Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) is an important globally distributed and highly contagious pathogen that has restricted cell tropism in vivo and in vitro. In the present study, we found that annexin A2 (ANXA2) is upregulated expressed in porcine alveolar macrophages infected with PRRSV. Additionally, PRRSV replication was significantly suppressed after reducing ANXA2 expression in Marc-145 cells using siRNA. Bioinformatics analysis indicated that ANXA2 may be relevant to vimentin, a cellular cytoskeleton component that is thought to be involved in the infectivity and replication of PRRSV. Co-immunoprecipitation assays and confocal analysis confirmed that ANXA2 interacts with vimentin, with further experiments indicating that the B domain (109–174 aa) of ANXA2 contributes to this interaction. Importantly, neither ANXA2 nor vimentin alone could bind to PRRSV and only in the presence of ANXA2 could vimentin interact with the N protein of PRRSV. No binding to the GP2, GP3, GP5, nor M proteins of PRRSV was observed. In conclusion, ANXA2 can interact with vimentin and enhance PRRSV growth. This contributes to the regulation of PRRSV replication in infected cells and may have implications for the future antiviral strategies.
Similar content being viewed by others
Introduction
Porcine reproductive and respiratory syndrome (PRRS), caused by the PRRS virus (PRRSV), is one of the most economically important diseases affecting the global pig farming industry. It is characterized by late term gestation reproductive failure in sows and general respiratory symptoms in pigs of all ages and sexes [1,2,3,4]. PRRSV is a member of the Nidovirales order (Arteriviridae family) and consists of an enveloped 15 kb positive-strand RNA genome containing nine open reading frames (ORFs) [5]. ORF1a and ORF1b encode viral replicase polyproteins, while ORF2a, ORF2b, and ORFs 3–7, encode the viral structural proteins GP2, E, GP3, GP4, GP5, M, and N, respectively. All of these structural proteins are required for the PRRSV replication [6,7,8].
Annexin A2 (ANXA2) is a member of the annexin family of calcium-dependent proteins expressed in many cells [9], which share structural and functional features [10]. ANXA2 is also a multifunctional protein involved in many biological processes, including endocytosis, exocytosis, membrane domain organization, extracellular receptor activity, signal transduction, protein assembly, transcription and mRNA transport, as well as numerous pathologies [11,12,13,14,15]. For example, ANXA2 is involved in the production of classical swine fever virus infectious particles [16]. ANXA2 was identified as a novel host factor contributing to the formation of infectious HCV particles [17]. Vimentin, a cellular cytoskeleton component, also plays an important role in the infection process of PRRSV as the anti-vimentin mAb has been shown to block PRRSV infection [18, 19]. Vimentin is also thought to be involved in PRRSV replication and transportation of the virus into cells by forming a complex with other intermediate filament components [20]. However, the relationship between ANXA2 and vimentin is unknown.
In previous experiment, we examined PAMs infected with the highly pathogenic PRRSV HuN4 strain. We found that ANXA2 was differentially expressed in infected cells and bioinformatics analysis indicated that ANXA2 may associate with vimentin. To investigate the role of ANXA2 in PRRSV replication and the relationship of ANXA2 and vimentin, RNA interference assay showed that ANXA2 could promote PRRSV infection. In addition, we had confirmed that ANXA2 could interact with the vimentin for the first time, and ANXA2 together with vimentin, form a complex that could bind to PRRSV N protein. These findings contribute to understanding the cellular proteins on the role of regulating PRRSV replication and may have implications for the future control of this important disease.
Materials and methods
Cells and virus
PAMs were harvested from 4-week old PRRSV-negative piglets in sterilized phosphate-buffered saline (PBS) by alveolar lavage and then maintained in RPMI-1640 medium. The animal experiments in this study were conducted with recommendations in the Chinese Regulations of Laboratory Animals—The Guidelines for the Care of Laboratory Animals (Ministry of Science and Technology of People’s Republic of China) and Laboratory Animal-Requirements of Environment and Housing Facilities (GB14925-2010, National Laboratory Animal Standardization Technical Committee). Marc-145 cells and human embryonic kidney (HEK) 293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% fetal bovine serum (FBS). PRRSV HuN4 (GenBank No. EF635006), a highly pathogenic PRRSV strain [21], was maintained in our laboratory.
