Abstract
Background
Hereditary spinocerebellar ataxias are a group of genetic neurological disorders that result in degeneration of the cerebellum and brainstem, leading to difficulty in controlling balance and muscle coordination.
Case presentation
A family affected by spinocerebellar ataxia was identified in Argentina and investigated using whole exome sequencing to determine the genetic etiology. The proband, a female white Hispanic aged 48, was noted to have slowly progressive gait ataxia, dysarthria, nystagmus, and moderate cerebellar atrophy. Whole exome sequencing was performed on three affected and two unaffected family members and revealed a dominant pathogenic variant, p.Gln127Arg (19:54392986 A>G), in the protein kinase C gamma gene, and the family was diagnosed with spinocerebellar ataxia type 14.
Conclusions
To our knowledge, no previous cases of spinocerebellar ataxia type 14 have been reported in Argentina, expanding the global presence of this neurological disorder. This diagnosis supports whole exome sequencing as a high-yield method for identifying coding variants causing cerebellar ataxias and emphasizes the importance of broadening the clinical availability of whole exome sequencing for undiagnosed patients and families.
Similar content being viewed by others
Background
Cerebellar ataxia is a degenerative neurological disease primarily involving the cerebellum, which coordinates movement, leading to an inability to control one’s balance, eye movements, and voluntary muscle coordination [1, 2]. Age of ataxia onset varies significantly, from childhood to adulthood. Hereditary primary ataxias include nearly 50 forms of autosomal dominant spinocerebellar ataxias and over 30 autosomal recessive cerebellar ataxias [3,4,5]. Degenerative hereditary ataxias cause progressive degeneration of fine motor skills, including speech and balance [2, 6]. Genetic ataxias are characterized by many different mutation types including nucleotide repeat expansions, point mutations, and copy number variations. Over 500 genes associated with cerebellar ataxia have already been identified [7, 8]. Spinocerebellar ataxia 14 (SCA14) is an inherited autosomal dominant cerebellar ataxia, characterized by cerebellar atrophy and occasionally dystonia [9, 10].
In this study, we identified a multigenerational family from Argentina with a dominant from of cerebellar ataxia that remained undiagnosed after an initial round of genetic testing for the most common ataxic disorders. Next-generation clinical exome sequencing allows for a broader genome-wide evaluation of ataxia genes that can improve diagnosis rate following initial testing [7, 8, 11]. We describe the genomic analysis of this family of Argentinian descent in which three affected siblings presenting with spinocerebellar ataxia were found to have spinocerebellar ataxia type 14 (SCA14) due to mutation of the protein kinase C gamma (PRKCG) gene. To date, cases of SCA1, SCA2, SCA3, SCA6, and SCA7 have been reported in Argentina [12, 13] but, to the best of our knowledge, no prior cases of SCA14 have been reported in this region, expanding the global prevalence of this disorder.
Case presentation
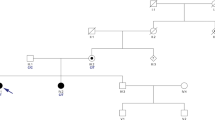
A multigenerational family (Fig. 1) affected with cerebellar ataxia was evaluated. The proband, individual II-2, a white Hispanic female, was seen initially at age 48 and showed slowly progressive gait ataxia, dysarthria, nystagmus, and moderate cerebellar atrophy on brain magnetic resonance imaging (MRI) examination. Disease onset was at age 19 years, with all symptoms present by the fourth decade, and her condition remained slowly progressive until her death at age 62 years. Initial genetic testing for common dominant genetic ataxias was performed on individual II-2, and although this individual was negative for SCA1, SCA3, SCA6, SCA7, SCA17, and dentatorubral-pallidoluysian atrophy (DRPLA), an inconclusive result was obtained for SCA2.
