Abstract
The presence of non-aureus staphylococci (NAS) in bovine rectal feces has recently been described. Similar to other mastitis causing pathogens, shedding of NAS in the environment could result in intramammary infection. The objective of this study was to investigate whether NAS strains present in feces can cause intramammary infection, likely via teat apex colonization. During a cross-sectional study in 5 dairy herds, samples were collected from the habitats quarter milk, teat apices, and rectal feces from 25%, 10%, and 25% of the lactating cows, respectively, with a cow serving as the source of one type of sample only. Samples from clinical mastitis cases were continuously collected during the 1-year study period as well. The 6 most prevalent NAS species, Staphylococcus (S.) chromogenes, S. cohnii, S. devriesei, S. equorum, S. haemolyticus, and S. hominis, were further subtyped by random amplification of polymorphic deoxyribonucleic acid polymerase chain reaction (RAPD-PCR), when the same NAS species was present in the same herd in the three habitats. For S. chromogenes, S. cohnii, S. devriesei, and S. haemolyticus, the same RAPD type was found in rectal feces, teat apices, and quarter milk, indicating that fecal NAS can infect the mammary gland. For S. hominis and S. equorum, we were unable to confirm the presence of the same RAPD types in the three habitats.
Similar content being viewed by others
Introduction
Non-aureus staphylococci (NAS), a heterogeneous group of bacteria, are the most frequently identified bacteria in bovine milk samples worldwide [1,2,3,4]. They are also colonizing the teat apex of both lactating and dry cows [5, 6] as well as the teat canal [7] and other body parts [8]. We recently reported that Staphylococcus (S.) agnetis, S. auricularis, S. chromogenes, S. cohnii, S. epidermidis, S. equorum, S. haemolyticus, S. hominis, S. kloosii, S. rostri, and S. xylosus can also be isolated from rectal feces [9], indicating cows are shedding NAS into the environment which could eventually give rise to the establishment of intramammary infection (IMI), most likely after colonization of the teat apices. In that respect, S. arlettae, S. auricularis, S. chromogenes, S. cohnii, S. devriesei, S. equorum, S. haemolyticus, S. hominis, and S. vitulinus were recovered from rectal feces as well as from teat apices and quarter milk samples collected in the same herds (Wuytack et al. unpublished observations).
Intramammary infection typically occurs when bacteria enter the mammary gland through the teat canal. Quirk et al. [7] documented that teat canal colonization with S. chromogenes, S. epidermidis, S. equorum, S. haemolyticus, S. hyicus, and S. simulans was associated with IMI although strain-typing was not performed. NAS originating from the teat apices have been reported as a potential cause of IMI, as well. E.g. teat apex colonization with S. chromogenes, S. equorum, and S. haemolyticus has been described as a risk factor for NAS IMI based on species-level identification [10,11,12]. To date, a possible link between the presence of NAS species in feces and on teat apices and IMI has not been studied using a strain-typing approach.
Therefore, the purpose of this study was to investigate whether fecal NAS are potential causes of NAS IMI, likely via colonization of the teat apices.
Materials and methods
Herds, cows, and samples
Samples were obtained from 5 commercial dairy farms, all of them participating in the local dairy herd improvement program in Flanders with an interval of 4 to 6 weeks between 2 test-days (CRV, Arnhem, the Netherlands). The study was conducted during a 1-year period from March 2017 to March 2018. During the study period, herds had an average number of 75 lactating cows (ranging from 50 to 106) and a yearly milk production of 9304 kg milk (ranging from 8646 to 11 564). Bulk milk data were retrieved from MCC Flanders (Lier, Belgium). The geometric mean of the monthly bulk milk somatic cell count (SCC) was 106 000 cells/mL milk during the study (ranging from 77 000 to 148 000).
Quarter milk, teat apex, and rectal fecal samples were collected during a single cross-sectional sampling in each herd. Dairy cows were sampled once, either for quarter milk, teat apex swabs, or rectal feces. Quarter milk samples (total n = 334) were randomly obtained from approximately 25% of the lactating cows and collected using the guidelines of the National Mastitis Council (NMC) [13]. The SCC of the quarter milk samples was determined with a Direct Cell Counter (DCC, DeLaval, Tumba, Sweden) and used to stratify the quarters as healthy quarters (SCC ≤ 50 000 cells/mL milk) or as quarters with subclinical mastitis (SCC > 50 000 cells/mL). The teat apices (total n = 192) of a total number equal to approximately 10% of the lactating cows, were randomly sampled with a dry cotton swab (Copan, Novolab, Belgium) as described by De Vliegher et al. [14]. Both lactating and dry cows were sampled with a distribution of 1/3 and 2/3, respectively, and the teat apices of the lactating cows were sampled twice, before and after milking. Rectal fecal samples (total n = 80) were randomly obtained from approximately 25% of the lactating cows, based on the guarded sampling technique as used by Wuytack et al. [9]. A cow could serve as the source of one type of sample (quarter milk, teat apex, rectal feces) only.
