Abstract
Background
The aims of this study were to determine the optimal tracer kinetic model for [11C]-meta- hydroxyephedrine ([11C]HED) and to evaluate the performance of several simplified methods.
Methods
Thirty patients underwent dynamic 60-min [11C]HED scans with online arterial blood sampling. Single-tissue and both reversible and irreversible two-tissue models were fitted to the data using the metabolite-corrected arterial input function. For each model, reliable fits were defined as those yielding outcome parameters with a coefficient of variation (CoV) <25%. The optimal model was determined using Akaike and Schwarz criteria and the F-test, together with the number of reliable fits. Simulations were performed to study accuracy and precision of each model. Finally, quantitative results obtained using a population-averaged metabolite correction were evaluated, and simplified retention index (RI) and standardized uptake value (SUV) results were compared with quantitative volume of distribution (VT) data.
Results
The reversible two-tissue model was preferred in 75.8% of all segments, based on the Akaike information criterion. However, VT derived using the single-tissue model correlated highly with that of the two-tissue model (r2 = 0.94, intraclass correlation coefficient (ICC) = 0.96) and showed higher precision (CoV of 24.6% and 89.2% for single- and two-tissue models, respectively, at 20% noise). In addition, the single-tissue model yielded reliable fits in 94.6% of all segments as compared with 77.1% for the reversible two-tissue model. A population-averaged metabolite correction could not be used in approximately 20% of the patients because of large biases in VT. RI and SUV can provide misleading results because of non-linear relationships with VT.
Conclusions
Although the reversible two-tissue model provided the best fits, the single-tissue model was more robust and results obtained were similar. Therefore, the single-tissue model was preferred. RI showed a non-linear correlation with VT, and therefore, care has to be taken when using RI as a quantitative measure.
Similar content being viewed by others
Background
Recently, non-invasive imaging of sympathetic innervation of the myocardium using PET [1]-[5] or SPECT [6]-[8] has gained interest based on its ability to predict life-threatening ventricular arrhythmias [9]-[11] and to assess whether implantable cardioverter defibrillator (ICD) therapy is appropriate [12],[13]. It has been shown that sympathetic nerve terminals in the myocardium are more sensitive to ischemic damage than cardiomyocytes [9],[14]-[21]. In addition, the area of innervation defects often exceeds the area of non-viable scar tissue [9],[11],[16],[17]. More importantly, it has been shown that areas of viable myocardium, but with impaired innervation, are related to inducible ventricular tachycardias originating from these areas [22]. Therefore, non-invasive imaging of sympathetic innervation may play a key role in risk stratification and treatment planning in ischemic cardiomyopathy.
Sympathetic innervation can be measured using the PET tracers [18 F]-6-fluorodopamine [2], [11C] epinephrine [1],[3] or [11C]-meta-hydroxyephedrine ([11C]HED) [4],[5], with [11C]HED being used most often. Analysis of [11C]HED and [11C] epinephrine data often has been performed using the retention index (RI) [5], a semi-quantitative parameter that can be derived relatively easy. In general, it is obtained by normalizing late activity concentrations to the integral of the blood time-activity curve. However, RI can be sensitive to cardiac motion, partial volume effects and contribution of intravascular activity and spill-over and therefore may yield inaccurate results. More quantitative results can be obtained by using compartment models, in which correction factors for blood volume and spill-over of activity from the blood can be applied. Interestingly, RI has never been validated using a direct comparison with quantitative results obtained from a full tracer kinetic analysis. Furthermore, blood time-activity curves both with [3],[22] and without [23]-[25] corrections for radioactive metabolites of [11C]HED have been used for RI, making it difficult to compare studies. On the other hand, another semi-quantitative measure, standardized uptake value (SUV), could provide a further simplification relative to RI, as it does not require measurement of blood time-activity curves, nor dynamic scanning.
Only few studies [26],[27] have performed quantitative analysis of [11C]HED data using a single-tissue compartment model, but no systematic assessment of the best tracer kinetic model was performed. Therefore, the aims of this study were to determine the optimal tracer kinetic model for analysing [11C]HED data and to assess the validity of several simplifications, such as RI, SUV and a population-averaged metabolite correction.
Methods
Patient population
Thirty patients (mean age 67 years, range 43 to 80; 26 males) with ischemic cardiomyopathy and a left ventricular ejection fraction below 35%, as determined by cardiac magnetic resonance imaging (MRI), were included. Ischemic cardiomyopathy was defined as having one or more stenoses >50% on coronary angiography and delayed contrast enhancement on cardiac MRI. The study was approved by the Medical Ethics Review Committee of the VU University Medical Center, and all participants gave written informed consent prior to inclusion.
Synthesis of [11C]HED
Radiolabelled [11C]HED was synthesized according to a GMP-compliant procedure (licence nr. 108879 F), using a modification of the method of Van Dort and Tluczek [28]. The starting material, metaraminol (3.0 μmol, 0.5 mg, purchased from ABX, Radeberg, Germany), was dissolved in 100 μL of acetonitrile. [11C] Methyl triflate was added to this solution at room temperature, and after reacting for 1 min, this mixture was diluted with 1 mL of 0.1 M ammonium formate in water. The resulting mixture was injected onto a Luna C18 5 μm 250*10 mm high-performance liquid chromatography (HPLC) column (Phenomenex, Torrance, CA, USA), which was eluted with a 95/5 0.1 M ammonium formate/acetonitrile mixture. The product, [11C]HED, was eluted at 10 min. This fraction was collected in 60 mL of water for injection. The total solution was passed over a preconditioned (using 5 mL of sterile ethanol (96%) and subsequently 10 mL of water for injection) Oasis WCX cartridge (Waters, Milford, MA, USA). The cartridge was washed with 20 mL of water for injection, and subsequently, the product was eluted from the cartridge with 1.0 mL of sterile ethanol (96%) and 10.0 mL of a sterile and pyrogen-free citrate buffer (3.8 mM citric acid, 5.4 mM sodium citrate, 5.1 mM sodium acetate; pH 5.2). This final mixture was passed over a Millex-GV 0.22 μm filter (Millipore, Billerica, MA, USA), yielding a sterile, isotonic and pyrogen-free solution of 1.6 to 5.1 GBq of [11C]HED. The product was analysed using a GraceSmart RP18 5 μm 250*4.6 mm HPLC column (Grace, Columbia, MD, USA), which was eluted with 95/5 0.1 M NaH2PO4 in a water/acetonitrile mixture. The radiochemical purity was >99% and the specific activity was 54 to 239 GBq • μmol-1 at the end of synthesis (N = 30).