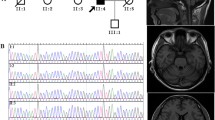
Pedigree. Index patient is individual II-2. Affected individuals are shown by shaded symbols. Deceased individuals are indicated by a line. Genotypes of the PRKCG gene are shown. G represents the reference while A is the p.Gln127Arg mutation. Affected family members and their corresponding phenotypes are indicated. *Gait ataxia, limb ataxia, horizontal nystagmus, ophthalmoplegia. ^Hyper-reflexia
Written informed consent was obtained from the family for participation in this study and publication of this case report. Both affected and unaffected family members consented for genetic analysis and this study was approved by the University of California, Los Angeles (UCLA) Institutional Review Board. Whole exome sequencing was performed and bioinformatically evaluated to assess for sequence and copy number variants as well as repeat expansions. Burrows-Wheeler Aligner was used to map the computed sequencing reads to the human genome [14]. Corresponding sequence alignment measures were quantified using Picard Tools [15], and repeated reads were marked. Variants were evaluated using VarSeq v2.2.1 (Golden Helix, Inc; http://www.goldenhelix.com). Variants were further filtered to exclude those with a minor allele frequency greater than 2% using the Genome Aggregation Database (gnomAD) public database [16]. This was due to the low likelihood of variants with a minor allele frequency greater than 2% causing Mendelian disease. Variants that had a disproportionately high allele frequency or allele count were eliminated from further annotation and classification. A list of keywords describing the patients and phenotypes that they presented with was then identified and used to produce a prioritized list of genes. This gene list was based on the clinical information found in the Online Mendelian Inheritance in Man database [17] and the Human Gene Mutation Database (HGMD) Professional Version [18]. Genetic variants detected within the genes in this list were evaluated initially prior to reviewing the remainder of the genomic variants. The HGMD and ClinVar [19] databases were used to assist annotation of these variants as pathogenic or likely pathogenic. Classification of variants was based on the guidelines provided by the American College of Medical Genetics and Genomics [20]. Detection of short tandem repeat (STR) expansions was performed using ExpansionHunter [21]. A Z score for each locus was calculated and used to determine how significant the number of repeats was by comparing it with the mean number of repeats at that locus. Loci of interest were filtered to only include those with a Z-score greater than two and were assessed on the basis of their segregation with disease status. Copy number variation (CNV) analysis was performed using WES data with the copy number inference from exome reads (CoNIFER) [22] and exome hidden Markov Model (XHMM) [23] tools. CNV calls were evaluated using the Database of Genomic Variants [24] and the DECIPHER [25] database based on reported pathogenicity and phenotypes.
Exome sequencing was initially performed on five family members of this multigenerational family, consisting of an unaffected parent, three affected siblings, and one unaffected sibling. As the family had previous inconclusive results when tested for SCA2, their exome data was further analyzed for repeat expansions using the ExpansionHunter tool, and no ATXN2 repeat expansions were detected. No pathogenic copy number variants were detected following copy number variant analysis using the CoNIFER and XHMM tools. Sequencing analysis revealed a pathogenic variant in the PRKCG gene that was identified in all three affected siblings (Fig. 1). This PRKCG variant (19:54,392,986 A>G, p.Gln127Arg) is a single nucleotide missense variant not found in the gnomAD public database of human variation. This specific PRKCG variant has been identified as causing ataxia in multiple families, all with phenotypes consistent with spinocerebellar ataxia 14 [26]. Pathogenicity was determined using standard ACMG criteria (PS1, PS3, PS4, PM1, PM2, PP1, PP4) [20]. Clinical features of the examined family members (Fig. 1) are presented in Table 1. The general ataxia clinical phenotype in this family is consistent with the SCA14 phenotype [9, 10, 26].
To our knowledge, this is the first identified case of SCA14 in Argentina. However, it remained possible that ancestors of this family may have migrated to Argentina from other locations, such as Europe or Japan, where SCA14 is more common [27,28,29,30,31,32]. To assess the origins of this family, a genetic ancestry analysis was performed using principal component analysis of data from the 1000 Genomes Project. The resulting ancestry map was investigated to visualize clustering of this family with designated superpopulations. Although precise identification is not possible by this method, individuals from this family clustered most consistently with the Admixed American superpopulation (Fig. 2), as evidenced by the most nonsignificant p-value (0.34) and a Z-score most closely approaching zero (0.9467) (Table 2). The Admixed American superpopulation contains individuals from regions within South America, thus supporting ancestral origins of this family to this region of the world.