During the entire study period, quarter milk samples of all clinical mastitis cases (total n = 103) were collected secundum artem by the producers.
All samples were transported under cooled conditions (4 °C) to the Mastitis and Milk Quality Research Laboratory (Faculty of Veterinary Medicine, Ghent University, Merelbeke, Belgium).
Bacterial isolates
Quarter milk samples were plated with a 0.01 mL loop. Rectal feces were first diluted [1 g in 9 mL of 0.9 physiological salt solution (9 g NaCl/L, Eurovet, Bladel, The Netherlands)] and then plated with a 0.01 mL loop. All milk samples were streaked on aesculin blood agar (Oxoid, Aalst, Belgium), MacConkey agar (Oxoid), and the semi-selective agar, mannitol salt agar (MSA, Chapman medium, Oxoid) for the recovery of NAS isolates. Samples from teat apices or rectal feces were only streaked on MSA to prevent overgrowth of bacteria which are frequently observed [15, 16]. All plates were examined at 24 and 48 h after incubation at 37 °C. All phenotypically different colonies (≥ 1 colony) were analyzed following the NMC procedure guidelines for bacteriological identification [13]. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-Tof) was used to identify the Staphylococcus isolates at the species level [17]. When no identification with the MALDI-Tof could be obtained, rpoB, and/or 16S rRNA gene sequencing was performed [18]. All isolates originating from the MSA plates and phenotypically identified as Staphylococcus spp. were stored at −80 °C.
Random amplification of polymorphic DNA-analysis
A selection of isolates of the 6 most prevalent NAS species (S. chromogenes, S. cohnii, S. devriesei, S. equorum, S. haemolyticus, and S. hominis) was analyzed for further subtyping by random amplification of polymorphic DNA polymerase chain reaction (RAPD-PCR). The selection was based on the following criteria: the isolates had to (1) originate from the same herd, (2) belong to the same NAS species, and (3) be present in the 3 different habitats (quarter milk, teat apices, and rectal feces). If more than 20 teat apex isolates from the same NAS species were available, only 10 isolates were selected. In that case, teat apex isolates originating from different quarters and cows and yielding the highest number of colonies were selected.
Deoxyribonucleic acid (DNA) was extracted with the use of the commercial DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. All sets of extractions were run with a negative control consisting of kit reagents with no sample to control for kit or environmental contamination. Aliquots of the DNA samples were stored at −20 °C until further analysis.
The RAPD-PCR was performed as described by Wuytack et al. [9], using the primer D11344 with the following PCR conditions: 4 cycles of 94 °C at 5 min, 36 °C at 5 min, and 72 °C at 5 min and 30 cycles of 94 °C at 1 min, 36 °C at 1 min, and 72 °C at 2 min. Isolates belonging to the same NAS species, originating from the same farm were analyzed together, in the same PCR run. Images were imported in BioNumerics version 7.6.3 (Applied Maths, Sint-Martens-Latem, Belgium) and analyzed with the Dice similarity coefficient and the unweighted pair group method with arithmetic mean (UPGMA) with an optimization set at 0.5% and position tolerance at 1.0%. Isolates with the same banding pattern, number, and size of bands, were considered the same RAPD type and given an arbitrary lower case letter.
Results
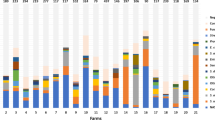
The isolates used in the current study are part of a larger study (Wuytack et al., unpublished) comparing the NAS distribution in milk, on teat apices, and in rectal feces in multiple commercial dairy herds, comprising a total of 1228 isolates (365 from milk, 830 from teat apices, and 33 from rectal feces, respectively). Nine NAS species were present in quarter milk, on teat apices, and in rectal feces in the same herd, namely S. arlettae, S. auricularis, S. chromogenes, S. cohnii, S. devriesei, S. equorum, S. haemolyticus, S. hominis, and S. vitulinus. Other species were isolated from only one habitat, namely S. hyicus and S. warneri in quarter milk and S. agnetis, S. kloosii, S. pettenkoferi, and S. rostri on teat apices. None of the NAS species were solely present in rectal feces. In total, 194 NAS isolates (70 from quarter milk, 103 from teat apices, and 21 from rectal feces) from the larger collection of 1228 isolates belonging to the species S. chromogenes, S. cohnii, S. devriesei, S. equorum, S. haemolyticus, and S. hominis were selected for RAPD-PCR (Table 1).
Among the 22 S. chromogenes isolates, originating from herd 1 only, 7 different fingerprints (a-g, 60% similarity) were identified (Figure 1). Five RAPD types (a, b, c, d, and f) were identified more than once. RAPD type d (n = 6) was isolated from all the habitats, i.e. quarter milk (as cause of both subclinical and clinical mastitis, n = 4), teat apices (n = 1), and rectal feces (n = 1). On the other hand, type f was only detected in milk, as cause of both subclinical and clinical mastitis. In cow F, multiple isolates of S. chromogenes (n = 4) were found in the same quarter (clinical mastitis) over a time period of 32 days, belonging to 3 different RAPD types.