Scanning protocol
Studies were performed on a GEMINI TF-64 PET/CT scanner (Philips Healthcare, Best, the Netherlands) [29]. A 60-min emission scan was started simultaneously with the injection of 370 MBq [11C]HED, administered as a 5-mL bolus (0.8 mL • s-1) followed by a 35-mL saline flush (2 mL • s-1). This emission scan was followed immediately by a respiration-averaged low-dose CT (LD-CT) scan (55 mAs, rotation time 1.5 s, pitch 0.825, collimation 16 × 0.625, acquiring 20 cm in 37 s compared to 5 s for a regular LD-CT) during normal breathing.
Images were reconstructed into 36 frames (1 × 10, 8 × 5, 4 × 10, 3 × 20, 5 × 30, 5 × 60, 4 × 150, 4 × 300, 2 × 600 s) using the 3D row action maximum likelihood algorithm and applying all appropriate corrections for scanner normalization, dead time, decay, scatter, randoms and attenuation based on the corresponding LD-CT scan.
Blood sampling
All patients received an indwelling radial artery catheter for arterial blood sampling during the dynamic emission scan. Using an online detection system [30], arterial blood was withdrawn continuously at a rate of 5 mL • min-1 during the first 5 min and 1.7 mL • min-1 thereafter. In addition, 7-mL arterial samples were collected manually at 2.5, 5, 10, 15, 20, 30, 40 and 60 min post injection and analysed for plasma and whole blood activity and the presence of radiolabelled metabolites, similarly as described in [31]. For each sample, activity concentrations in plasma and whole blood were determined. Furthermore, plasma was analysed for radiolabelled metabolites of [11C]HED by solid-phase extraction (SPE). In brief, 1 mL of plasma was diluted with 2 mL water and loaded onto an activated Oasis WCX cartridge (Waters, Milford, MA, USA, 3 mL, 60 mg). First, the cartridge was washed with 3 mL of 1% acetonitrile in water and subsequently eluted with 3 mL of 1 M HCl/ethanol (95/5). Radioactivity in all three fractions (plasma, 1% acetonitrile in water, 1 M HCl/ethanol) was measured. The first two fractions represent polar radiolabelled metabolites of [11C]HED and the third intact [11C]HED. No other metabolites were observed in the third fraction, and therefore, further analysis using HPLC was not required.
Input functions
Blood sampler data were corrected for delay and dispersion by fitting the early part of the sampler curve to the ascending aorta (AA) time-activity curve (TAC) using a single-tissue compartment model with additional parameters for delay and dispersion constants [32]. The AA was chosen as it was shown to provide more reproducible curves than either the left ventricle or the left atrium [33]. This region was obtained by drawing 1-cm-diameter circular regions of interest (ROIs) over the AA in at least five consecutive transaxial images of an early frame of the dynamic scan, i.e. the frame in which the first pass of the bolus through the AA was best visible [34]. To avoid partial volume effects, ROIs were placed in the centre of the AA, with ROI borders at least 5 mm from the edge of the AA. Next, the resulting delay- and dispersion-corrected sampler curve was calibrated using the manually drawn blood samples. Plasma/whole blood ratios derived from the manual blood samples were fitted to a sigmoid function, as described previously [35]. Similarly, parent fractions derived from the same samples were also fitted to a sigmoid function. Finally, the plasma blood sampler input function (BSIF) CP(t) was obtained by multiplying the delay- and dispersion-corrected and calibrated blood TAC with both these sigmoid functions.
In addition to these individual corrections, a population-averaged correction for both plasma/whole blood ratios and parent fractions was obtained using data from ten randomly selected patients, which was applied to all delay- and dispersion-corrected and calibrated blood TAC.
To correct for spill-over from the right ventricle (RV), a set of ROIs was placed over the RV cavity in five consecutive transaxial planes, with ROI boundaries at least 1 cm away from the RV wall to avoid spill-over of myocardial activity. These ROIs were combined in an RV VOI, which was then transferred to the full dynamic image sequence to obtain the right ventricular time-activity curve.
Data analysis
Using software developed in-house within MATLAB 7.0.4 (MathWorks, Natick, MA, USA), 16 myocardial segment VOIs were drawn on the final frame of the dynamic scan according to the 17-segment model of the American Heart Association, excluding the apex [36]. This VOI template was projected onto the entire dynamic emission scan to extract segment TACs. Segment TACs were fitted to a single-tissue compartment model (1T2k) and both an irreversible (2T3k) and reversible (2T4k) two-tissue compartment model using non-linear least squares regression. No additional compartments for metabolites within the heart were used, as presence of metabolites in the heart was found to be negligible in a previous preclinical study [4]. Corrections for spill-over from left and right ventricular cavities were included in all models as follows, based on the findings in the Appendix:
in which CPET(t) represents the measured concentration, CT(t) the myocardial tissue concentration, CA(t) the arterial whole blood concentration and CRV(t) the right ventricular cavity concentration. In Equation 1, RV activity is treated as spill-over and the VA contribution is assumed to be due to spill-over from the left ventricular cavity, rather than due to arterial activity within the myocardium. Clearly, it is not possible to distinguish between contributions from true myocardial blood volume and spill-over from the left ventricle, but previously, it has been shown that assuming VA to be due to spill-over rather than to myocardial arterial blood volume provided more stable results for [15O] water [37], which was confirmed by simulation studies for [11C]HED (Appendix).