Genetic Ancestry Assessment. Plot shows the principal component analysis of the family in this report compared with subpopulations from the 1000 Genomes Project. AFR Africans (circle), AMR Admixed Americans (triangle), ARG-FAM Argentinian family in this report (+), EAS East Asians (x), EUR Europeans (diamond), SAS South Asians (inverted triangle)
Discussion and conclusions
Exome sequencing was performed on an undiagnosed multigenerational Argentinian family affected by spinocerebellar ataxia. This revealed a pathogenic variant in the PRKCG gene, associated with spinocerebellar ataxia type 14 (SCA14). PRKCG encodes protein kinase C gamma, a serine/threonine kinase found in the Purkinje cells of the cerebellum that contributes to neuronal functions including synapse morphology, receptor turnover, and cytoskeletal integrity [33, 34]. The p.Gln127Arg identified in this family maps to the C1 regulatory domain of PRKCG, a common location for SCA14-causing mutations [33, 34]. This domain interacts with second messengers and regulates activation and membrane translocation of the protein [34]. Although the precise mechanism of pathogenesis remains unknown, one study has suggested that SCA14 mutations, including p.Gln127Arg, may promote amyloidogenesis [33].
SCA14-causing mutations in the PRKCG gene have been reported in Europe (including France, Germany, the Netherlands, and Scandinavia) and Japan [27,28,29,30,31,32]. Although multiple cases of SCA1, SCA2, SCA3, SCA6, and SCA7 in Argentinian families have been published [12, 13], to our knowledge this is the first example of SCA14 in this population. We further determined the ancestral origins of the family to be most consistent with the Admixed American superpopulation in the 1000 Genomes project data, thus expanding the prevalence of SCA14 to South America. This diagnosis emphasizes the importance and value of WES in identifying coding variants in cerebellar ataxias. WES has been recommended as a crucial step in the diagnosis of rare genetic disorders due to its broad efficacy [35], with a 17–35% success rate for neurological diseases [11] and a diagnosis rate of 25–50% for cerebellar ataxia [7, 8]. In this case, despite remaining undiagnosed following initial clinical and genetic testing, exome analysis obtained a definitive diagnosis. This underscores the importance of improving access to genetic diagnostic testing in developing countries that may have limited clinical resources or where access to such testing is impeded by cost to the patient. Access to WES could dramatically improve the diagnosis of rare genetic disorders affecting the populations in these locations. The cost of next-generation sequencing is rapidly decreasing and continued bioinformatic advances are anticipated to make this a cost-effective comprehensive single test for the diagnosis of genetically complex disorders such as ataxia [8, 36]. In developing countries, more modern technologies may be quickly adopted, in some cases by bypassing older technologies entirely [37], and this may be a potential means to closing this diagnostic testing gap. This study exemplifies the need for continuous research and analysis of modern next-generation sequencing methods and making them accessible to patients worldwide.
Availability of data and materials
The research data generated and analyzed in this study is not available or shared publicly to protect patient privacy but is available from the corresponding author on reasonable request.
Abbreviations
- CoNIFER:
-
Copy number inference from exome reads
- DRPLA:
-
Dentatorubral-pallidoluysian atrophy
- gnomAD:
-
Genome Aggregation Database
- HGMD:
-
Human Gene Mutation Database
- PRKCG:
-
Protein kinase C gamma
- SCA14:
-
Spinocerebellar ataxia type 14
- STR:
-
Short tandem repeats
- WES:
-
Whole exome sequencing
- XHMM:
-
Exome hidden Markov Model
References
Jung I, Kim JS. Abnormal eye movements in parkinsonism and movement disorders. J Mov Disord. 2019;12(1):1–13. https://doi.org/10.14802/jmd.18034.
Klockgether T, Mariotti C, Paulson HL. Spinocerebellar ataxia. Nat Rev Dis Primers. 2019;5(1):24. https://doi.org/10.1038/s41572-019-0074-3.
Teive HA, Ashizawa T. Primary and secondary ataxias. Curr Opin Neurol. 2015;28(4):413–22. https://doi.org/10.1097/WCO.0000000000000227.
