Abstract
Background
Chromosome 13q deletion syndrome shows variable clinical features related to the different potential breakpoints in chromosome 13q. The severely malformed phenotype is known to be associated with the deletion of a critical region in 13q32. However, esophageal atresia is a rare symptom and the relevant region is unknown. Thus, determining the association between accurate breakpoints and new clinical features is essential.
Case presentation
A 28-year-old Japanese primigravid woman was referred for fetal growth restriction, absence of a gastric bubble, cerebellar hypoplasia, overlapping fingers, and polyhydramnios at 31 weeks gestation. At 38 + 0 weeks, she delivered a 1774 g female infant. The infant presented with isolated esophageal atresia (Gross type A), Dandy–Walker malformation, right microphthalmia, left coloboma, overlapping fingers, pleurocentrum in the thoracic vertebrae, reduced anogenital distance, and hearing loss. Her karyotype was diagnosed as 46,XX,del(13)(q32.1–qter) by amniocentesis, but array comparative genomic hybridization after birth revealed the deletion of 13q31.3–qter. At 48 days after birth, the infant underwent surgery for esophageal atresia and was later discharged from the hospital at 7 months of age.
Conclusion
This case report and the literature reviews supports the previous findings on the pathological roles of haploinsufficiency of the ZIC2/ZIC5 in Dandy–Walker malformation and the EFBN2 haploinsufficiency in eye malformation and hearing loss. Furthermore, the possible involvement of IRS2, COLA1, and COLA2 in eye malformation were identified. This is the first case of 13q deletion syndrome with esophageal atresia (Gross A), but it may be a symptom of VATER/VACTER association (vertebral defects, anorectal malformations, cardiac defects, tracheoesophageal fistula with or without esophageal atresia, renal malformations, and limb defects), as in the previous cases. These symptoms might also be associated with EFBN2 haploinsufficiency, although further research is required.
Similar content being viewed by others
Background
Genotype–phenotype analyses have revealed several critical regions related to specific anomalies in 13q deletion syndrome [1]. Brown et al. categorized 13q deletions into three groups: group 1, proximal deletions not extending into q32; group 2, more distal deletions including at least part of q32; and group 3, most distal deletions involving q33–34 [2]. They also suggested that the severely malformed 13q phenotype results from the deletion of a critical region in 13q32 [3], which was later verified by others [4, 5]. However, genotype–phenotype correlations are still not completely understood. Since 13q deletion syndrome is exceedingly rare and presents as multiple phenotypic symptoms, the collection and analysis of findings regarding genotype–phenotype correlations are necessary to better understand this condition.
In the present report, the patient was an infant with a prenatal diagnosis of 13q deletion syndrome, defined as the deletion of 13q32.1–qter, consistent with the most severe form of group 2 deletions. The neonate exhibited several related symptoms, including fetal growth restriction (FGR), congenital anomalies of microphthalmia, Dandy–Walker malformation (DWM), overlapping finger, and esophageal atresia. Esophageal atresia is a rare symptom. The comparative genomic hybridization (CGH) array revealed a breakpoint of 13q31.3.