Dendrogram of the RAPD fingerprints of 22Staphylococcus chromogenesisolates, originating from herd 1.1Cows were assigned an arbitrary upper case letter, independently of the other dendrograms. 2RAPD types were assigned an arbitrary lower case letter based on the clustering. HQuarter milk samples with a SCC ≤ 50 000 cells/mL milk (healthy). SQuarter milk samples with a SCC > 50 000 cells/mL milk (subclinical mastitis). CQuarter milk samples from a quarter with clinical signs of mastitis (clinical mastitis).
Seventeen S. cohnii isolates originating from herd 2 only, were divided over 4 RAPD types (a–d, 86.1% similarity, Figure 2), with a, c, and d being identified more than once. Isolates of RAPD type a (n = 5) were isolated from quarter milk with a low SCC, teat apices, and rectal feces from 5 different cows, whereas type b and c were only detected on teat apices. RAPD type d (n = 9) was mainly found on either 2 or 3 teat apices from 3 different cows and 2 quarter milk samples, but not in feces (Figure 2).
Dendrogram of the RAPD fingerprints of 17Staphylococcus cohniiisolates, originating from herd 2.1Cows were assigned an arbitrary upper case letter, independently of the other dendrograms. 2RAPD types were assigned an arbitrary lower case letter based on the clustering. HQuarter milk samples with a SCC ≤ 50 000 cells/mL milk (healthy). SQuarter milk samples with a SCC > 50 000 cells/mL milk (subclinical mastitis). C Quarter milk samples from a quarter with clinical signs of mastitis (clinical mastitis). CQuarter milk samples from a quarter with clinical signs of mastitis (clinical mastitis).
Random amplification of polymorphic DNA divided the 22 isolates of S. devriesei from herd 4 in 4 RAPD types (a–d, 66% similarity, Figure 3), with a, b, and d being identified more than once. RAPD type a was only colonizing teat apices whereas b was isolated 12 times from the three habitats, i.e. quarter milk both with a low and an elevated SCC, teat apices, and rectal feces. Cow A and J harbored multiple RAPD types in one quarter milk and one teat apex sample, respectively (Table 2).
Dendrogram of the RAPD fingerprints of 22Staphylococcus devrieseiisolates, originating from herd 4.1Cows were assigned an arbitrary upper case letter, independently of the other dendrograms. 2RAPD types were assigned an arbitrary lower case letter based on the clustering. HQuarter milk samples with a SCC ≤ 50 000 cells/mL milk (healthy). SQuarter milk samples with a SCC > 50 000 cells/mL milk (subclinical mastitis). C Quarter milk samples from a quarter with clinical signs of mastitis (clinical mastitis).
The 12 S. equorum isolates originating from herd 2 only, were divided in 6 RAPD types (e–f, 27.8% similarity, Figure 4), with a and d being identified more than once. None of the RAPD types was isolated from all the habitats. Type a (n = 2) was found in quarter milk (clinical mastitis) and rectal feces while type d (n = 6) was found in quarter milk from healthy quarters and on teat apices. All 4 teat apices of cow I were positive for S. equorum, belonging to RAPD type d or e.
Dendrogram of the RAPD fingerprints of 12Staphylococcus equorumisolates, originating from herd 2.1Cows were assigned an arbitrary upper case letter, independently of the other dendrograms. 2RAPD types were assigned an arbitrary lower case letter based on the clustering. HQuarter milk samples with a SCC ≤ 50 000 cells/mL milk (healthy). SQuarter milk samples with a SCC > 50 000 cells/mL milk (subclinical mastitis). CQuarter milk samples from a quarter with clinical signs of mastitis (clinical mastitis).
Staphylococcus haemolyticus was isolated from all habitats from two herds, i.e. herd 1 (n = 23, 27.3% similarity), with a, c, e, f, and g being identified more than once, and herd 5 (n = 16, 20.2% similarity), with a and c, being identified more than once. In herd 1, the isolates belonged to 7 different RAPD fingerprints (a–g, Figure 5). The isolates originating from the rectal feces (n = 2) belonged to the same RAPD type f, originating from cow A. RAPD type f was not retrieved from quarter milk or teat apices. The S. haemolyticus isolates originating from quarter milk or teat apices were divided in 6 other RAPD types. Cow H and I have multiple quarters harboring 1 or 2 S. haemolyticus RAPD types, while cow C, J, and K had multiple teat apices positive for one or two RAPD types (Figure 5). In herd 5, S. haemolyticus RAPD type c (n = 12) was found in quarter milk both with low SCC or clinical signs, teat apices, and rectal feces (Figure 6). Cow C and E had more than one teat apex positive for RAPD type c. Cow D has 2 teat apices positive for either RAPD type a or c.