Two-dimensional difference gel electrophoresis (2D-DIGE) and image analysis
PAMs were infected with PRRSV HuN4 at a MOI of 0.01. At 48 hours post-infection (hpi), cells were harvested, treated, and separated by 2D-DIGE as previously described [22, 23]. Briefly, samples were actively rehydrated in 24 cm precast immobilized pH gradient (IPG) strips (GE Healthcare, Chicago, IL, USA) for 12 h using an Ettan IPGphor three system (GE Healthcare). Isoelectric focusing (IEF) was then performed for a total of 76 kV h. Once the IEF was completed, the IPG strips were equilibrated in equilibration buffer supplemented with 1% DTT for 15 min and washed for another 15 min. Separation in the second dimension was performed using 12.5% SDS-PAGE. After SDS-PAGE, gels were scanned on a Typhoon TRIO Scanner (GE Healthcare). Analysis of the 2D-DIGE was performed using DeCyder 6.5 software (GE Healthcare), according to the manufacturer’s recommendations.
Identification by mass spectrometry and Western blotting
Differentially expressed protein spots of interest were manually excised, treated, and identified by mass spectrometry using protocols and methodology from a previous study [22]. The expression levels of ANXA2 in HuN4 infected PAMs at 48 hpi were determined via Western blotting using a commercial polyclonal rabbit anti-ANXA2 mAb (Cell Signaling Technology, Danvers, MA, USA).
Bioinformatics analysis
STRING, an open source web-based tool consisting of established and predicted protein interactions, was utilized to analyze the protein–protein interaction networks, as described previously [24]. This tool integrates biomolecular interaction networks using high throughput expression results, and other molecular states, in a unified conceptual framework.
Knockdown and overexpression of ANXA2
Small interfering RNAs (siRNA) targeting the ANXA2 gene (siANXA2) were synthesized by GenePharma (Shanghai, China). The siRNA sequences (5′-GGACAUUAUUUCGGAUACATT-3′) used have been previously reported and validated [25]. The non-target control siRNA (siNC) sequence (5′-UUCUCCGAACGUGUCACGUTT-3′) was confirmed to have no matches in either the viral or Chlorocebus aethiops (green monkey) genome. Marc-145 cells were transfected with either 100 nM siANXA2 or siNC using X-tremeGene siRNA Transfection Reagent (Roche, Basel, Switzerland). At 36 hours post-transfection (hpt), the transfected cells were infected with PRRSV HuN4 at a MOI of 0.01. After 1 h, the cells were washed and incubated at 37 °C. At 24 and 48 hpi, the cell lysates were harvested for analysis by Western blotting. Viral RNA copy number and viral titers in the cell culture supernatants were also determined [26].
To construct a stable cell line overexpressing ANXA2, 293T cells grown in 10-cm cell dishes were co-transfected with 20 μg pFUGW-ANXA2 or pFUGW empty vector, together with 15 μg of pSPAX2 and 6 μg of pMD2G. The recombinant lentiviruses were harvested at 48 hpt and centrifugation at 6000 × g for 20 min at 4 °C using an Amicon Ultra-15 Centrifugal Filter Unit (Millipore, Burlington, MA, USA). The Marc-145 cells were then transduced with recombinant lentiviruses. At 36 hpt, the Marc-145 cells were infected with PRRSV HuN4 at a MOI of 0.01. At 48 hpi, cell culture supernatants and cell lysates were harvested for analysis.
Construction of the vimentin, ANXA2, and truncated-ANXA2 mutant plasmids
The full ANXA2 and vimentin gene sequences were amplified and cloned into an empty pCMV-HA vector (Clontech, Mountain View, CA, USA) to generate pANXA2-Flag and pVIM-HA, respectively. In order to identify the binding domain of ANXA2, various domain positions in ANXA2 were analyzed. According to the positions of the four putative domains, five truncated ANXA2 fragments were designed and amplified by PCR. The primers for amplifying these fragments are listed in Table 1.
Confocal microscopy
PAMs and Marc-145 cells at 70% confluence were washed three times and then fixed with 4% paraformaldehyde at room temperature for 30 min, blocked with 5% skimmed milk for 2 h at room temperature, and then incubated with the respective antibodies and then examined using a Leica SP2 confocal system (Leica Microsystems, Wetzlar, Germany).
Co-immunoprecipitation (Co-IP) and Western blotting
PAMs and Marc-145 cells were grown on 10-cm dishes and then harvested and washed three times with cold PBS. The cells were then lysed with 1% Triton X-100 containing protease inhibitor cocktail (Roche) on ice for 1 h. The cell lysates were then centrifuged at 13 000 × g for 10 min at 4 °C. The supernatants were precleared with Protein A/G PLUS-Agarose (Santa Cruz Biotechnology, Dallas, TX, USA) at 4 °C for 2 h and then incubated with anti-ANXA2 mAb with rotation at 4 °C for 2 h. The immunoprecipitates were washed five times with 1% Triton X-100 and bound proteins were analyzed by Western blotting.