Mundwiler A, Shakkottai VG. Autosomal-dominant cerebellar ataxias. Handb Clin Neurol. 2018;147:173–85. https://doi.org/10.1016/B978-0-444-63233-3.00012-9.
Fogel BL. Autosomal-recessive cerebellar ataxias. Handb Clin Neurol. 2018;147:187–209. https://doi.org/10.1016/B978-0-444-63233-3.00013-0.
Bushart DD, Chopra R, Singh V, Murphy GG, Wulff H, Shakkottai VG. Targeting potassium channels to treat cerebellar ataxia. Ann Clin Transl Neurol. 2018;5(3):297–314. https://doi.org/10.1002/acn3.527.
Fogel BL, Lee H, Deignan JL, Strom SP, Kantarci S, Wang X, Quintero-Rivera F, Vilain E, Grody WW, Perlman S, Geschwind DH, Nelson SF. Exome sequencing in the clinical diagnosis of sporadic or familial cerebellar ataxia. JAMA Neurol. 2014;71(10):1237–46. https://doi.org/10.1001/jamaneurol.2014.1944.
Ngo KJ, Rexach JE, Lee H, Petty LE, Perlman S, Valera JM, Deignan JL, Mao Y, Aker M, Posey JE, Jhangiani SN, Coban-Akdemir ZH, Boerwinkle E, Muzny D, Nelson AB, Hassin-Baer S, Poke G, Neas K, Geschwind MD, Grody WW, Gibbs R, Geschwind DH, Lupski JR, Below JE, Nelson SF, Fogel BL. A diagnostic ceiling for exome sequencing in cerebellar ataxia and related neurological disorders. Hum Mutat. 2020;41(2):487–501. https://doi.org/10.1002/humu.23946.
Chen DH, Bird TD, Raskind WH. Spinocerebellar Ataxia Type 14. In: Adam MP, Everman DB, Mirzaa GM, Pagon RA, Wallace SE, Bean LJH, Gripp KW, Amemiya A, editors. GeneReviews((R)). Seattle (WA); 2020.
Schmitz-Hubsch T, Lux S, Bauer P, Brandt AU, Schlapakow E, Greschus S, Scheel M, Gartner H, Kirlangic ME, Gras V, Timmann D, Synofzik M, Giorgetti A, Carloni P, Shah JN, Schols L, Kopp U, Bussenius L, Oberwahrenbrock T, Zimmermann H, Pfueller C, Kadas EM, Ronnefarth M, Grosch AS, Endres M, Amunts K, Paul F, Doss S, Minnerop M. Spinocerebellar ataxia type 14: refining clinicogenetic diagnosis in a rare adult-onset disorder. Ann Clin Transl Neurol. 2021;8(4):774–89. https://doi.org/10.1002/acn3.51315.
Rexach J, Lee H, Martinez-Agosto JA, Nemeth AH, Fogel BL. Clinical application of next-generation sequencing to the practice of neurology. Lancet Neurol. 2019;18(5):492–503. https://doi.org/10.1016/S1474-4422(19)30033-X.
Teive HAG, Meira AT, Camargo CHF, Munhoz RP. The geographic diversity of spinocerebellar ataxias (SCAs) in the Americas: a systematic review. Mov Disord Clin Pract. 2019;6(7):531–40. https://doi.org/10.1002/mdc3.12822.
Rosa AL, Molina I, Kowaljow V, Conde CB. Brisk deep-tendon reflexes as a distinctive phenotype in an Argentinean spinocerebellar ataxia type 2 pedigree. Mov Disord. 2006;21(1):66–8. https://doi.org/10.1002/mds.20636.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. https://doi.org/10.1093/bioinformatics/btp324.