Dendrogram of the RAP fingerprints of 23Staphylococcus haemolyticusisolates, originating from herd 1.1Cows were assigned an arbitrary upper case letter, independently of the other dendrograms. 2RAPD types were assigned an arbitrary lower case letter based on the clustering. HQuarter milk samples with a SCC ≤ 50 000 cells/mL milk (healthy). SQuarter milk samples with a SCC > 50 000 cells/mL milk (subclinical mastitis). CQuarter milk samples from a quarter with clinical signs of mastitis (clinical mastitis).
Dendrogram of the RAPD fingerprints of 16Staphylococcus haemolyticusisolates, originating from herd 5.1Cows were assigned an arbitrary upper case letter, independently of the other dendrograms. 2RAPD types were assigned an arbitrary lower case letter based on the clustering. HQuarter milk samples with a SCC ≤ 50 000 cells/mL milk (healthy). SQuarter milk samples with a SCC > 50 000 cells/mL milk (subclinical mastitis).
The 82 S. hominis isolates were found in quarter milk, teat apices, and rectal feces on all but herd 5 (Figures 7, 8, 9, 10). In herd 1, type c, e, and f were identified more than once, whereas this were type d, e, and h, type a and c, and type a, c, and e in herd 2, herd 3, and herd 4, respectively. The isolates were divided in 5 to 13 different RAPD types in the 4 herds with a similarity of 57.9%, 38.9%, 27.7%, and 37.8% in herd 1, 2, 3, and 4, respectively. The S. hominis isolates originating from rectal feces belonged to 1, 4, 1, and 2 RAPD types, on herd 1, 2, 3, and 4, respectively. Seven of those 8 types were habitat-specific, only RAPD type e from herd 2 consisted of isolates from both teat apices and rectal feces. In herd 1, RAPD type c and e were found in either quarter milk from clinical mastitis or teat apex samples only. The left hind quarter milk sample of cow B was positive for both RAPD type c and d. Cow H had one or two S. hominis types on all 4 teat apices. In herd 2, RAPD type h was recovered from teat apices only. Cow B, E, F, and G, had one or 2 RAPD types in the rectal fecal samples. Cow C, D, I, and J had 3 or 4 teat apices either positive for one or 2 RAPD types. Type a from herd 3 and type c from herd 4 were was recovered both from quarter milk and teat apices. As tabulated in Table 2, Cow G (herd 3) and cow K (herd 4) had one to 3 S. hominis RAPD types on multiple teat apices.
Dendrogram of the RAPD fingerprints of 18Staphylococcus hominisisolates, originating from herd 1.1Cows were assigned an arbitrary upper case letter, independently of the other dendrograms. 2RAPD types were assigned an arbitrary lower case letter based on the clustering. HQuarter milk samples with a SCC ≤ 50 000 cells/mL milk (healthy). SQuarter milk samples with a SCC > 50 000 cells/mL milk (subclinical mastitis). CQuarter milk samples from a quarter with clinical signs of mastitis (clinical mastitis).
Dendrogram of the RAPD fingerprints of 29Staphylococcus hominisisolates, originating from herd 2.1Cows were assigned an arbitrary upper case letter, independently of the other dendrograms. 2RAPD types were assigned an arbitrary lower case letter based on the clustering. HQuarter milk samples with a SCC ≤ 50 000 cells/mL milk (healthy). SQuarter milk samples with a SCC > 50 000 cells/mL milk (subclinical mastitis).
Dendrogram of the RAPD fingerprints of 13Staphylococcus hominisisolates, originating from herd 3.1Cows were assigned an arbitrary upper case letter, independently of the other dendrograms. 2RAPD types were assigned an arbitrary lower case letter based on the clustering. HQuarter milk samples with a SCC ≤ 50 000 cells/mL milk (healthy). SQuarter milk samples with a SCC > 50 000 cells/mL milk (subclinical mastitis). CQuarter milk samples from a quarter with clinical signs of mastitis (clinical mastitis).
Dendrogram of the RAPD fingerprints of 22Staphylococcus hominisisolates, originating from herd 4.1Cows were assigned an arbitrary upper case letter, independently of the other dendrograms. 2RAPD types were assigned an arbitrary lower case letter based on the clustering. HQuarter milk samples with a SCC ≤ 50 000 cells/mL milk (healthy). SQuarter milk samples with a SCC > 50 000 cells/mL milk (subclinical mastitis). CQuarter milk samples from a quarter with clinical signs of mastitis (clinical mastitis).