Co-IP assays were also performed using exogenous ANXA2 or vimentin transfected 293T cells. The 293T cells were co-transfected with pVIM-HA and pANXA2-Flag, or with pVIM-HA and plasmids expressing the ANXA2 mutants (8 μg each), using X-tremeGENE HP DNA Transfection Reagent (Roche). At 48 hpt, the transfected cells were harvested, and lysed with 1% Triton X-100 containing protease inhibitor cocktail on ice for 1 h. Finally, the cell lysate was centrifuged at 13 000 × g for 10 min at 4 °C. The supernatants were transferred to a tube containing equilibrated anti-HA affinity gel beads (Sigma-Aldrich) or anti-Flag affinity gel beads (Sigma-Aldrich) and vortexed briefly and incubated with gentle mixing for 2 h at 4 °C. In the experiment of ANXA2 is required for vimentin binding to N, the supernatants obtained from HEK 293T transfected with plasmids expressing tagged VIM and ANXA2 proteins alternatively mixed with PRRSV proteins for 2 h at 4 °C and then transferred to a tube containing equilibrated anti-Flag affinity gel beads. The mixtures were then vortexed briefly and incubated with gentle mixing for 2 h at 4 °C. Finally, the immunoprecipitates were washed five times and then analyzed by immunoblotting.
Preparation of PRRSV proteins
Marc-145 cells were infected with PRRSV HuN4 (MOI = 0.01), and the cell cultures were harvested at 84 hpi. After freezing and thawing three times, the cellular debris was removed by centrifugation at 8000 × g for 10 min at 4 °C. The supernatant was centrifuged at 111 000 × g for 2 h at 4 °C, and the pellets were resuspended in PBS. Subsequently, the PRRSV proteins were extracted using mild extraction buffer 1% Triton X-100 containing protease inhibitor cocktail.
Real-time reverse transcriptase PCR
Total RNA from PRRSV-infected cells was extracted using a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) and then reverse transcribed with a random primer RT kit (TaKaRa, Kusatsu, Japan). The products were used as a template for quantifying PRRSV RNA copy number using a previously validated TaqMan fluorescent quantitative PCR method [26].
Virus titration
Marc-145 cells, grown in 96-well plates, were infected with tenfold serial dilutions of PRRSV HuN4. After 1 h incubation at 37 °C, the supernatants were replaced with fresh DMEM containing 2% FBS. Viral titers were determined using endpoint dilution analysis at 5 dpi. The Reed–Muench method was used to determine the 50% tissue culture infected dose (TCID50) [27].
Statistical analysis
Data are expressed as mean ± standard deviation (SD). Differences between groups were examined for statistical significance using Student’s t tests. An unadjusted P value of < 0.05 was considered statistically significant.
Results
ANXA2 expression is higher in PAMs infected with PRRSV HuN4
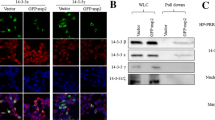
Proteins from infected and uninfected cells were separated using 2D-DIGE. Using an independent two-way ANOVA, 15 of the most significant spots were selected and sent for identification using MALDI-TOF/TOF mass spectrometry. Fourteen of the spots were successfully identified, with 11 of the 14 matching their homologous protein in databases (Table 2). Of these 11 differentially expressed proteins, ANXA2 was found to be upregulated when compared with uninfected PAM cells (Figures 1A–C). Subsequent bioinformatics analysis indicated that ANXA2 was potentially involved in a protein interaction network that included several other proteins, including vimentin (Figure 1D). The results indicated that ANXA2 is a differential protein and may be relevant to vimentin.
ANXA2 was identified as a differentially upregulated protein by 2D-DIGE/MS analysis. A PAMs were infected with PRRSV HuN4 at a MOI of 0.01. At 48 hpi, cells were harvested and analyzed by 2D-DIGE. The 2D-DIGE profiles of differentially expressed protein levels in either the uninfected control PAM or HuN4-infected PAM. The 3D spot intensity represents the differentially expressed ANXA2 protein. B ANXA2 was identified using Western blotting. PAMs were infected with PRRSV HuN4 at a MOI of 0.01. At 48 hpi, cells were harvested for analysis by Western blotting. C The relative intensity ratios of ANXA2. D Bioinformatics analysis about ANXA2 was conducted using STRING and the interaction network revealed that ANXA2 and other proteins (including vimentin) form a protein interaction network.