Case presentation
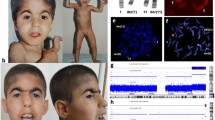
A 28-year-old Japanese primigravid woman was referred at 31 weeks of gestation for abnormal ultrasound findings including FGR, absence of a gastric bubble, and polyhydramnios. She had no history of abortion or complications. The couple had no family history of genetic diseases. The fetus showed an estimated fetal body weight of −3.2 standard deviations at the first ultrasound survey, had a score of 26 on the amniotic fluid index, exhibited no gastric bubble, and had cerebellar hypoplasia (Fig. 1a, b, arrows) and overlapping fingers. Microphthalmia was also detected in the right eye on fetal magnetic resonance imaging (MRI) (Fig. 1b). Amniocentesis was performed, and Giemsa banding (Fig. 2a) and fluorescence in situ hybridization (Fig. 2b) revealed a 46,XX,del(13)(q32.1–qter) karyotype. At 38 + 0 weeks gestation, an infant girl weighing 1774 g was born and examined by pediatric doctors. She presented with isolated esophageal atresia (Gross type A), DWM, right microphthalmia, left coloboma, overlapping fingers, pleurocentrum in the thoracic vertebrae, reduced anogenital distance, and hearing loss. At 48 days after birth, the infant underwent radical surgery for esophageal atresia and was discharged from the hospital uneventfully at 7 months age. The CGH array (SurePrint G3 human CGH 1 × 1 M; Agilent Technologies, CA, USA) using villus sampling after birth revealed that the region stretching from 13q31.3 to the terminus was deleted (22.12 Mb; from 92,973,314 to 115,097,664 bp, GRCh37, Fig. 2c). The parents both had normal karyotypes, indicating that the infant’s 13q deletion was de novo. The couple later had three healthy children.
Chromosomal analyses. a Results of Giemsa banding by amniocentesis. The arrow indicates the abnormal chromosome 13, as del(13)(q32.1). b The result of fluorescent in situ hybridization by amniocentesis. The normal chromosome 13 had both 13q14 (blue) and 13qter (yellow), but no signal indicative of 13qter was detected on the chromosome del(13)(q32.1). c Array CGH analysis reveals the deleted region of 13q31.2–qter (92973314–115097664). The right panel shows an enlarged view of the gene deletion site in the left panel
Discussion and conclusion
Esophageal atresia is a rare phenotype of 13q deletion syndrome. CGH array analysis determined that the breakdown point was 13q31.3. The patient also exhibited various phenotypes that matched with group 2 of 13q deletion syndrome, including overlapping finger, DWM, right microphthalmia, left coloboma, hearing loss, reduced anogenital distance, and pleurocentrum in the thoracic vertebrae (Fig. 3). She had no cardiac anomaly.
The relation between haploinsufficiency of EFNB2 and vertebral defects (V), anorectal malformations (A), cardiac defects (C), tracheoesophageal fistula with or without esophageal atresia (TE), renal malformations (R), and limb defects (L) (VATER/VACTERL) associations in 13q deletion syndrome have recently been reported [6,7,8]. Esophageal atresia without tracheoesophageal fistula (Gross A), vertebra anomalies, and reduced anogenital distance in the present patient also indicate VATER/VACTERL association-like symptoms. We found no previous reports demonstrating esophageal atresia (Gross A) with 13q deletion syndrome, but esophageal atresia (Gross A) might be a symptom of VATER/VACTER [9]. Our review found three 13q deletion patients with esophageal atresia, including the present patient (Table 1) [8, 10]. All of these patients showed partial symptoms of VATER/VACTER. Therefore, the present patient and our literature review suggest that 13q deletion syndrome with esophageal atresia would show VATER/VACTERL association-like symptoms. Two of the three patients had 13.32q deletion, which results in the haploinsufficiency of EFNB2. However, the specific region associated with esophageal fistula/atresia remains undetermined. Therefore, further studies are needed to clarify the association between insufficiency of the 13q region and tracheoesophageal fistula/atresia. For this purpose, it would be helpful to determine the deleted region in detail using the CGH array.
Finger anomalies are related to loss of the GPC5 gene in 13q31.3 [5] and the GPC6 gene in 13q31.3–q32.1 [11], consistent with the present patient. However, one report showed that limb abnormalities may not be related to any specific genomic region [12].
DWM is well known to be associated with haploinsufficiency of the ZIC2 and ZIC5 located in the 13q32.3 [4, 12,13,14,15], which is consistent with the findings of the present patient. In addition, the 13q33.1 may also be associated with DWM [14], which is also consistent with our patients.