Discussion
This is the first study investigating whether fecal NAS are potential causes of bovine IMI, using RAPD-PCR. For S. chromogenes, S. devriesei, and S. haemolyticus (herd 5), the same RAPD fingerprints were found in rectal feces, teat apices, and quarter milk suggesting that NAS originating from rectal feces can cause IMI. Although the teat apex and teat canal form the main entryway to the udder for microorganisms [19], some authors suggest the existence of a so-called entero-mammary pathway [20]. Transfer of bacteria from the gut via intracellular transport to the mammary gland has been proven for mice and suggested for humans [21,22,23] yet the immune system of the mammary gland and gut is not as closely linked in ruminants as in monogastric species [24] making the hypothesis that NAS (or other bacteria) can reach the mammary gland without entering through the teat canal less likely [25]. A study specifically designed to test this hypothesis, would require the confirmation of the transfer of viable bacteria from the gut to peripheral lymph nodes and mammary gland via (migrating leukocytes in) the lymphatic and/or blood circulation. For S. cohnii, the same strains present in rectal feces and on teat apices were only found in milk samples from healthy quarters in the absence of inflammation. Either these strains should be seen as commensals, or the milk samples were contaminated when samples were taken [26].
For S. hominis, S. equorum, and S. haemolyticus (herd 1), we were unable to confirm the presence of the same strains in the three different habitats. This could reflect species differences or be due to the selection criteria and limited number of RAPD typed isolates per NAS species. Per herd only a random selection of lactating cows was sampled and each cow was sampled once for either quarter milk, teat apices, or rectal feces. Also, not all isolates originating from the teat apices were included in the RAPD analysis which could have influenced our results as well.
The diversity degree for S. chromogenes in this study is comparable to the genetic diversity reported by others [27,28,29], although a lower degree of diversity was presented by Shimizu et al. [30] and Piessens et al. [31]. For S. cohnii, high similarity scores were found indicating a low genetic diversity unlike prior research [29]. The diversity for S. devriesei is comparable to the findings of Supré et al. [32]. For S. equorum, a wide genetic diversity has been previously described for isolates originating from sausages and their environment [33]. Our findings corroborate with earlier studies that reported S. haemolyticus and S. hominis are highly diverse species and adaptable to various conditions [12, 29, 31]. The differences in reported diversity between studies can also be explained by the limited number and origin of isolates and the used techniques.
For S. haemolyticus (herd 1) and S. hominis, fecal isolates might be more adapted to survival in rectal feces than in quarter milk or on teat apices as only 1 out of 8 RAPD types divided over 4 herds was retrieved from both teat apices and rectal feces. For S. chromogenes, S. devriesei, S. equorum, S. haemolyticus (herd 5), and S. hominis, one or more strains were solely found in either quarter milk or on teat apices suggesting some strains within species could be more udder-adapted while others have no or a lower ability to colonize the mammary gland. Strain differences within NAS species have actually been confirmed before. E.g. we reported that S. chromogenes originating from a persistent IMI displays a better in vitro bacterial growth in conditions mimicking the mammary gland and in vivo bacterial growth in the mammary glands of mice and dairy cows, compared to S. chromogenes isolated from the teat apex [9, 34, 35]. Differences in inflammatory response in both mice and dairy cows between strains of the same species and growth inhibition of other major mastitis pathogens have been demonstrated as well [35, 36], indicating it is relevant to subtype NAS in future research efforts.
Some NAS species tend to be more relevant for the udder health, e.g. S. chromogenes [8, 37, 38]. This was substantiated in the current study as in herd 1, only milk samples from quarters with a SCC > 50 000 cells/mL milk and clinical signs harbored S. chromogenes. Also, the same strain was isolated from a quarter with subclinical mastitis and a quarter with clinical signs, substantiating that not only the virulence potential of the pathogen but also host factors might influence the severity of mastitis. Staphylococcus hominis was reguraly isolated from quarters with subclinical and clinical mastitis in herd 1 and 4. These results deviate from those reported by Jenkins et al. [29] and Condas et al. [39]. While S. cohnii was found more often in healthy quarters (SCC ≤ 50 000 cells/mL milk) which is in agreement with Supré et al. [37], suggesting a potential commensal nature of this species or sample contamination as S. cohnii was also isolated from one clinical mastitis case.
Random amplification of polymorphic DNA is a rapid and easy to perform technique that is commonly used for comparing bacterial communities with consistent results compared to other techniques such as restriction fragment length polymorphism, amplified fragment typing methods, and pulsed-field gel electrophoresis (PFGE) [40,41,42,43]. Still, it is known that RAPD can lack some discriminatory power for some species [31]. For specific species, e.g. S. haemolyticus and S. simulans, the combination with other techniques such as PFGE and amplified fragment typing methods could improve the detection of variation between isolates [31, 44]. Still, it is hard to compare fingerprint-based methods between laboratories [45]. The low mutation rate of housekeeping genes makes multilocus sequence typing more suitable, but it is currently only available for S. epidermidis, S. haemolyticus, S. hominis, S. lugdunensis, and S. pseudintermedius [46,47,48,49,50]. Whole genome sequencing provides a theoretically optimal resolution, but is, momentarily, less suitable for larger collections of isolates and routine analyses [51].