ANXA2 is critical for PRRSV replication
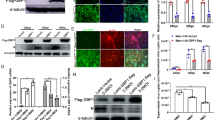
Specific siRNAs were used to down-regulate the expression of ANXA2 in Marc-145 cells. Western blotting confirmed that expression of ANXA2 and the PRRSV N protein were decreased relative to cells treated with a non-targeting siNC, transfection reagent alone (mock treated), and untreated (no treatment; NT) controls at 48 hpi (Figures 2A–C). In addition, viral genome copy numbers in the ANXA2 knockdown cells decreased at 24 and 48 hpi relative to the controls (Figure 2D). Similarly, PRRSV titers decreased after ANXA2 knockdown in the Marc-145 cells (Figure 2E). Together, these results show that knockdown of ANXA2 negatively modulated PRRSV reproduction. In comparison, Western blotting of overexpressing ANXA2 in Marc-145 cells demonstrated that the abundance of the protein had increased, although N protein expression levels and PRRSV genome copy number remained unchanged when compared to the controls (data not shown).
ANXA2 knockdown by siRNA and its effect on PRRSV multiplication. A Reduced ANXA2 expression by siRNA. Marc-145 cells transfected with 100 nM siANXA2 or a non-targeting control (siNC), transfection reagent (mock treatment), or left untreated (no treatment; NT) for 36 h were infected with PRRSV HuN4 at a MOI of 0.01 for 24 and 48 h, and then collected the samples for Western blotting using mouse anti-β actin monoclonal antibody (GenScript), rabbit anti-ANXA2 monoclonal antibody (Cell Signaling Technology) and mouse anti-PRRSV N protein monoclonal antibody. B The relative intensity ratios of N protein and C ANXA2 in a Western blot showing successful knockdown. D PRRSV replication in ANXA2 knockdown cells obtained from cell culture supernatants treated with PRRSV HuN4 was assessed by RNA copy number by quantitative real-time RT-PCR assay and E PRRSV protein titers. *P < 0.05; **P < 0.01.
Confocal analysis of ANXA2 and vimentin
Bioinformatics analysis suggested that ANXA2 may interact with vimentin in an interactomic network. To determine if ANXA2 interacts as a partner of vimentin, confocal analysis was performed using PAMs and Marc-145 cells to reveal that ANXA2 co-localized with vimentin (Figure 3), suggesting an interaction. In addition, in ANXA2-Flag and vimentin-HA co-transfected 293T cells, co-localization was also observed (data not shown).
Co-localization of ANXA2 and vimentin in PAMs, Marc-145 cells. PAMs and Marc-145 cells at 70% confluence were washed, fixed, blocked with 5% skimmed milk and then incubated with rabbit anti-ANXA2 mAb (Cell Signaling Technology) or mouse anti-vimentin mAb (Cell Signaling Technology) at 4 °C overnight and then incubated for 1 h with goat anti-rabbit IgG-FITC antibodies or goat anti-mouse IgG-TRITC antibodies. Finally, the cells were then counter-stained with DAPI and subjected to confocal assay. Fluorescent staining of ANXA2 (green) and vimentin (red) in PAMs and Marc-145 cells, counterstained with DAPI (blue). The scale bar indicates 10 µm.
Co-immunoprecipitation (Co-IP) of ANXA2 and vimentin
To further confirm that an interaction existed between ANXA2 and vimentin, endogenous Co-IP assays were performed using PAMs and Marc-145 cells. The results showed that intracellular ANXA2 precipitated with endogenous vimentin in both PAMs and Marc-145 cells (Figure 4). In a subsequent exogenous Co-IP assay, ANXA2-Flag and vimentin-HA proteins expressed in 293T cells were employed. And the results revealed that exogenous ANXA2-Flag and vimentin-HA also could specifically interact with either vimentin-HA or ANXA2-Flag, respectively (Figure 5). Together, these results confirmed that ANXA2 is a binding partner of vimentin.
Interaction between ANXA2-Flag and vimentin-HA in 293T cells. 293T cells co-transfected with pCMV-ANXA2-Flag and pCMV-vimentin-HA were harvested at 48 h and subjected to co-IP assay using mouse A anti-HA affinity gel or B anti-Flag affinity gel using. The precipitated proteins were analyzed by Western blotting using antibodies against Flag or HA.