Differences in variances of measured activity in each frame were accounted for by including the following frame weighing factors W during the fitting process:
in which L is the length of each frame, C the total number of counts of each frame and f the decay correction factor of each frame. Weights were normalized such that the average of all weighing factors was equal to unity.
Outcome parameters for each model are listed in Table 1 and were defined as in [38]. With the exception of VS, all parameters were used as independent variables during the fitting process.
To exclude outliers and unreliable fits, standard deviations of outcome parameters were estimated. Only outcome parameters with a coefficient of variation (CoV; standard deviation divided by the final outcome parameter) <25% were used for further analysis. For each model, the number of fits with stable parameters was used as an outcome measure.
For each segment, the best fit was determined using the Akaike information criterion (AIC [39]), the Schwarz criterion (SC [40]) and the F-test. AIC, SC and the F-statistic were defined according to Equations 3, 4 and 5, respectively:
in which N is the number of frames (36 in the present study), WSSE the weighted squared sum of errors, using the weights of Equation 2, and p the total number of parameters for each model, i.e. 4, 5 and 6 for 1T2k, 2T3k and 2T4k, respectively, including spill-over fractions. The subscripts 1 and 2 in Equation 5 represent the model with the highest (2) and the lowest (1) number of parameters. For the F-test, an F-statistic with a p value <0.05 was considered to correspond to a significant improvement in the goodness of fit.
Correlations between volumes of distribution (VT) of various models and between individual and population-averaged metabolite corrections were assessed using linear regressions. Agreement between different VT and metabolite corrections were assessed using intraclass correlation coefficients (ICC). For validation of the population-averaged metabolite correction, patients who were used to define this population average were not included in the comparison between individual and population corrections.
Finally, RI and SUV were calculated using the last 10 min of the tissue TAC normalized to the integral of the parent plasma TAC (RIP) or of the uncorrected whole blood TAC (RIWB) and to the injected dose divided by patient weight (SUV), respectively.
Simulations
Simulations were performed using input functions of a randomly selected patient. Tissue TACs (CPET(t)) were calculated using the reversible two-tissue model for healthy, scar and viable but denervated tissues. Based on clinical data obtained after a pilot analysis of five patients, values for K1, k2, k3 and k4 were set at 0.6 mL • cm-3 • min-1, 0.15 min-1, 0.3 min-1 and 0.02 min-1 for healthy tissue, at 0.2 mL • cm-3 • min-1, 0.1 min-1, 0.15 min-1 and 0.05 min-1 for scar tissue, and at 0.6 mL • cm-3 • min-1, 0.15 min-1, 0.15 min-1 and 0.05 min-1 for viable but denervated tissue, respectively. An arterial spill-over fraction of 0.3 and a right ventricular spill-over fraction of 0.1 (dimensionless) were assumed in all settings. Different levels of Gaussian noise (1%, 5%, 10% and 20%, representing noise on the whole LV level, the coronary level, the segmental level and the voxel level, respectively) were added to tissue TACs, after which they were fitted using non-linear least squares to each of the three compartment models. To correct for differences in variance of activity in each frame, noise was weighted according to Equation 2 and the average noise of the final six frames was set to the aforementioned values of noise levels. For each noise level, this process was repeated 1,000 times. Finally, average values of the various outcome parameters together with their CoV were obtained. Bias in derived parameters was obtained by comparing all obtained parameters with those estimated using the reversible two-tissue model.
Results
Compartment models
Clinical data
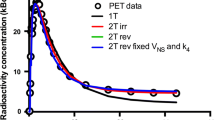
For one patient, online sampling could not be performed due to technical problems, and this patient was excluded from the study. Typical examples of myocardial TACs with high and low uptake, together with corresponding fits, are shown in Figure 1. Based on AIC, the 2T4k, 2T3k and 1T2k models were preferred in 364 (75.8%), 79 (16.4%) and 37 (7.7%) segments, respectively. Based on SC, this was 324 (67.5%), 99 (20.6%) and 57 (11.9%), respectively. Finally, the F-test showed a significantly improved fit when using the 2T4k model instead of both 2T3k and 1T2k models in 88.5% and 100% of all segments, respectively, indicating that the 2T4k model is the most appropriate model. Both 1T2k and 2T3k show systematic errors in the obtained fits (runs test p value <0.001 for both). No systematic error in 2T4k fits was observed (p = 0.06).
In 76.9%, 96.9% and 99.6% of all segments, K1 was fitted with a CoV <25% for 2T4k, 2T3k and 1T2k models, respectively. For 2T4k-derived VT (VT,2t), VS and BPND, 2T3k-derived K i and 1T2k-derived VT (VT,1t), a CoV <25% was obtained in 77.1%, 77.1%, 23.3%, 86.0% and 94.6% of all segments, respectively. Due to the very low number of BPND values with a sufficiently low CoV, BPND was excluded from further analysis.
Simulations
Simulations showed that, based on both AIC and SC, preference for the simpler single-tissue model increased with increasing noise levels (Table 2). Tables 3 and 4 provide accuracy and precision of VT,1t, K i , VT,2t and K1 estimates for increasing noise levels. In general, accuracy and precision of VT,2t and K1 derived using 2T4k decreased rapidly with increasing noise levels. At increasing noise levels, the best accuracy and precision were obtained for VT,1t followed by K i , especially for scar and viable but denervated tissue. The 1T2k model also provided the highest precision of K1, especially for tissues with low uptake, and the lowest noise-induced bias.
Single-tissue vs two-tissue compartment model
The correlation between VT,1t and VT,2t is shown in Figure 2 for clinical data. Both correlation (r2 = 0.94, slope = 0.84) and agreement (ICC = 0.96) between VT,1t and VT,2t were high. VT,1t was significantly lower than VT,2t, as indicated by the slope, which was significantly different from 1 (p < 0.001). These results indicate that VT obtained with the 1T2k model yields the same information as that obtained with the 2T4k model, albeit with a consistent underestimation. Consequently, based on its better precision, VT,1t was used to validate simplifications.