For S. chromogenes, S. cohnii, S. devriesei, and S. haemolyticus, the same RAPD type was found in rectal feces, teat apices, and quarter milk originating from the same herd, suggesting fecal NAS can infect the bovine mammary gland, at least for some species. For S. hominis and S. equorum, we were unable to confirm the presence of the same RAPD strain in the three habitats.
Availability of data and materials
The data on which the conclusions of the manuscript rely are presented in the main paper.
Abbreviations
- DNA:
-
deoxyribonucleic acid
- IMI:
-
intramammary infection
- MALDI-Tof:
-
matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- MSA:
-
mannitol salt agar
- n :
-
number
- NAS:
-
non-aureus staphylococci
- PFGE:
-
pulsed-field gel electrophoresis
- RAPD-PCR:
-
random amplification of polymorphic deoxyribonucleic acid polymerase chain reaction
- S. :
-
Staphylococcus
- SCC:
-
quarter somatic cell count
References
Pyörälä S, Taponen S (2009) Coagulase-negative staphylococci-emerging mastitis pathogens. Vet Microbiol 134:3–8
Condas LAZ, De Buck J, Nobrega DB, Carson DA, Naushad S, De Vliegher S, Zadoks RN, Middleton JR, Dufour S, Kastelic JP, Barkema HW (2017) Prevalence of non-aureus staphylococci species causing intramammary infections in Canadian dairy herds. J Dairy Sci 100:5592–5612
Sampimon OC, Barkema HW, Berends IM, Sol J, Lam TJ (2009) Prevalence and herd-level risk factors for intramammary infection with coagulase-negative staphylococci in Dutch dairy herds. Vet Microbiol 134:37–44
De Vliegher S, Fox LK, Piepers S, McDougall S, Barkema HW (2012) Invited review: mastitis in dairy heifers: nature of the disease, potential impact, prevention, and control. J Dairy Sci 95:1025–1040
De Visscher A, Piepers S, Haesebrouck F, De Vliegher S (2016) Teat apex colonization with coagulase-negative Staphylococcus species before parturition: distribution and species-specific risk factors. J Dairy Sci 99:1427–1439
De Visscher A, Supré K, Haesebrouck F, Zadoks RN, Piessens V, Van Coillie E, Piepers S, De Vliegher S (2014) Further evidence for the existence of environmental and host-associated species of coagulase-negative staphylococci in dairy cattle. Vet Microbiol 172:466–474
Quirk T, Fox LK, Hancock DD, Capper J, Wenz J, Park J (2012) Intramammary infections and teat canal colonization with coagulase-negative staphylococci after postmilking teat disinfection: species-specific responses. J Dairy Sci 95:1906–1912
Adkins PRF, Dufour S, Spain JN, Calcutt MJ, Reilly TJ, Stewart GC, Middleton JR (2018) Molecular characterization of non-aureus Staphylococcus spp. from heifer intramammary infections and body sites. J Dairy Sci 101:5388–5403
Wuytack A, De Visscher A, Piepers S, Boyen F, Haesebrouck F, De Vliegher S (2019) Non-aureus staphylococci in fecal samples of dairy cows: first report and phenotypic and genotypic characterization. J Dairy Sci 102:9345–9359
De Visscher A, Piepers S, Haesebrouck F, De Vliegher S (2016) Intramammary infection with coagulase-negative staphylococci at parturition: species-specific prevalence, risk factors, and effect on udder health. J Dairy Sci 99:6457–6469
Mahmmod YS, Klaas IC, Svennesen L, Pedersen K, Ingmer H (2018) Communications of Staphylococcus aureus and non-aureus Staphylococcus species from bovine intramammary infections and teat apex colonization. J Dairy Sci 101:7322–7333
Leroy F, Van Coillie E, Braem G, Piessens V, Verbist B, De Vuyst L, De Vliegher S (2015) Short communication: subtyping of Staphylococcus haemolyticus isolates from milk and corresponding teat apices to verify the potential teat-skin origin of intramammary infections in dairy cows. J Dairy Sci 98:7893–7898
Hogan JS, Gonzalez RN, Harmon RJ, Nickerson SC, Oliver SP, Pankey JW, Smith KL (1999) Laboratory handbook on bovine mastitis, 2nd edn. National Mastitis Council, Madison
De Vliegher S, Laevens H, Devriese LA, Opsomer G, Leroy JLM, Barkema HW, de Kruif A (2003) Prepartum teat apex colonization with Staphylococcus chromogenes in dairy heifers is associated with low somatic cell count in early lactation. Vet Microbiol 92:245–252
De Visscher A, Haesebrouck F, Piepers S, Vanderhaeghen W, Supré K, Leroy F, Van Coillie E, De Vliegher S (2013) Assessment of the suitability of mannitol salt agar for growing bovine-associated coagulase-negative staphylococci and its use under field conditions. Res Vet Sci 95:347–351