The B domain of ANXA2 is required for the ANXA2-vimentin interaction
To map the region of ANXA2 required for vimentin binding, five plasmids expressing various Flag-tagged truncated ANXA2 mutants were constructed (Figure 6A). These plasmids were co-transfected with HA-tagged vimentin into HEK293T cells. Further Co-IP assays showed that vimentin interacted with ANXA2-Flag (1–174 aa), ANXA2-Flag (1–259 aa), and ANXA2-Flag (109–340 aa), but not with ANXA2-Flag (1–102 aa) nor ANXA2-Flag (192–340 aa) (Figure 6B). Finally, we attempted to directly establish whether the B domain alone could bind the ANXA2-vimentin complex. However, we were unable to detect the antigen–antibody complex on Western blots due to the small molecular weight of the fragment (7 kDa) and low expression levels (data not shown). Despite this, these findings indicate that the B domain (109–174 aa) of ANXA2 is required for the interaction between vimentin and ANXA2.
Interaction between vimentin and a series of ANXA2-Flag mutants. A Schematic representation of the predicted protein domains of ANXA2. The full-length ANXA2 protein and five truncated ANXA2 mutants were examined in this study. B 293T cells co-transfected with pVIM-HA and plasmids expressing the ANXA2 mutants were collected at 48 h and subjected to co-IP assay using mouse anti-HA affinity gel. The precipitated proteins were analyzed by Western blotting using antibodies against Flag or HA.
ANXA2 is required for vimentin binding to the PRRSV N protein
To investigate the role that ANXA2 plays in the binding of vimentin to PRRSV, 293T cells were transfected individually or simultaneously with pVIM-HA and pANXA2-Flag. Next, the expressed protein(s) were allowed to bind to PRRSV proteins and then probed with a series of mAbs specific to various viral proteins (anti-GP2, anti-GP3, anti-GP5, anti-M protein, and anti-N protein). This showed that, while of all the mAbs recognized their corresponding PRRSV proteins (Figure 7A). In addition, ANXA2 and vimentin expressed individually did not interact with GP2, GP3, GP5, nor the M or N proteins of PRRSV (Figures 7B, C). Only when ANXA2 and vimentin exist together could interact with the N protein. No interaction was found between vimentin and GP2, GP3, GP5, or the M protein of PRRSV, even when expressed in tandem with ANXA2 (Figures 7D, E). These findings suggest that ANXA2 is an important co-factor involved in the binding of vimentin to the PRRSV N protein.
ANXA2/vimentin binding to PRRSV structural proteins. A Validation of the reactivity of PRRSV structural protein mAbs and the PRRSV proteins by SDS-PAGE. B The mixture of PRRSV protein incubated with the extraction of HEK293T transfected with plasmids expressing tagged ANXA2 protein and subjected to Co-IP. The precipitated proteins were performed using anti-GP2, anti-GP3, anti-GP5, anti-M, and anti-N mAbs. C The binding of vimentin to PRRSV structural proteins. The mixture of PRRSV protein incubated with the extraction of HEK293T transfected with plasmids expressing tagged vimentin protein and then subjected to Co-IP. D The binding of the ANXA2-vimentin complex to PRRSV structural proteins. The mixture of PRRSV protein mixed with ANXA2-vimentin complex, and then performed the Co-IP assay. The triangle (black up-pointing triangle) highlights the position of the PRRSV N protein. E The mixture of PRRSV protein mixed with HEK293T transfected with plasmids expressing tagged ANXA2 and vimentin protein or control vector, and then performed Co-IP assay using anti-Flag affinity gel. The precipitated proteins were tested by Western blotting using the corresponding antibodies.
Discussion
Here, we have shown that ANXA2 can promote the replication of PRRSV. Moreover, ANXA2 was confirmed that could interact with vimentin for the first time. In addition, the ANXA2-vimentin complex has the ability to bind to the PRRSV N protein. Individually, neither ANXA2 nor vimentin can bind to PRRSV, suggesting that ANXA2 is a necessary accessory molecule for vimentin binding. Since the N protein isn’t exposed on the surface of PRRS virion, the ANXA2 may not be involved in the entry process, and ANXA2/vimentin interaction may be important cellular factors for PRRSV multiplication. The interaction between NSP2 and vimentin and N protein was well confirmed [19, 20]. Stable overexpression of ANXA2 in Marc-145 cells did not enhance the infectivity nor multiplication of PRRSV. Although this supports data from a previous study [25], the reasons for this are unclear and may be related to endogenous levels of vimentin.