The Broad Institute. Picard Tools. https://broadinstitute.github.io/picard/. Accessed May 2022
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, Walters RK, Tashman K, Farjoun Y, Banks E, Poterba T, Wang A, Seed C, Whiffin N, Chong JX, Samocha KE, Pierce-Hoffman E, Zappala Z, O’Donnell-Luria AH, Minikel EV, Weisburd B, Lek M, Ware JS, Vittal C, Armean IM, Bergelson L, Cibulskis K, Connolly KM, Covarrubias M, Donnelly S, Ferriera S, Gabriel S, Gentry J, Gupta N, Jeandet T, Kaplan D, Llanwarne C, Munshi R, Novod S, Petrillo N, Roazen D, Ruano-Rubio V, Saltzman A, Schleicher M, Soto J, Tibbetts K, Tolonen C, Wade G, Talkowski ME, Database C, Neale BM, Daly MJ, MacArthur DG. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–43. https://doi.org/10.1038/s41586-020-2308-7.
Online Mendelian Inheritance in Man, OMIM. [Internet]. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD). [accessed November 2022]. World Wide Web URL: http://omim.org/.
Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, Hussain M, Phillips AD, Cooper DN. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet. 2017;136(6):665–77. https://doi.org/10.1007/s00439-017-1779-6.
Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, Jang W, Katz K, Ovetsky M, Riley G, Sethi A, Tully R, Villamarin-Salomon R, Rubinstein W, Maglott DR. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–8. https://doi.org/10.1093/nar/gkv1222.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, Committee ALQA. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. https://doi.org/10.1038/gim.2015.30.
Dolzhenko E, Deshpande V, Schlesinger F, Krusche P, Petrovski R, Chen S, Emig-Agius D, Gross A, Narzisi G, Bowman B, Scheffler K, van Vugt J, French C, Sanchis-Juan A, Ibanez K, Tucci A, Lajoie BR, Veldink JH, Raymond FL, Taft RJ, Bentley DR, Eberle MA. ExpansionHunter: a sequence-graph-based tool to analyze variation in short tandem repeat regions. Bioinformatics. 2019;35(22):4754–6. https://doi.org/10.1093/bioinformatics/btz431.
Krumm N, Sudmant PH, Ko A, O’Roak BJ, Malig M, Coe BP, Project NES, Quinlan AR, Nickerson DA, Eichler EE. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22(8):1525–32. https://doi.org/10.1101/gr.138115.112.
Fromer M, Purcell SM. Using XHMM software to detect copy number variation in whole-exome sequencing data. Curr Protoc Hum Genet. 2014;81(23):1–1. https://doi.org/10.1002/0471142905.hg0723s81.
MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–92. https://doi.org/10.1093/nar/gkt958.
Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Van Vooren S, Moreau Y, Pettett RM, Carter NP. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet. 2009;84(4):524–33. https://doi.org/10.1016/j.ajhg.2009.03.010.
Yabe I, Sasaki H, Chen DH, Raskind WH, Bird TD, Yamashita I, Tsuji S, Kikuchi S, Tashiro K. Spinocerebellar ataxia type 14 caused by a mutation in protein kinase C gamma. Arch Neurol. 2003;60(12):1749–51. https://doi.org/10.1001/archneur.60.12.1749.
Yamashita I, Sasaki H, Yabe I, Fukazawa T, Nogoshi S, Komeichi K, Takada A, Shiraishi K, Takiyama Y, Nishizawa M, Kaneko J, Tanaka H, Tsuji S, Tashiro K. A novel locus for dominant cerebellar ataxia (SCA14) maps to a 10.2-cM interval flanked by D19S206 and D19S605 on chromosome 19q13.4-qter. Ann Neurol. 2000;48(2):156–63. https://doi.org/10.1002/1531-8249(200008)48:2<156::aid-ana4>3.0.co;2-9.
van de Warrenburg BP, Verbeek DS, Piersma SJ, Hennekam FA, Pearson PL, Knoers NV, Kremer HP, Sinke RJ. Identification of a novel SCA14 mutation in a Dutch autosomal dominant cerebellar ataxia family. Neurology. 2003;61(12):1760–5. https://doi.org/10.1212/01.wnl.0000098883.79421.73.
Stevanin G, Hahn V, Lohmann E, Bouslam N, Gouttard M, Soumphonphakdy C, Welter ML, Ollagnon-Roman E, Lemainque A, Ruberg M, Brice A, Durr A. Mutation in the catalytic domain of protein kinase C gamma and extension of the phenotype associated with spinocerebellar ataxia type 14. Arch Neurol. 2004;61(8):1242–8. https://doi.org/10.1001/archneur.61.8.1242.