Lugsomya K, Tummaruk P, Hampson DJ, Prapasarakul N (2012) Development of a modified selective medium to enhance the recovery rate of Brachyspira hyodysenteriae and other porcine intestinal spirochaetes from faeces. Lett Appl Microbiol 54:330–335
Cameron M, Perry J, Middleton JR, Chaffer M, Lewis J, Keefe GP (2018) Short communication: evaluation of MALDI-TOF mass spectrometry and a custom reference spectra expanded database for the identification of bovine-associated coagulase-negative staphylococci. J Dairy Sci 101:590–595
Supré K, De Vliegher S, Sampimon OC, Zadoks RN, Vaneechoutte M, Baele M, De Graef E, Piepers S, Haesebrouck F (2009) Technical note: use of transfer RNA-intergenic spacer PCR combined with capillary electrophoresis to identify coagulase-negative Staphylococcus species originating from bovine milk and teat apices. J Dairy Sci 92:3204–3210
Plastridge WN (1958) Bovine mastitis—a review. J Dairy Sci 41:1141–1181
Young W, Hine BC, Wallace OA, Callaghan M, Bibiloni R (2015) Transfer of intestinal bacterial components to mammary secretions in the cow. PeerJ 3:e888
Perez PF, Dore J, Leclerc M, Levenez F, Benyacoub J, Serrant P, Segura-Roggero I, Schiffrin EJ, Donnet-Hughes A (2007) Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics 119:e724–e732
Fernandez L, Langa S, Martin V, Maldonado A, Jimenez E, Martin R, Rodriguez JM (2013) The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res 69:1–10
Rodriguez JM (2014) The origin of human milk bacteria: is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv Nutr 5:779–784
Kehrli ME Jr, Harp JA (2001) Immunity in the mammary gland. Vet Clin North Am Food Anim Pract 17(495–516):vi
Rainard P (2017) Mammary microbiota of dairy ruminants: fact or fiction? Vet Res 48:25
Hiitio H, Simojoki H, Kalmus P, Holopainen J, Pyörälä S, Taponen S (2016) The effect of sampling technique on PCR-based bacteriological results of bovine milk samples. J Dairy Sci 99:6532–6541
Gillespie BE, Headrick SI, Boonyayatra S, Oliver SP (2009) Prevalence and persistence of coagulase-negative Staphylococcus species in three dairy research herds. Vet Microbiol 134:65–72
Rajala-Schultz PJ, Torres AH, DeGraves FJ, Gebreyes WA, Patchanee P (2009) Antimicrobial resistance and genotypic characterization of coagulase-negative staphylococci over the dry period. Vet Microbiol 134:55–64
Jenkins SN, Okello E, Rossitto PV, Lehenbauer TW, Champagne J, Penedo MCT, Arruda AG, Godden S, Rapnicki P, Gorden PJ, Timms LL, Aly SS (2019) Molecular epidemiology of coagulase-negative Staphylococcus species isolated at different lactation stages from dairy cattle in the United States. PeerJ 7:e6749
Shimizu A, Kloos WE, Berkhoff HA, George CG, Ballard DN (1997) Pulsed-field gel electrophoresis of Staphylococcus hyicus and Staphylococcus chromogenes genomic DNA and its taxonomic, epidemiologic and ecologic applications in veterinary medicine. J Vet Med Sci 59:443–450
Piessens V, De Vliegher S, Verbist B, Braem G, Van Nuffel A, De Vuyst L, Heyndrickx M, Van Coillie E (2012) Intra-species diversity and epidemiology varies among coagulase-negative Staphylococcus species causing bovine intramammary infections. Vet Microbiol 155:62–71
Supré K, De Vliegher S, Cleenwerck I, Engelbeen K, Van Trappen S, Piepers S, Sampimon OC, Zadoks RN, De Vos P, Haesebrouck F (2010) Staphylococcus devriesei sp. nov., isolated from teat apices and milk of dairy cows. Int J Syst Evol Microbiol 60:2739–2744
Leroy S, Lebert I, Chacornac JP, Chavant P, Bernardi T, Talon R (2009) Genetic diversity and biofilm formation of Staphylococcus equorum isolated from naturally fermented sausages and their manufacturing environment. Int J Food Microbiol 134:46–51
Souza FN, Piepers S, Della Libera AMMP, Heinemann MB, Cerqueira MMOP, De Vliegher S (2016) Interaction between bovine-associated coagulase-negative staphylococci species and strains and bovine mammary epithelial cells reflects differences in ecology and epidemiological behavior. J Dairy Sci 99:2867–2874
Piccart K, Verbeke J, De Visscher A, Piepers S, Haesebrouck F, De Vliegher S (2016) Local host response following an intramammary challenge with Staphylococcus fleurettii and different strains of Staphylococcus chromogenes in dairy heifers. Vet Res 47:56
Breyne K, De Vliegher S, De Visscher A, Piepers S, Meyer E (2015) Technical note: a pilot study using a mouse mastitis model to study differences between bovine associated coagulase-negative staphylococci. J Dairy Sci 98:1090–1100