Previous reports suggested that eye malformation might be associated with the EFNB2 gene [4], and deletion of the 13q32 region [2]. Recently, the 13q33.3–q34 deletion (110,302,002–11,394,979, GPCh37) has been shown to be associated with microphthalmia or anophthalmia with/without coloboma in 15 patients [16], and 13 genes were encoded in the region: IRS2[110405042–110438930], COL4A1[110801310–110959504], COL4A2[110959631–111165556], CARS2[111293757–111358862], ING1[111364970–111375686], SOX1[112721463–112726020], ATP11A[113344352–113541482], MCF2L[113623528–113754056], F7[113760102–113774999], F10[113777113–113803843], PROZ[113812962–113826700], PCID2[113831850–113862983] and CUL4A[113862507–113921422] (GpCh37) [7]. In our review of these genes, IRS2 was reported to be involved in retina function [17, 18]. COLA1 and/or COLA2 mutations are involved in ocular defects [19,20,21,22]. The present patient and our literature review suggest that IRS2, COLA1, and COLA2 in the 13q33.3–q34 would also have a pathological role in eve malformation, including microphthalmia or anophthalmia with/without coloboma.
It has been reported that EFNB2 haploinsufficiency [23,24,25] or 13q32 deletion [15], is involved in hearing loss, although deafness has been reported in the patients with deletions of the 13q13.1–q14.3 and 13q12.3–q21.1 [26]. Furthermore, EFNB2 is involved in the morphogenesis of the endolymphatic sac and duct epithelia in the mouse inner ear, which requires normal hearing [27]. Our data also supports the role of the EFNB2 haploinsufficiency in hearing loss.
The present patient showed no cardiac defect without the distal 13q34 region, which has been reported to be associated with cardiac defect [7, 28].
This study presented a case that indicated an association between esophageal atresia, a rare phenotype, with deletion of the 13q31.3–qter region. The present case and the literature review suggest that it is part of VATER/VACTERL association-like symptoms and suggests the association between haploinsufficiency of EFNB2 and VATER/VACTERL association-like symptoms. These findings support the previous findings on pathological roles of haploinsufficiency of ZIC2/ZIC5 on DWM, and EFBN2 haploinsufficiency on eye malformation and hearing loss. Furthermore, we identify the possible involvement of IRS2, COLA1, and COLA2 in eye malformation. We hope that these findings will help identify the causal genes of various phenotypes of 13q deletion and provide more precise information during prenatal counseling, although further accumulation of such reports is required.
Availability of data and materials
Not applicable.
References
Allderdice PW, Davis JG, Miller OJ, Klinger HP, Warburton D, Miller DA, Allen FH Jr, Abrams CA, McGilvray E. The 13q-deletion syndrome. Am J Hum Genet. 1969;21:499–512.
Brown S, Gersen S, Anyane-Yeboa K, Warburton D. Preliminary definition of a “critical region” of chromosome 13 in q32: report of 14 cases with 13q deletions and review of the literature. Am J Med Genet. 1993;45:52–9.
Brown S, Russo J, Chitayat D, Warburton D. The 13q- syndrome: the molecular definition of a critical deletion region in band 13q32. Am J Hum Genet. 1995;57:859–66.
Ballarati L, Rossi E, Bonati MT, Gimelli S, Maraschio P, Finelli P, Giglio S, Lapi E, Bedeschi MF, Guerneri S, et al. 13q Deletion and central nervous system anomalies: further insights from karyotype-phenotype analyses of 14 patients. J Med Genet. 2007;44: e60.
Quelin C, Bendavid C, Dubourg C, de la Rochebrochard C, Lucas J, Henry C, Jaillard S, Loget P, Loeuillet L, Lacombe D, et al. Twelve new patients with 13q deletion syndrome: genotype-phenotype analyses in progress. Eur J Med Genet. 2009;52:41–6.