Simplifications
Figure 3 shows mean parent [11C]HED fractions and plasma/whole blood ratios as a function of time. The corresponding standard deviations indicate substantial variation between patients. Figure 4 shows a scatter plot of VT,1t calculated with individual and population-averaged metabolite corrections. Correlation (r2 = 0.89) and agreement (ICC = 0.94) were high and the slope of the linear fit was 0.98. However, there was significant bias in several patients when the population-averaged metabolite correction was used, ranging from -28% to 34% for individual patients.
Figure 5 shows relationships of RIP, RIWB and SUV with VT,1t. There was a clear and strong correlation of RIP and RIWB with VT,1t (r2 = 0.77 for RIP and r2 = 0.76 for RIwb). Correlation of SUV with VT,1t was significantly lower (r2 = 0.62). It is clear, however, that all three relationships were non-linear.
RI P (A), RI WB (B) and SUV (C) as a function of corresponding V T,1t . RIP, RIWB and SUV were obtained by normalizing radioactivity concentrations between 50 and 60 min post injection to the total amount of [11C]HED in plasma (RIP), total amount of radioactivity in whole blood (RIWB) and injected dose per patient weight (SUV), respectively.
Discussion
In the present study, the optimal tracer kinetic model for kinetic analysis of [11C]HED scans was identified and several commonly used simplifications were evaluated. First, Akaike and Schwarz criteria clearly indicated that, for both clinical and simulated (Table 2) data, the reversible two-tissue model was the preferred model for describing [11C]HED kinetics. Similar results were obtained when using the F-test, which is insensitive to any potential bias introduced in AIC and SC due to the scaling factors as presented in Equations 3 and 4. This confirms that, based on goodness of fit, the reversible two-tissue model was the optimal model. Simulations, on the other hand, also indicated that with increasing noise levels (starting at 5% noise, corresponding with noise at the level of the coronary territory), precision of all parameter estimates using this model rapidly decreased (Table 3) and that, for higher noise levels, model preference shifted towards models with fewer parameters (Table 2). In addition, for clinical data, the number of reliable estimates was lower for the 2T4k parameters VT,2t, VS and BPND (77.1, 77.1 and 23.3%, respectively) than for K i derived using the 2T3k model (86.0%) and VT,1t obtained using the 1T2k model (94.6%). The large number of estimates with high uncertainty indicates that the use of the reversible two-tissue model may not be feasible in routine clinical practice, as it would mean discarding over 20% of all data. In addition, most estimates with high uncertainty in 2T4k parameters were in regions with reduced [11C]HED uptake, indicating that the 2T4k model is especially vulnerable in diseased myocardium, which significantly limits the applicability of this model.
For the reversible two-tissue model, the total volume of distribution can be separated in specific and non-specific volumes of distribution. In this study, the total volume of distribution approximated the specific volume of distribution and enabled the use of the simpler single-tissue model, as the contribution of the non-specific compartment was very small, if not negligible. As shown in Figure 2, VT,1t yields similar results to VT,2t, indicating that the single-tissue model yields similar information to the reversible two-tissue model. In addition, precision was much higher for VT,1t than for VT,2t (Tables 3 and 4) and noise-induced biases were lower for VT,1t. Since the single-tissue model is simpler, more stable and less sensitive to noise compared with the reversible two-tissue model, it is preferred for further use, especially for smaller regions or regions with relatively low uptake.
In this study, arterial activity was assumed to be spill-over from the arterial blood pool, rather than activity originating from arterial blood within the tissue. Both assumptions are physiologically not fully correct and ideally, a combination should be used. However, differentiating between contributions from spill-over and myocardial blood volume is impossible, as kinetics of both contributions are identical. Previously, it has been shown that, in a comparison of MBF as measured with [15O] water against labelled microspheres, assuming arterial activity to be spill-over yields more accurate results [37], at least for [15O] water. A simulation study (see Figure 6 in Appendix) showed that the same is true for HED, although a bias was found in both implementations. Bias was smallest when arterial activity was assumed to be due to spill-over. In addition, arterial blood volume fraction within myocardial tissue is relatively constant in the myocardium [41], and therefore, it is expected that bias in the spill-over implementation, which is dependent on the magnitude of this blood volume fraction, is more consistent than bias in the tissue blood volume implementation, which depends on the recovery of the scanner and segment size.
The use of HED as an innervation tracer has recently been debated [42], where it was suggested that HED is a flow-limited rather than innervation-limited tracer. This was suggested to lead to the impossibility to fit HED data using standard kinetic modelling techniques and make HED uptake insensitive to early changes in innervation. However, when performing tracer kinetic analyses, parameters representing innervation (i.e. K i , VT, VS and BPND) can be obtained together with parameters representing flow and extraction (i.e. K1). This enables separating flow effects from innervation effects by studying both K1 and, when the 1T2k model is used, VT. Furthermore, in this study we show that, utilizing plasma input functions fully corrected for radiolabelled metabolites and plasma-to-blood concentration ratios, HED could reliably be fitted using a 2T4k model or, with slightly poorer fits, using a 1T2k model, which is in contrast to the suggestion in [42].
The non-linear relation between RI and VT, however, does indicate that for RI it might be the case that early changes are not readily detected. Indeed, when a sensitivity analysis is performed for changes in k3, which represents HED uptake through the norepinephrine transporter, using K1, k2 and k4 of 0.6 mL • cm-3 • min-1, 0.15 min-1 and 0.02 min-1, respectively, and gradually decreasing k3, RI showed a lower and non-linear sensitivity to changes in innervation (see Additional file 1). On the other hand, VT,1t showed good sensitivity to changes in innervation, where decreases in VT,1t were similar to decreases in k3, confirming that VT,1t is a more reliable measure of innervation than RI. Nevertheless, sensitivity of RI to changes in k3 may still be sufficient for routine clinical use as large reductions in k3 are readily detected by RI. It is also important to note that another recent study [43] showed that HED was able to detect early changes in innervation, confirming that HED is likely to represent innervation even when using RI.