Supporting our study, both ANXA2 and vimentin have been shown to play a role in viral disease, although ours is the first to demonstrate that the two proteins interact. Vimentin has previously been demonstrated to act as an attachment receptor for enterovirus 71 [28] and can also interact with dengue virus nonstructural protein 4A, a component of the viral replication complex [29]. ANXA2 is a member of the annexin superfamily of proteins that are calcium-dependent phospholipid binding proteins responsible for mediating essential several biological processes, including membrane trafficking, endosome formation, and vesicle aggregation [16, 30]. Consequently, ANXA2 itself is found in a wide variety of cells and tissues and is a key host factor involved in many diverse cellular processes, including vesicle trafficking [31], membrane fusion [32], and exocytosis [33]. ANXA2 has been found to be involved in various viral infections, including cytomegalovirus [34], hepatitis C virus [17], and infectious bursal disease virus [35]. ANXA2 also has an important role during human immunodeficiency virus (HIV) type 1 entry, assembly, and budding [36]. In classical swine fever virus infections, ANXA2 has been shown to be involved in several events, such as viral attachment and entry, replication, assembly, and the budding and release of viral particles from host cells [37,38,39]. ANXA2 is also a potential receptor for respiratory syncytial virus in human epithelial cells [40]. Here, we have shown that ANXA2 is an important cellular factor for vimentin and PRRSV replication.
Vimentin has also been identified as an important part of the PRRSV receptor complex [19, 41, 42]. Notably, it has been reported that vimentin indirectly interacts with nonstructural protein 2 (NSP2) of PRRSV in the presence of N protein [20] and experiments using vimentin bound to PRRSV N protein and anti-vimentin antibodies showed that PRRSV activity could be blocked [19]. Additionally, the interaction between the N protein, NSP9, and cellular DHX9 was found to regulate viral RNA synthesis [43] and NSP2 has been shown to form a complex with vimentin using the N protein as an intermediate [20]. A limitation of our study is that interactions between ANXA2-vimentin and GP4, and various NSP proteins, could not be examined due to the lack of available mAbs. Further investigation is therefore required to establish if there is binding between ANXA2, vimentin, and either GP4 or NSPs, in addition to binding to the N protein.
In conclusion, our data show for the first time that ANXA2 is an essential molecule for the binding of vimentin and that the ANXA2 B domain of contributes to this interaction. Furthermore, the ANXA2-vimentin complex has the ability to bind the PRRSV N protein. Our work provides important information concerning the regulation of PRRSV replication and may lead to better control efforts to manage this economically important disease.
Change history
05 October 2018
In the original publication of this article [1], the author found the brand of vimentin antibody was wrong in Fig. 3. The legend of Fig. 3, ‘mouse anti-vimentin mAb (Cell Signaling Technology) at 4 °C overnight’ should be ‘mouse anti-vimentin mAb (Sigma-Aldrich) at 4 °C overnight’.
Abbreviations
- PRRSV:
-
porcine reproductive and respiratory syndrome virus
- ANXA2:
-
annexin A2
- PAMs:
-
porcine alveolar macrophages
- ORFs:
-
open reading frames
- PBS:
-
phosphate-buffered saline
- DMEM:
-
Dulbecco’s modified Eagle’s medium
- FBS:
-
fetal bovine serum
- 2D-DIGE:
-
two dimension difference gel electrophoresis
- hpi:
-
hours post-infection
- IEF:
-
isoelectric focusing
- TCID50 :
-
50% tissue culture infected dose
References
Wensvoort G, Terpstra C, Pol JM, ter Laak EA, Bloemraad M, de Kluyver EP, Kragten C, van Buiten L, den Besten A, Wagenaar F (1991) Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet Quart 13:121–130
Collins JE, Benfield DA, Christianson WT, Harris L, Hennings JC, Shaw DP, Goyal SM, McCullough S, Morrison RB, Joo HS (1992) Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J Vet Diagn Invest 4:117–126
Pejsak Z, Stadejek T, Markowska-Daniel I (1997) Clinical signs and economic losses caused by porcine reproductive and respiratory syndrome virus in a large breeding farm. Vet Microbiol 55:317–322