Morita H, Yoshida K, Suzuki K, Ikeda SI. A Japanese case of SCA14 with the Gly128Asp mutation. J Hum Genet. 2006;51(12):1118–21. https://doi.org/10.1007/s10038-006-0063-8.
Wieczorek S, Arning L, Gizewski ER, Alheite I, Timmann D. Benign SCA14 phenotype in a German patient associated with a missense mutation in exon 3 of the PRKCG gene. Mov Disord. 2007;22(14):2135–6. https://doi.org/10.1002/mds.21673.
Koht J, Stevanin G, Durr A, Mundwiller E, Brice A, Tallaksen CM. SCA14 in Norway, two families with autosomal dominant cerebellar ataxia and a novel mutation in the PRKCG gene. Acta Neurol Scand. 2012;125(2):116–22. https://doi.org/10.1111/j.1600-0404.2011.01504.x.
Takahashi H, Adachi N, Shirafuji T, Danno S, Ueyama T, Vendruscolo M, Shuvaev AN, Sugimoto T, Seki T, Hamada D, Irie K, Hirai H, Sakai N, Saito N. Identification and characterization of PKCgamma, a kinase associated with SCA14, as an amyloidogenic protein. Hum Mol Genet. 2015;24(2):525–39. https://doi.org/10.1093/hmg/ddu472.
Wong MMK, Hoekstra SD, Vowles J, Watson LM, Fuller G, Nemeth AH, Cowley SA, Ansorge O, Talbot K, Becker EBE. Neurodegeneration in SCA14 is associated with increased PKCgamma kinase activity, mislocalization and aggregation. Acta Neuropathol Commun. 2018;6(1):99. https://doi.org/10.1186/s40478-018-0600-7.
Sainio MT, Aaltio J, Hyttinen V, Kortelainen M, Ojanen S, Paetau A, Tienari P, Ylikallio E, Auranen M, Tyynismaa H. Effectiveness of clinical exome sequencing in adult patients with difficult-to-diagnose neurological disorders. Acta Neurol Scand. 2022;145(1):63–72. https://doi.org/10.1111/ane.13522.
Ibanez K, Polke J, Hagelstrom RT, Dolzhenko E, Pasko D, Thomas ERA, Daugherty LC, Kasperaviciute D, Smith KR. Lancet Neurol. 2022;21(3):234–45. https://doi.org/10.1016/S1474-4422(21)00462-2.
James J. Leapfrogging in mobile telephony: a measure for comparing country performance. Technol Forecast Soc. 2009;76(7):991–8. https://doi.org/10.1016/j.techfore.2008.09.002.
Acknowledgements
The authors would like to thank the patients and their family for their contributions to this study. SMP was supported by a PhD-student training fellowship from CONICET (Argentina). ALR is Principal Investigator of CONICET (Argentina)
Funding
Funding for this study was provided by generous donations to the University of California (BLF).
Author information
Authors and Affiliations
Contributions
ND performed data analysis and wrote the manuscript. KJN performed data analysis. SMP coordinated the study in Argentina and collected and prepared all DNA samples. ALR performed clinical evaluation of patients. BLF was responsible for concept and design, provided supervision, and wrote the manuscript. All authors contributed to review of the manuscript and all authors gave approval for publication. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The University of California, Los Angeles Institutional Review Board approved this study.
Consent for publication
Written informed consent was obtained from the patients for publication of this case report and any accompanying images. A copy of the written consents is available for review by the Editor-in-Chief of this journal.
Competing interests
Dr. Fogel reports funding from the National Ataxia Foundation and the National Institutes of Health. The remaining authors have no conflicts of interest to disclose.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Duggirala, N., Ngo, K.J., Pagnoni, S.M. et al. Spinocerebellar ataxia type 14 (SCA14) in an Argentinian family: a case report. J Med Case Reports 17, 168 (2023). https://doi.org/10.1186/s13256-023-03897-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13256-023-03897-y