Potentially, a population-averaged metabolite correction could, in combination with an image-derived input function, make arterial cannulation obsolete. There was, however, substantial variation in individual metabolism (Figure 3), resulting in large errors of up to 30% in fitted parameters when using a population-averaged metabolite correction (Figure 4) for approximately 20% of the patients. Clearly, for absolute quantification, a population-averaged metabolite correction is not feasible, at least not in the case of [11C]HED, as a method that produces inaccurate results in as much as 20% of the cases should be discarded. However, when relative differences within a single patient are of interest, e.g. when assessing defect areas (percentage of LV with VT below a reference value), population-averaged metabolite corrections can be considered. Using venous samples for metabolite correction might be an alternative for both relative analysis and absolute quantification, but this requires further validation.
In addition, the use of an image-derived input function would be preferred over the use of online blood sampling. However, image-derived input functions require validation for each individual radiotracer and preliminary results for [11C]HED [44] have shown that late activity concentrations were overestimated in the image-derived input functions, which is in line with observations in other studies that made use of a combination of image-derived input function and blood sampler data [26],[27]. Given the fact that an image-derived input function can be used for other tracers such as [15O] water [45] and [18 F] FDG [33], the particular biodistribution of HED with its high contrast between the myocardium and the blood at later time points, together with high liver uptake, will require improved reconstruction methods and/or scatter correction algorithms before an image-derived input function can be used for [11C]HED.
A reasonable correlation was found between VT,1t and either RIP, RIWB or SUV (r2 = 0.77, 0.76 and 0.62, respectively). These correlations suggest that RIP, RIWB and SUV may be used to distinguish innervated from denervated regions of the myocardium. However, it should be noted that relationships were non-linear (Figure 5). Consequently, these simplified measures will be less sensitive in detecting early changes in regions with high innervation, e.g. in early disease, or for monitoring response during treatment. In these cases, the use of quantitative measures is recommended. On the other hand, for routine clinical (diagnostic) use, RI might be sufficient, as large reductions in HED uptake are reflected in large reductions in RI. In addition, simplified methods may lead to biased estimates of defect sizes, as these usually are derived using cut-off values, which are based on a percentage of uptake in a reference region within the heart. Due to the non-linear relationships, cut-off values for RIP, RIWB and SUV will be different from those for VT,1t and, consequently, defect areas may differ significantly.
In the present study, RI was based on uptake between 50 and 60 min post injection, whilst 30 to 40 min is more commonly used. When RI was calculated using uptake from 30 to 40 min post injection, similar results were obtained (r2 = 0.72 and r2 = 0.73 for RIP and RIWB, respectively). For RIP, this correlation was slightly, but significantly, lower than that for the 50 to 60 min uptake period (p = 0.043). For RIWB, this difference was not statistically significant (p = 0.071). Although correlation with VT,1t was still reasonable, based on these findings, it is recommended to use later times for measuring RI due to the slow kinetics of HED.
Finally, it is important to note that the aforementioned results and interpretations are made from a modelling point of view, i.e. which parameter describes the measured HED activity best. This might be different from which parameter actually describes myocardial presynaptic activity best. Ideally, a blocking study or animal study should be performed to compare the obtained parameters with a gold standard measurement. In addition, the performance of the quantitative parameters used in the present study needs to be assessed in clinical studies, as the added clinical value of VT over simplified measures such as RIP still needs to be defined. Whilst simulations (see Additional file 1) show that quantitative measures are more sensitive to changes in k3, the rate constant reflecting norepinephrine transport, RI also showed significant correlation with k3, suggesting that RI may be sufficiently sensitive for measuring (changes in) innervation. Further clinical validation studies using RIP, RIWB, SUV and VT are warranted.
Conclusions
Although a reversible two-tissue compartment model best describes [11C]HED kinetics, a single-tissue compartment model is preferred for routine clinical studies, as it is more robust at clinically relevant noise levels and, at the same time, provides VT results that are highly correlated with those obtained with the two-tissue compartment model. Simplified measures, such as RI and SUV, showed good correlation with fully quantitative results and may be used to detect regions of denervation, although the non-linear relationship with VT might limit their application in, for instance, monitoring response to treatment.
Authors' contributions
HJH performed the analyses and drafted the manuscript. SdH and MTR included and scanned the patients. PK and CPA participated in the design of the study. RCS was responsible for blood sampling and metabolite analysis. ADW was responsible for tracer synthesis. AAL, MCH and ML helped with analyses, discussions and drafting of the manuscript. All authors read and approved the final manuscript.
Appendix
Contribution of arterial activity measured within a myocardial region is typically the combination of two different effects: a physical blood volume fraction within the myocardium and spill-over of activity from the left ventricular cavity into the myocardium. This can be represented as follows:
in which CT(t) represents the HED concentrations in the myocardial tissue itself whilst CPET(t), CA(t) and CRV(t) represent the measured regional, arterial and right ventricular blood activity concentrations, respectively. VA represents the actual arterial blood volume fraction in the tissue, and VLV and VRV represent the spill-over fractions from the left and the right ventricular cavity, respectively. During compartment modelling, it is not possible to distinguish VA from VLV, and therefore, the estimated arterial activity measured within a myocardial region can be interpreted in two different ways:
Interpretation SO (spill-over, Equation A2) assumes all measured arterial activity in the tissue regions to be due to spill-over from the left ventricular cavity, and a correction factor VA • CA(t) is used to correct for this spill-over fraction. In contrast, interpretation BV (blood volume, Equation A3) assumes all arterial activity in the tissue regions to be due to intravascular activity originating from the arterial blood volume within the myocardium itself. Since measured K1 represents K1 in the myocardial tissue only, a correction factor for tissue fraction has to be applied in model BV, in addition to the correction factor VA • CA(t). This tissue fraction is estimated as 1 - VA. In reality, measured activity originates from both spill-over from the left ventricular cavity and myocardial arterial blood, but it is impossible to distinguish between both contributions and, in practice, only a single correction can be applied. It is important to note that the actual fits obtained using Equations A2 and A3 are identical and only the interpretation of K1 and consequently VT is different.