Dworschak GC, Draaken M, Marcelis C, de Blaauw I, Pfundt R, van Rooij IA, Bartels E, Hilger A, Jenetzky E, Schmiedeke E, et al. De novo 13q deletions in two patients with mild anorectal malformations as part of VATER/VACTERL and VATER/VACTERL-like association and analysis of EFNB2 in patients with anorectal malformations. Am J Med Genet A. 2013;161a:3035–41.
He X, Shen H, Fu H, Feng C, Liu Z, Jin Y, Mao J. Reduced anogenital distance, hematuria and left renal hypoplasia in a patient with 13q33.1–34 deletion: case report and literature review. BMC Pediatr. 2020;20:327.
Walsh LE, Vance GH, Weaver DD. Distal 13q Deletion Syndrome and the VACTERL association: case report, literature review, and possible implications. Am J Med Genet. 2001;98:137–44.
Guptha S, Shumate C, Scheuerle AE. Likelihood of meeting defined VATER/VACTERL phenotype in infants with esophageal atresia with or without tracheoesophageal fistula. Am J Med Genet A. 2019;179:2202–6.
Jackson J, Robson L, Meagher S, Watson G, Smith A. How accurate does rapid fetal karyotyping need to be? Case of unbalanced t(13;18). Prenat Diagn. 1993;13:767–70.
van der Zwaag PA, Dijkhuizen T, Gerssen-Schoorl KB, Colijn AW, Broens PM, Flapper BC, van Ravenswaaij-Arts CM. An interstitial duplication of chromosome 13q31.3q32.1 further delineates the critical region for postaxial polydactyly type A2. Eur J Med Genet. 2010;53:45–9.
Bellucco FT, Rodrigues de Oliveira-Junior H, Santos Guilherme R, Bragagnolo S, Alvarez Perez AB, Ayres Meloni V, Melaragno MI. Deletion of chromosome 13 due to different rearrangements and impact on phenotype. Mol Syndromol. 2019;10:139–46.
Alp MY, Çebi AH, Seyhan S, Cansu A, Aydin H, Ikbal M. 225 MB deletion OF 13q31.1–q34 associated with HPE, DWM, and HSCR: a case report and redefining the smallest deleted regions. Genet Couns. 2016;27:43–9.
Kirchhoff M, Bisgaard AM, Stoeva R, Dimitrov B, Gillessen-Kaesbach G, Fryns JP, Rose H, Grozdanova L, Ivanov I, Keymolen K, et al. Phenotype and 244k array-CGH characterization of chromosome 13q deletions: an update of the phenotypic map of 13q21.1-qter. Am J Med Genet A. 2009;149a:894–905.
Mademont-Soler I, Morales C, Armengol L, Soler A, Sánchez A. Description of the smallest critical region for Dandy-Walker malformation in chromosome 13 in a girl with a cryptic deletion related to t(6;13)(q23;q32). Am J Med Genet A. 2010;152a:2308–12.
Boerkoel PK, Dixon K, Fitzsimons C, Shen Y, Huynh S, Schlade-Bartusiak K, Culibrk L, Chan S, Boerkoel CF, Jones SJM, Chin HL. Long-read genome sequencing resolves a complex 13q structural variant associated with syndromic anophthalmia. Am J Med Genet A. 2022;188:1589–94.
Albert-Fort M, Hombrebueno JR, Pons-Vazquez S, Sanz-Gonzalez S, Diaz-Llopis M, Pinazo-Durán MD. Retinal neurodegenerative changes in the adult insulin receptor substrate-2 deficient mouse. Exp Eye Res. 2014;124:1–10.
Iglesias-Osma MC, Blanco EJ, Carretero-Hernández M, Catalano-Iniesta L, García-Barrado MJ, Sánchez-Robledo V, Blázquez JL, Carretero J. The lack of Irs2 induces changes in the immunocytochemical expression of aromatase in the mouse retina. Ann Anat. 2022;239: 151726.