Nieuwenhuis N, Duinhof TF, van Nes A (2012) Economic analysis of outbreaks of porcine reproductive and respiratory syndrome virus in nine sow herds. Vet Rec 170:225
Meulenberg JJ, Petersen den Besten A, de Kluyver E, van Nieuwstadt A, Wensvoort G, Moormann RJ (1997) Molecular characterization of Lelystad virus. Vet Microbiol 55:197–202
Wieringa R, de Vries AAF, van der Meulen J, Godeke GJ, Onderwater JJM, van Tol H, Koerten HK, Mommaas AM, Snijder EJ, Rottier PJM (2004) Structural protein requirements in equine arteritis virus assembly. J Virol 78:13019–13027
Wissink EH, Kroese MV, van Wijk HA, Rijsewijk FA, Meulenberg JJ, Rottier PJ (2005) Envelope protein requirements for the assembly of infectious virions of porcine reproductive and respiratory syndrome virus. J Virol 79:12495–12506
Music N, Gagnon CA (2010) The role of porcine reproductive and respiratory syndrome (PRRS) virus structural and non-structural proteins in virus pathogenesis. Anim Health Res Rev 11:135–163
Gerke V, Moss SE (2002) Annexins: from structure to function. Physiol Rev 82:331–371
Moss SE, Morgan RO (2004) The annexins. Genome Biol 5:219
Gerke V, Creutz CE, Moss SE (2005) Annexins: linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol 6:449–461
Hedhli N, Falcone DJ, Huang B, Cesarman-Maus G, Kraemer R, Zhai H, Tsirka SE, Santambrogio L, Hajjar KA (2012) The annexin A2/S100A10 system in health and disease: emerging paradigms. J Biomed Biotechnol 2012:406273
Vedeler A, Hollas H, Grindheim AK, Raddum AM (2012) Multiple roles of annexin A2 in post-transcriptional regulation of gene expression. Curr Protein Pept Sci 13:401–412
Hayes MJ, Moss SE (2004) Annexins and disease. Biochem Biophys Res Commun 322:1166–1170
Grindheim AK, Saraste J, Vedeler A (2017) Protein phosphorylation and its role in the regulation of Annexin A2 function. Biochim Biophys Acta 1861:2515–2529
Sheng C, Liu X, Jiang Q, Xu B, Zhou C, Wang Y, Chen J, Xiao M (2015) Annexin A2 is involved in the production of classical swine fever virus infectious particles. J Gen Virol 96:1027–1032
Backes P, Quinkert D, Reiss S, Binder M, Zayas M, Rescher U, Gerke V, Bartenschlager R, Lohmann V (2010) Role of annexin A2 in the production of infectious hepatitis C virus particles. J Virol 84:5775–5789
Ding Z, Li ZJ, Zhang XD, Li YG, Liu CJ, Zhang YP, Li Y (2012) Proteomic alteration of Marc-145 cells and PAMs after infection by porcine reproductive and respiratory syndrome virus. Vet Immunol Immunopathol 145:206–213
Kim JK, Fahad AM, Shanmukhappa K, Kapil S (2006) Defining the cellular target(s) of porcine reproductive and respiratory syndrome virus blocking monoclonal antibody 7G10. J Virol 80:689–696
Song T, Fang L, Wang D, Zhang R, Zeng S, An K, Chen H, Xiao S (2016) Quantitative interactome reveals that porcine reproductive and respiratory syndrome virus nonstructural protein 2 forms a complex with viral nucleocapsid protein and cellular vimentin. J Proteomics 142:70–81
Tong GZ, Zhou YJ, Hao XF, Tian ZJ, An TQ, Qiu HJ (2007) Highly pathogenic porcine reproductive and respiratory syndrome, China. Emerg Infect Dis 13:1434–1436
Yang Y, An T, Gong D, Li D, Peng J, Leng C, Yuan Z, Tong G, Tian Z, Zhang D (2012) Identification of porcine serum proteins modified in response to HP-PRRSV HuN4 infection by two-dimensional differential gel electrophoresis. Vet Microbiol 158:237–246
Zhou S, Yi T, Liu R, Bian C, Qi X, He X, Wang K, Li J, Zhao X, Huang C, Wei Y (2012) Proteomics identification of annexin A2 as a key mediator in the metastasis and proangiogenesis of endometrial cells in human adenomyosis. Mol Cell Proteomics 11(M112):017988
Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C (2009) STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37:D412–D416
Li J, Guo D, Huang L, Yin M, Liu Q, Wang Y, Yang C, Liu Y, Zhang L, Tian Z, Cai X, Yu L, Weng C (2014) The interaction between host Annexin A2 and viral Nsp9 is beneficial for replication of porcine reproductive and respiratory syndrome virus. Virus Res 189:106–113
Liu Y, Shi W, Zhou E, Wang S, Hu S, Cai X, Rong F, Wu J, Xu M, Xu M, Li L (2010) Dynamic changes in inflammatory cytokines in pigs infected with highly pathogenic porcine reproductive and respiratory syndrome virus. Clin Vaccine Immunol 17:1439–1445
Reed LJ, Muench H (1938) A simple method of estimating fifty percent endpoints. Am J Epidemiol 27:493–497