To define the optimal model implementation, a simulation study was performed for both model interpretations. Three tissue curves (i.e. healthy tissue, scar tissue and viable but denervated tissue) were defined, using the same parameters as for the noise simulations. VRV was 0.1 for all simulated tissue curves. For each of the three tissue curves, arterial activity was simulated to be due to both spill-over from the left ventricular cavity (VLV) and arterial blood volume in the myocardium (VA) using Equation A1.
The total contribution of arterial blood activity was set to 0.3, based on clinical data (average VA for all segments 0.30 ± 0.10, 0.28 ± 0.10 and 0.25 ± 0.11 for 1T2k, 2T3k and 2T4k models, respectively), and VRV was set to 0.1, again based on clinical data (average VRV for all segments 0.07 ± 0.08, 0.06 ± 0.07 and 0.06 ± 0.07 for 1T2k, 2T3k and 2T4k models, respectively, and for septal segments alone 0.14 ± 0.09, 0.13 ± 0.09 and 0.13 ± 0.09 for 1T2k, 2T3k and 2T4k models, respectively). For each tissue curve, a simulation was performed with VA set to 0.1, 0.15 and 0.2 and VLV to 0.2, 0.15 and 0.1, respectively. In addition, for each tissue curve, VA, VLV and VRV were set to 0 and a fit was performed to obtain estimates without any confounding effects of VA, VLV and VRV, and these values were used as reference values. Then, the obtained time-activity curves were fitted using both Equations A2 and A3 and the reversible one- and two-tissue compartment models, and the obtained K1 and VT were compared with the reference values. For healthy tissue, bias in the obtained results is shown in Figure 6. Similar results were obtained for scar and viable but denervated tissue. As expected, bias was observed for both BV and SO. In general, magnitude and variation in bias were larger for BV than for SO, indicating that estimates obtained with SO are more stable and closer to the true simulated tissue values. For these reasons, SO was considered to be the most robust model interpretation. In addition, VA is relatively constant in the myocardium (around 10% [41]), and therefore, it is likely that spill-over has a larger and more variable contribution to VA. Therefore, it is expected that bias in SO, which is dependent on the magnitude of VA, is also relatively constant. In contrast, bias in BV is dependent on the magnitude of VLV, which depends on scanner resolution, segment size and thickness of the myocardial wall, and therefore, it is expected to vary between patients and scans. For these reasons, SO was used in the present study.
Bias due to interpretation of blood activity. Bias in % for estimates of K1(A) and VT(B) derived using 1T2k (red) and 2T4k (blue) models using interpretations BV (continuous lines) and SO (dashed lines) for healthy tissue. Please note that the sum of VA and VSO was set to 0.3 for all simulated tissue curves.
Additional file
References
Chakraborty PK, Gildersleeve DL, Jewett DM, Toorongian SA, Kilbourn MR, Schwaiger M, Wieland DM: High yield synthesis of high specific activity R-(-)-[11C] epinephrine for routine PET studies in humans. Nucl Med Biol 1993, 20: 939–944. 10.1016/0969-8051(93)90094-B
Goldstein DS, Chang PC, Eisenhofer G, Miletich R, Finn R, Bacher J, Kirk KL, Bacharach S, Kopin IJ: Positron emission tomographic imaging of cardiac sympathetic innervation and function. Circulation 1990, 81: 1606–1621. 10.1161/01.CIR.81.5.1606
Munch G, Nguyen NT, Nekolla S, Ziegler S, Muzik O, Chakraborty P, Wieland DM, Schwaiger M: Evaluation of sympathetic nerve terminals with [(11) C] epinephrine and [(11) C] hydroxyephedrine and positron emission tomography. Circulation 2000, 101: 516–523. 10.1161/01.CIR.101.5.516
Rosenspire KC, Haka MS, Van Dort ME, Jewett DM, Gildersleeve DL, Schwaiger M, Wieland DM: Synthesis and preliminary evaluation of carbon-11-meta-hydroxyephedrine: a false transmitter agent for heart neuronal imaging. J Nucl Med 1990, 31: 1328–1334.
Schwaiger M, Kalff V, Rosenspire K, Haka MS, Molina E, Hutchins GD, Deeb M, Wolfe E Jr, Wieland DM: Noninvasive evaluation of sympathetic nervous system in human heart by positron emission tomography. Circulation 1990, 82: 457–464. 10.1161/01.CIR.82.2.457
Dae MW, O'Connell JW, Botvinick EH, Ahearn T, Yee E, Huberty JP, Mori H, Chin MC, Hattner RS, Herre JM: Scintigraphic assessment of regional cardiac adrenergic innervation. Circulation 1989, 79: 634–644. 10.1161/01.CIR.79.3.634
Kline RC, Swanson DP, Wieland DM, Thrall JH, Gross MD, Pitt B, Beierwaltes WH: Myocardial imaging in man with I-123 meta-iodobenzylguanidine. J Nucl Med 1981, 22: 129–132.
Wieland DM, Wu J, Brown LE, Mangner TJ, Swanson DP, Beierwaltes WH: Radiolabeled adrenergic neuron-blocking agents: adrenomedullary imaging with [131I] iodobenzylguanidine. J Nucl Med 1980, 21: 349–353.
