Abstract
Background
The Autism Sequencing Consortium identified 102 high-confidence autism spectrum disorder (ASD) genes, showing that individuals with ASD and with potentially damaging single nucleotide variation (pdSNV) in these genes had lower cognitive levels and delayed age at walking, when compared to ASD participants without pdSNV. Here, we made use of a Swedish sample of individuals with ASD (called PAGES, for Population-Based Autism Genetics & Environment Study) to evaluate the frequency of pdSNV and their impact on medical and psychiatric phenotypes, using an epidemiological frame and universal health reporting. We then combine findings with those for potentially damaging copy number variation (pdCNV).
Methods
SNV and CNV calls were generated from whole-exome sequencing and chromosome microarray data, respectively. Birth and medical register data were used to collect phenotypes.
Results
Of 808 individuals assessed by sequencing, 69 (9%) had pdSNV in the 102 ASC genes, and 144 (18%) had pdSNV in the 102 ASC genes or in a larger set of curated neurodevelopmental genes (from the Deciphering Developmental Disorders study, the gene2phenotype database, and the Radboud University gene lists). Three or more individuals had pdSNV in GRIN2B, POGZ, SATB1, DYNC1H1, SCN8A, or CREBBP. In comparison, out of the 996 individuals from whom CNV were called, 105 (11%) carried one or more pdCNV, including four or more individuals with CNV in the recurrent 15q11q13, 22q11.2, and 16p11.2 loci. Carriers of pdSNV were more likely to have intellectual disability (ID) and epilepsy, while carriers of pdCNV showed increased rates of congenital anomalies and scholastic skill disorders. Carriers of either pdSNV or pdCNV were more likely to have ID, scholastic skill disorders, and epilepsy.
Limitations
The cohort only included individuals with autistic disorder, the more severe form of ASD, and phenotypes are defined from medical registers. Not all genes studied are definitively ASD genes, and we did not have de novo information to aid in classification.
Conclusions
In this epidemiological sample, rare pdSNV were more common than pdCNV and the combined yield of potentially damaging variation was substantial at 27%. The results provide compelling rationale for the use of high-throughout sequencing as part of routine clinical workup for ASD and support the development of precision medicine in ASD.
Similar content being viewed by others
Background
Autism spectrum disorder (ASD) is a childhood-onset neurological and developmental disorder that affects more than 1% of the population [1]. The affected individuals can have lifelong impairments in social interaction, communication, and adaptive functioning. In the Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM-IV) [2], severity across the ASD spectrum was reflected by different terms, from a mild form called Asperger's syndrome, to the severest form called autistic disorder. In 2013, in the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-5) [3], autistic disorder, Asperger's syndrome and additional pervasive developmental disorder diagnoses were replaced with the umbrella diagnosis of ASD, with severity specifiers for social communication and for restricted interests and repetitive behaviors.
ASD has a complex genetic architecture with both rare and common variation contributing to risk. While common variation accounts for the majority of genetic liability for autism, rare variation, often de novo, accounts for substantial individual liability [4]. Numerous studies have identified de novo and inherited SNV and CNV associated with ASD [5,6,7]. Given that CNV has been easier to identify in both affected and unaffected populations, there is a large literature describing medical findings associated with CNV, as well as reliable estimates of the frequency of potentially damaging copy number variation (pdCNV) [5, 8,9,10,11,12,13,14,15,16,17]. Less is known about potentially damaging single nucleotide variation (pdSNV).
In the most extensive whole-exome sequencing study to date, the Autism Sequencing Consortium (ASC) identified 102 genes that, when carrying specific types of deleterious variants, are strongly associated with risk for ASD [13]. The same study showed that individuals with ASD and pdSNV in these 102 genes showed lower IQs and greater delays in walking, on average, as compared to individuals with ASD without pdSNV. However, clinical information was restricted in that study. Over 5% of all ASD participants carried pdSNV, although the ascertainment of the cohorts that make up the ASC study were almost universally convenience samples, so these rates are hard to generalize.
The objective of the current work is to extend the comparison of comorbid medical findings in individuals diagnosed with ASD with or without pdSNV or pdCNV, making use of a Swedish epidemiological sample called Population-Based Autism Genetics and Environment Study (PAGES) [18]. By incorporating robust and relatively unbiased phenotype data obtained from the Swedish national register, we compare the phenotypes of those with ASD with potentially damaging variation (PDV)—pdCNV or pdSNV, and those with ASD without a PDV. In addition, because sample collection was carried out in an epidemiological framework, we are able to describe the genetic architecture of PDV in ASD on a population level, including estimates of rates of genetic findings in ASD. In the companion study by Klei et al. [19], the inter-related role of common variation and PDV in ASD risk is explored in the PAGES sample.