Supré K, Haesebrouck F, Zadoks RN, Vaneechoutte M, Piepers S, De Vliegher S (2011) Some coagulase-negative Staphylococcus species affect udder health more than others. J Dairy Sci 94:2329–2340
De Visscher A, Piepers S, Supre K, Haesebrouck F, De Vliegher S (2015) Short communication: species group-specific predictors at the cow and quarter level for intramammary infection with coagulase-negative staphylococci in dairy cattle throughout lactation. J Dairy Sci 98:5448–5453
Condas LAZ, De Buck J, Nobrega DB, Carson DA, Roy JP, Keefe GP, DeVries TJ, Middleton JR, Dufour S, Barkema HW (2017) Distribution of non-aureus staphylococci species in udder quarters with low and high somatic cell count, and clinical mastitis. J Dairy Sci 100:5613–5627
Paffetti D, Scotti C, Gnocchi S, Fancelli S, Bazzicalupo M (1996) Genetic diversity of an Italian Rhizobium meliloti population from different Medicago sativa varieties. Appl Environ Microbiol 62:2279–2285
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breeding 2:225–238
Gonzalez-Rey C, Belin AM, Jorbeck H, Norman M, Krovacek K, Henriques B, Kallenius G, Svenson SB (2003) RAPD-PCR and PFGE as tools in the investigation of an outbreak of beta-haemolytic Streptococcus group A in a Swedish hospital. Comp Immunol Microb 26:25–35
Casey AL, Worthington T, Caddick JM, Hilton AC, Lambert PA, Elliott TS (2006) RAPD for the typing of coagulase-negative staphylococci implicated in catheter-related bloodstream infection. J Infect 52:282–289
Chiang YC, Lai CH, Lin CW, Chang CY, Tsen HY (2014) Improvement of strain discrimination by combination of superantigen profiles, PFGE, and RAPD for Staphylococcus aureus isolates from clinical Samples and food-poisoning cases. Foodborne Pathog Dis 11:468–477
Murchan S, Kaufmann ME, Deplano A, de Ryck R, Struelens M, Zinn CE, Fussing V, Salmenlinna S, Vuopio-Varkila J, El Solh N, Cuny C, Witte W, Tassios PT, Legakis N, van Leeuwen W, van Belkum A, Vindel A, Laconcha I, Garaizar J, Haeggman S, Olsson-Liljequist B, Ransjo U, Coombes G, Cookson B (2003) Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J Clin Microbiol 41:1574–1585
Thomas JC, Vargas MR, Miragaia M, Peacock SJ, Archer GL, Enright MC (2007) Improved multilocus sequence typing scheme for Staphylococcus epidermidis. J Clin Microbiol 45:616–619
Solyman SM, Black CC, Duim B, Perreten V, van Duijkeren E, Wagenaar JA, Eberlein LC, Sadeghi LN, Videla R, Bemis DA, Kania SA (2013) Multilocus sequence typing for characterization of Staphylococcus pseudintermedius. J Clin Microbiol 51:306–310
Zhang LF, Thomas JC, Miragaia M, Bouchami O, Chaves F, d’Azevedo PA, Aanensen DM, de Lencastre H, Gray BM, Robinson DA (2013) Multilocus sequence typing and further genetic characterization of the enigmatic pathogen, Staphylococcus hominis. PLoS One 8:e66496
Chassain B, Lemee L, Didi J, Thiberge JM, Brisse S, Pons JL, Pestel-Caron M (2012) Multilocus sequence typing analysis of staphylococcus lugdunensis implies a clonal population structure. J Clin Microbiol 50:3003–3009
Kornienko M, Ilina E, Lubasovskaya L, Priputnevich T, Falova O, Sukhikh G, Govorun V (2016) Analysis of nosocomial Staphylococcus haemolyticus by MLST and MALDI-TOF mass spectrometry. Infect Genet Evol 39:99–105
Cunningham SA, Chia N, Jeraldo PR, Quest DJ, Johnson JA, Boxrud DJ, Taylor AJ, Chen J, Jenkins GD, Drucker TM, Nelson H, Patel R (2017) Comparison of whole-genome sequencing methods for analysis of three methicillin-resistant Staphylococcus aureus outbreaks. J Clin Microbiol 55:1946–1953
Acknowledgements
The authors thank Kristien Mertens (Department of Reproduction, Obstetrics, and Herd Health, Faculty of Veterinary Medicine, Ghent University, Merelbeke, Belgium) for the technical support.
Author information
Authors and Affiliations
Contributions
AW performed the data sampling and laboratory analyses, analyzed the data, and contributed to drafting and editing the manuscript; ADV contributed to the study design and edited the manuscript; SP contributed to the study design and edited the manuscript; FH edited the manuscript; SDV contributed to the study design and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wuytack, A., De Visscher, A., Piepers, S. et al. Fecal non-aureus Staphylococci are a potential cause of bovine intramammary infection. Vet Res 51, 32 (2020). https://doi.org/10.1186/s13567-020-00761-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13567-020-00761-5