Jacobson AF, Senior R, Cerqueira MD, Wong ND, Thomas GS, Lopez VA, Agostini D, Weiland F, Chandna H, Narula J: Myocardial iodine-123 meta-iodobenzylguanidine imaging and cardiac events in heart failure. Results of the prospective ADMIRE-HF (AdreView Myocardial Imaging for Risk Evaluation in Heart Failure) study. J Am Coll Cardiol 2010, 55: 2212–2221. 10.1016/j.jacc.2010.01.014
Paul M, Schafers M, Kies P, Acil T, Schafers K, Breithardt G, Schober O, Wichter T: Impact of sympathetic innervation on recurrent life-threatening arrhythmias in the follow-up of patients with idiopathic ventricular fibrillation. Eur J Nucl Med Mol Imaging 2006, 33: 866–870. 10.1007/s00259-005-0061-7
Yukinaka M, Nomura M, Ito S, Nakaya Y: Mismatch between myocardial accumulation of 123I-MIBG and 99mTc-MIBI and late ventricular potentials in patients after myocardial infarction: association with the development of ventricular arrhythmias. Am Heart J 1998, 136: 859–867. 10.1016/S0002-8703(98)70132-2
Boogers MJ, Borleffs CJ, Henneman MM, van Bommel RJ, van Ramshorst J, Boersma E, Dibbets-Schneider P, Stokkel MP, van der Wall EE, Schalij MJ, Bax JJ: Cardiac sympathetic denervation assessed with 123-iodine metaiodobenzylguanidine imaging predicts ventricular arrhythmias in implantable cardioverter-defibrillator patients. J Am Coll Cardiol 2010, 55: 2769–2777. 10.1016/j.jacc.2009.12.066
Nishisato K, Hashimoto A, Nakata T, Doi T, Yamamoto H, Nagahara D, Shimoshige S, Yuda S, Tsuchihashi K, Shimamoto K: Impaired cardiac sympathetic innervation and myocardial perfusion are related to lethal arrhythmia: quantification of cardiac tracers in patients with ICDs. J Nucl Med 2010, 51: 1241–1249. 10.2967/jnumed.110.074971
Allman KC, Stevens MJ, Wieland DM, Hutchins GD, Wolfe ER Jr, Greene DA, Schwaiger M: Noninvasive assessment of cardiac diabetic neuropathy by carbon-11 hydroxyephedrine and positron emission tomography. J Am Coll Cardiol 1993, 22: 1425–1432. 10.1016/0735-1097(93)90553-D
Bengel FM, Barthel P, Matsunari I, Schmidt G, Schwaiger M: Kinetics of 123I-MIBG after acute myocardial infarction and reperfusion therapy. J Nucl Med 1999, 40: 904–910.
Inoue H, Zipes DP: Results of sympathetic denervation in the canine heart: supersensitivity that may be arrhythmogenic. Circulation 1987, 75: 877–887. 10.1161/01.CIR.75.4.877
Kammerling JJ, Green FJ, Watanabe AM, Inoue H, Barber MJ, Henry DP, Zipes DP: Denervation supersensitivity of refractoriness in noninfarcted areas apical to transmural myocardial infarction. Circulation 1987, 76: 383–393. 10.1161/01.CIR.76.2.383
Luisi AJ Jr, Suzuki G, Dekemp R, Haka MS, Toorongian SA, Canty JM Jr, Fallavollita JA: Regional 11C-hydroxyephedrine retention in hibernating myocardium: chronic inhomogeneity of sympathetic innervation in the absence of infarction. J Nucl Med 2005, 46: 1368–1374.
Matsunari I, Schricke U, Bengel FM, Haase HU, Barthel P, Schmidt G, Nekolla SG, Schoemig A, Schwaiger M: Extent of cardiac sympathetic neuronal damage is determined by the area of ischemia in patients with acute coronary syndromes. Circulation 2000, 101: 2579–2585. 10.1161/01.CIR.101.22.2579
Schwaiger M, Guibourg H, Rosenspire K, McClanahan T, Gallagher K, Hutchins G, Wieland DM: Effect of regional myocardial ischemia on sympathetic nervous system as assessed by fluorine-18-metaraminol. J Nucl Med 1990, 31: 1352–1357.
Simoes MV, Barthel P, Matsunari I, Nekolla SG, Schomig A, Schwaiger M, Schmidt G, Bengel FM: Presence of sympathetically denervated but viable myocardium and its electrophysiologic correlates after early revascularised, acute myocardial infarction. Eur Heart J 2004, 25: 551–557. 10.1016/j.ehj.2004.02.016
Sasano T, Abraham MR, Chang KC, Ashikaga H, Mills KJ, Holt DP, Hilton J, Nekolla SG, Dong J, Lardo AC, Halperin H, Dannals RF, Marbán E, Bengel FM: Abnormal sympathetic innervation of viable myocardium and the substrate of ventricular tachycardia after myocardial infarction. J Am Coll Cardiol 2008, 51: 2266–2275. 10.1016/j.jacc.2008.02.062
Fallavollita JA, Banas MD, Suzuki G, de Kemp RA, Sajjad M, Canty JM Jr: 11C-meta-hydroxyephedrine defects persist despite functional improvement in hibernating myocardium. J Nucl Cardiol 2010, 17: 85–96. 10.1007/s12350-009-9164-z
Hartmann F, Ziegler S, Nekolla S, Hadamitzky M, Seyfarth M, Richardt G, Schwaiger M: Regional patterns of myocardial sympathetic denervation in dilated cardiomyopathy: an analysis using carbon-11 hydroxyephedrine and positron emission tomography. Heart 1999, 81: 262–270.
Stevens MJ, Raffel DM, Allman KC, Dayanikli F, Ficaro E, Sandford T, Wieland DM, Pfeifer MA, Schwaiger M: Cardiac sympathetic dysinnervation in diabetes: implications for enhanced cardiovascular risk. Circulation 1998, 98: 961–968. 10.1161/01.CIR.98.10.961
Rimoldi OE, Drake-Holland AJ, Noble MI, Camici PG: Basal and hyperaemic myocardial blood flow in regionally denervated canine hearts: an in vivo study with positron emission tomography. Eur J Nucl Med Mol Imaging 2007, 34: 197–205. 10.1007/s00259-006-0233-0
Schafers M, Dutka D, Rhodes CG, Lammertsma AA, Hermansen F, Schober O, Camici PG: Myocardial presynaptic and postsynaptic autonomic dysfunction in hypertrophic cardiomyopathy. Circ Res 1998, 82: 57–62. 10.1161/01.RES.82.1.57
Van Dort ME, Tluczek L: Synthesis and carbon-11 labeling of the stereoisomers of meta -hydroxyephedrine (HED) and meta -hydroxypseudoephedrine (HPED). J Label Compd Radiopharm 2000, 43: 603–612. 10.1002/(SICI)1099-1344(200005)43:6<603::AID-JLCR345>3.0.CO;2-#
Surti S, Kuhn A, Werner ME, Perkins AE, Kolthammer J, Karp JS: Performance of Philips Gemini TF PET/CT scanner with special consideration for its time-of-flight imaging capabilities. J Nucl Med 2007, 48: 471–480.