Du N, Cong H, Tian H, Zhang H, Zhang W, Song L, Tien P (2014) Cell surface vimentin is an attachment receptor for enterovirus 71. J Virol 88:5816–5833
Teo CS, Chu JJ (2014) Cellular vimentin regulates construction of dengue virus replication complexes through interaction with NS4A protein. J Virol 88:1897–1913
Drust DS, Creutz CE (1988) Aggregation of chromaffin granules by calpactin at micromolar levels of calcium. Nature 331:88–91
Jones PG, Moore GJ, Waisman DM (1992) A nonapeptide to the putative F-actin binding site of annexin-II tetramer inhibits its calcium-dependent activation of actin filament bundling. J Biol Chem 267:13993–13997
Lee DB, Jamgotchian N, Allen SG, Kan FW, Hale IL (2004) Annexin A2 heterotetramer: role in tight junction assembly. Am J Physiol Renal Physiol 287:F481–F491
Menke M, Ross M, Gerke V, Steinem C (2004) The molecular arrangement of membrane-bound annexin A2-S100A10 tetramer as revealed by scanning force microscopy. ChemBioChem 5:1003–1006
Wright JF, Kurosky A, Pryzdial EL, Wasi S (1995) Host cellular annexin II is associated with cytomegalovirus particles isolated from cultured human fibroblasts. J Virol 69:4784–4791
Ren X, Zhang L, Gao Y, Gao H, Wang Y, Liu C, Cui H, Zhang Y, Jiang L, Qi X, Wang X (2015) Binding chicken Anx2 is beneficial for infection with infectious bursal disease virus. Virus Res 210:232–240
Ma G, Greenwell-Wild T, Lei K, Jin W, Swisher J, Hardegen N, Wild CT, Wahl SM (2004) Secretory leukocyte protease inhibitor binds to annexin II, a cofactor for macrophage HIV-1 infection. J Exp Med 200:1337–1346
Yang Z, Shi Z, Guo H, Qu H, Zhang Y, Tu C (2015) Annexin 2 is a host protein binding to classical swine fever virus E2 glycoprotein and promoting viral growth in PK-15 cells. Virus Res 201:16–23
Sun JF, Jiang Y, Shi ZX, Yan YJ, Guo HC, He FC, Tu CC (2008) Proteomic alteration of PK-15 cells after infection by classical swine fever virus. J Proteome Res 7:5263–5269
Shi Z, Sun J, Guo H, Tu C (2009) Genomic expression profiling of peripheral blood leukocytes of pigs infected with highly virulent classical swine fever virus strain Shimen. J Gen Virol 90:1670–1680
Malhotra R, Ward M, Bright H, Priest R, Foster MR, Hurle M, Blair E, Bird M (2003) Isolation and characterisation of potential respiratory syncytial virus receptor(s) on epithelial cells. Microbes Infect 5:123–133
Shi C, Liu Y, Ding Y, Zhang Y, Zhang J (2015) PRRSV receptors and their roles in virus infection. Arch Microbiol 197:503–512
Zhang Q, Yoo D (2015) PRRS virus receptors and their role for pathogenesis. Vet Microbiol 177:229–241
Liu L, Tian J, Nan H, Tian M, Li Y, Xu X, Huang B, Zhou E, Hiscox JA, Chen H (2016) Porcine reproductive and respiratory syndrome virus nucleocapsid protein interacts with Nsp9 and cellular DHX9 to regulate viral RNA synthesis. J Virol 90:5384–5398
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
XBC, YQY, and TQA conceived and designed the experiments; XBC and YQY performed the experiments; XBC wrote the paper; XBC, YQY, JC-Gao, and KZ analyzed the data; JC-Guo, CY, CGJ, ZJT, XHC and GZT offered suggestions; TQA revised the manuscript and coordinated the research. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to thank Dr Yan-Jun Zhou for providing us with mouse anti-PRRSV mAbs.
Ethics approval and consent to participate
The animal experiments in this study were conducted with recommendations in the Chinese Regulations of Laboratory Animals—The Guidelines for the Care of Laboratory Animals (Ministry of Science and Technology of People’s Republic of China) and Laboratory Animal-Requirements of Environment and Housing Facilities (GB14925-2010, National Laboratory Animal Standardization Technical Committee).
Funding
The study was supported by grants from the National Natural Science Foundation of China (31270045), the Foundation for Science and Technology Innovative Talents of Harbin (2016RAQXJ142), the Heilongjiang Natural Science Fund for Distinguished Young Scholars (JC2017010), and the Foundation for Basic Scientific Research of Central Public Research Institutes (Y2017JC16). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Chang, XB., Yang, YQ., Gao, JC. et al. Annexin A2 binds to vimentin and contributes to porcine reproductive and respiratory syndrome virus multiplication. Vet Res 49, 75 (2018). https://doi.org/10.1186/s13567-018-0571-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13567-018-0571-5