Deml B, Reis LM, Maheshwari M, Griffis C, Bick D, Semina EV. Whole exome analysis identifies dominant COL4A1 mutations in patients with complex ocular phenotypes involving microphthalmia. Clin Genet. 2014;86:475–81.
Kuo DS, Labelle-Dumais C, Mao M, Jeanne M, Kauffman WB, Allen J, Favor J, Gould DB. Allelic heterogeneity contributes to variability in ocular dysgenesis, myopathy and brain malformations caused by Col4a1 and Col4a2 mutations. Hum Mol Genet. 2014;23:1709–22.
Mao M, Kiss M, Ou Y, Gould DB. Genetic dissection of anterior segment dysgenesis caused by a Col4a1 mutation in mouse. Dis Model Mech. 2017;10:475–85.
Matías-Pérez D, García-Montaño LA, Cruz-Aguilar M, García-Montalvo IA, Nava-Valdéz J, Barragán-Arevalo T, Villanueva-Mendoza C, Villarroel CE, Guadarrama-Vallejo C, la Cruz RV, et al. Identification of novel pathogenic variants and novel gene-phenotype correlations in Mexican subjects with microphthalmia and/or anophthalmia by next-generation sequencing. J Hum Genet. 2018;63:1169–80.
Chatmethakul T, Phaltas R, Minzes G, Martinez J, Bhat R. A rare co-occurrence of intestinal malrotation and Hirschsprung’s disease in a neonate with 13q21.31q33.1 interstitial deletion including the EDNRB gene. J Pediatr Genet. 2019;8:142–6.
Defourny J, Audouard C, Davy A, Thiry M. Efnb2 haploinsufficiency induces early gap junction plaque disassembly and endocytosis in the cochlea. Brain Res Bull. 2021;174:153–60.
Lévy J, Haye D, Marziliano N, Casu G, Guimiot F, Dupont C, Teissier N, Benzacken B, Gressens P, Pipiras E, et al. EFNB2 haploinsufficiency causes a syndromic neurodevelopmental disorder. Clin Genet. 2018;93:1141–7.
Mitter D, Ullmann R, Muradyan A, Klein-Hitpass L, Kanber D, Ounap K, Kaulisch M, Lohmann D. Genotype-phenotype correlations in patients with retinoblastoma and interstitial 13q deletions. Eur J Hum Genet. 2011;19:947–58.
Raft S, Andrade LR, Shao D, Akiyama H, Henkemeyer M, Wu DK. Ephrin-B2 governs morphogenesis of endolymphatic sac and duct epithelia in the mouse inner ear. Dev Biol. 2014;390:51–67.
Yang YF, Ai Q, Huang C, Chen JL, Wang J, Xie L, Zhang WZ, Yang JF, Tan ZP. A 1.1Mb deletion in distal 13q deletion syndrome region with congenital heart defect and postaxial polydactyly: additional support for a CHD locus at distal 13q34 region. Gene. 2013;528:51–4.
Acknowledgements
We would like to thank Editage (www.editage.com) for English language editing.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
TK and HT wrote the first draft of the manuscript. HT, YIt, TU, NN, KI, YIi, FK, and HK were involved in diagnosing and managing the patient (mother). YM and MH were involved in diagnosing and treatment of the patient (neonate). TK, MH, and HK critically analyzed and interpreted the patient data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Written informed consent was obtained from the patient to publish this case report and any accompanying images.
Consent for publication
Written informed consent was obtained from the patient to publish this case report and any accompanying images. A copy of the written consent is available for review by the Editor-in-Chief of this journal.
Competing interests
The authors have no competing interest to declare.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kotani, T., Tsuda, H., Ito, Y. et al. Prenatal diagnosis of distal 13q deletion syndrome in a fetus with esophageal atresia: a case report and review of the literature. J Med Case Reports 16, 481 (2022). https://doi.org/10.1186/s13256-022-03713-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13256-022-03713-z