Boellaard R, van Lingen A, van Balen SC, Hoving BG, Lammertsma AA: Characteristics of a new fully programmable blood sampling device for monitoring blood radioactivity during PET. Eur J Nucl Med 2001, 28: 81–89. 10.1007/s002590000405
Thackeray JT, Beanlands RS, Dasilva JN: Presence of specific 11C-meta-hydroxyephedrine retention in heart, lung, pancreas, and brown adipose tissue. J Nucl Med 2007, 48: 1733–1740. 10.2967/jnumed.107.043570
van den Hoff J, Burchert W, Muller-Schauenburg W, Meyer GJ, Hundeshagen H: Accurate local blood flow measurements with dynamic PET: fast determination of input function delay and dispersion by multilinear minimization. J Nucl Med 1993, 34: 1770–1777.
van der Weerdt AP, Klein LJ, Boellaard R, Visser CA, Visser FC, Lammertsma AA: Image-derived input functions for determination of MRGlu in cardiac (18) F-FDG PET scans. J Nucl Med 2001, 42: 1622–1629.
Lubberink M, Harms HJ, Halbmeijer R, de Haan S, Knaapen P, Lammertsma AA: Low-dose quantitative myocardial blood flow imaging using 15O-water and PET without attenuation correction. J Nucl Med 2010, 51: 575–580. 10.2967/jnumed.109.070748
Gunn RN, Sargent PA, Bench CJ, Rabiner EA, Osman S, Pike VW, Hume SP, Grasby PM, Lammertsma AA: Tracer kinetic modeling of the 5-HT1A receptor ligand [carbonyl-11C] WAY-100635 for PET. Neuroimage 1998, 8: 426–440. 10.1006/nimg.1998.0379
Cerqueira MD, Weissman NJ, Dilsizian V, Jacobs AK, Kaul S, Laskey WK, Pennell DJ, Rumberger JA, Ryan T, Verani MS: Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart. A statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Int J Cardiovasc Imaging 2002, 18: 539–542.
Lammertsma AA, De SR, Araujo LI, Jones T: Measurement of regional myocardial blood flow using C15O2 and positron emission tomography: comparison of tracer models. Clin Phys Physiol Meas 1992, 13: 1–20. 10.1088/0143-0815/13/1/001
Innis RB, Cunningham VJ, Delforge J, Fujita M, Gjedde A, Gunn RN, Holden J, Houle S, Huang SC, Ichise M, Iida H, Ito H, Kimura Y, Koeppe RA, Knudsen GM, Knuuti J, Lammertsma AA, Laruelle M, Logan J, Maguire RP, Mintun MA, Morris ED, Parsey R, Price JC, Slifstein M, Sossi V, Suhara T, Votaw JR, Wong DF, Carson RE: Consensus nomenclature for in vivo imaging of reversibly binding radioligands. J Cereb Blood Flow Metab 2007, 27: 1533–1539. 10.1038/sj.jcbfm.9600493
Akaike H: A new look at the statistical model identification. IEEE T Automat Contr 1974, 19: 716–723. 10.1109/TAC.1974.1100705
Schwarz G: Estimating the dimension of a model. Ann Statist 1978, 6: 461–464. 10.1214/aos/1176344136
Crystal GJ, Downey HF, Bashour FA: Small vessel and total coronary blood volume during intracoronary adenosine. Am J Physiol 1981, 241: H194-H201.
Raffel DM, Koeppe RA, Jung YW, Gu G, Jang KS, Sherman PS, Quesada CA: Quantification of cardiac sympathetic nerve density with N-11C-guanyl-meta-octopamine and tracer kinetic analysis. J Nucl Med 2013, 54: 1645–1652. 10.2967/jnumed.113.120659
Thackeray JT, de Kemp RA, Beanlands RS, Dasilva JN: Insulin restores myocardial presynaptic sympathetic neuronal integrity in insulin-resistant diabetic rats. J Nucl Cardiol 2013, 20: 845–856. 10.1007/s12350-013-9759-2
Harms HJ, de Haan S, Knaapen P, Huisman MC, Schuit RC, Windhorst AD, Allaart CP, Lammertsma AA, Lubberink M: Tracer kinetic analysis of myocardial [11C] hydroxyephedrine studies [abstract]. J Nucl Med 2011, 52: 254. 10.2967/jnumed.110.085431
Van der Veldt AAM, Hendrikse NH, Harms HJ, Comans EF, Postmos PE, Smit EF, Lammertsma AA, Lubberink M: Quantitative parametric perfusion images using 15O-labeled water and a clinical PET/CT scanner: test-retest variability in lung cancer. J Nucl Med 2010, 51: 1684–1690. 10.2967/jnumed.110.079137
Acknowledgements
The authors would like to thank Amina Elouahmani, Femke Jongsma, Judith van Es, Nazerah Sais, Nghi Pham, Robin Hemminga and Suzette van Balen for scanning the patients; Henri Greuter, Kevin Takkenkamp and Marissa Rongen for the blood analysis and Martien Mooijer, Jonas Eriksson, Anneloes Rijnders, Rolf van Kooij and Johan van den Hoek for the synthesis of [11C]HED.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
This work was supported financially by Philips Healthcare.
Electronic supplementary material
13550_2014_52_MOESM1_ESM.docx
Additional file 1: Figure S1.: Results of sensitivity analysis, showing relative change in k3 (x-axis, representing transport through the norepinephrine transporter) and resulting relative changes in fitted outcome measures. Dashed line represents line of identity. (DOCX 84 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits use, duplication, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Harms, H.J., de Haan, S., Knaapen, P. et al. Quantification of [11C]-meta-hydroxyephedrine uptake in human myocardium. EJNMMI Res 4, 52 (2014). https://doi.org/10.1186/s13550-014-0052-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13550-014-0052-4