019.108494
Esclatine A, Taddeo B, Evans L, Roizman B (2004) The herpes simplex virus 1 UL41 gene-dependent destabilization of cellular RNAs is selective and may be sequence-specific. Proc Natl Acad Sci USA 101:3603–3608. https://doi.org/10.1073/pnas.0400354101
Dauber B, Saffran HA, Smiley JR (2019) The herpes simplex virus host shutoff (VHS) RNase limits accumulation of double stranded RNA in infected cells: evidence for accelerated decay of duplex RNA. PLoS Pathog 15:e1008111. https://doi.org/10.1371/journal.ppat.1008111
Taddeo B, Sciortino MT, Zhang W, Roizman B (2007) Interaction of herpes simplex virus RNase with VP16 and VP22 is required for the accumulation of the protein but not for accumulation of mRNA. Proc Natl Acad Sci USA 104:12163–12168. https://doi.org/10.1073/pnas.0705245104
Acknowledgements
We thank our laboratory members who helped us to improve the research and the manuscript, with their skilful technical assistance, invaluable comments, and suggestions.
Funding
This work was supported by grants from the China Agriculture Research System of MOF and MARA, and the Sichuan Veterinary Medicine and Drug Innovation Group of the China Agricultural Research System (SCCXTD-2020-18).
Author information
Authors and Affiliations
Contributions
TH carried out the experiments and drafted the manuscript. MW and AC critically revised the manuscript and the experimental design. QY, YW, RJ, SC, DZ, ML, XZ, SZ, JH, BT, XO, SM, DS, QG, YY, LZ and YL helped with the experiments. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Animal Ethics Committee of Sichuan Agricultural University (2016–17). Experiments were conducted in accordance with approved guidelines.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
He, T., Wang, M., Cheng, A. et al. Duck plague virus UL41 protein inhibits RIG-I/MDA5-mediated duck IFN-β production via mRNA degradation activity. Vet Res 53, 22 (2022). https://doi.org/10.1186/s13567-022-01043-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13567-022-01043-y