Methods
Study population
In this study, we used data collected from study participants in PAGES, a large ongoing population-based cohort study in Sweden that started in 2012 with the overall aim to identify possible genetic and environmental risk factors for ASD [4]. The study was approved by the Regional Ethical Review Board in Stockholm, Sweden, and the Institutional Review Board at the Icahn School of Medicine at Mount Sinai, New York, USA. All individuals with a diagnosis of ASD according to the International Classification of Diseases (ICD) 9 and 10 criteria were identified in the Swedish National Patient Register. Our focus here is on autistic disorder, defined by ICD-9 codes 299.A/B/X and ICD-10 code F84.0. The eligible individuals were born in Sweden between 1960 and 1996 and followed up through 2011.
In PAGES, after a potential case was identified in the Swedish National Patient Register and the diagnosis confirmed, research nurses informed the family about the genetic study with a letter followed up with a phone call. Those interested in participating provided informed consent and biospecimens (blood in most cases). Information about sex, age at the time of diagnosis, date of admission and discharge, and diagnostic codes for intellectual functioning and psychiatric comorbidities were extracted from the Swedish National Patient Register after the consent form was signed. The date of the first registered ASD diagnosis was used as the diagnosis date.
In addition to the Swedish National Patient Register, the Multi-generation Register was also accessed, which allowed for the identification of family relations, as was the Swedish Medical Birth Register, which contained birth characteristics of all Swedish-born children since 1973 (including prenatal, perinatal and neonatal variables). For more information about the Swedish national registers, see [20].
DNA from 827 PAGES participants with ASD was subjected to whole exome sequencing by ASC [13]. In addition, 1,154 PAGES ASD samples were genotyped on either Infinium OmniExpress Exome V1 (n = 239, number of single nucleotide polymorphisms (SNPs): 951,117), V1.1 (n = 152, number of SNPs: 958,178), V1.2 (n = 553, number of SNPs: 964,193), V1.4 (n = 219, number of SNPs: 960,919), or the Infinium Global Screening Array (n = 82, number of SNPs: 700,078).
SNV calling
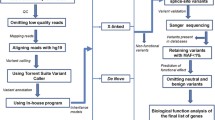
SNV was called using the Genome Analysis Toolkit [21] HaplotypeCaller package version 3.4 (for more details, see [13]). Rare SNV was defined as those absent from Genome Aggregation Database (gnomAD). Rare SNV in likely ASD and intellectual disability (ID) genes was classified as potentially damaging if the variant was either (1) a protein-truncating variant, or (2) a missense variant with a "Missense badness, PolyPhen-2, Constraint" (MPC) score > 2 [22].
While we initiated this study to extend the genome-wide ASC results, we recognize that the ASC gene list is both incomplete and will also include small numbers of false-positive findings. For this reason, and in response to reviews, we created a larger set of curated genes involved in ASD and/or other neurodevelopmental disorders. We made use of multiple data sources to define potential ASD genes. First, we used the 102 genes reported by the ASC in Satterstrom et al. [13]. Second, we created a developmental delay/ID gene list, relying on three sources of data. We began by incorporating the 94 genes reported in the Deciphering Developmental Disorders study in 2017 [23]. In addition, we accessed the gene2phenotype developmental disorders (DD) gene list [24] from [25] and the Radboud University Medical Center ID gene panel (version DG 2.18) from [26] on January 26, 2021. For these latter two gene lists, biallelic and imprinted genes were removed, and only genes with autosomal dominant or X-linked inheritance that were found in both lists were included in a combined list, to focus on genes clearly involved in neurodevelopmental disorders. The combined list from the Deciphering Developmental Disorders study, gene2phenotype, and Radboud University Medical Center is referred to as DGR, and included 560 genes (Additional File 1: Table S1). Hence, results are presented for three gene lists: (1) the 102 genes identified by the ASC (ASC102), (2) the independently derived but overlapping DGR gene list (n = 560); and (3) the union of the above two lists (ASC102 + DGR; n = 597 genes). We summarize findings for pdSNV in these gene lists.
CNV calling
CNV calls were generated from 1154 ASD samples genotyped on the Infinium OmniExpress Exome by PennCNV using hg19 genomic coordinates. Data and calls were cleaned using standard procedures in PennCNV (B Allele Frequency drift ≤ 0.01, |waviness factor |≤ 0.05, log R ratio SD ≤ 0.3). We combined neighboring CNV if the gap between them was less than or equal to 20% of the total length of the two adjacent CNV plus the gap. We excluded CNV with SNPs < 20, as well as CNV with at least 50% reciprocal overlap with previously described common CNV regions according to the Database of Genomic Variants v10.
We first developed a list of CNV that had prior strong evidence for being associated with a genomic neurodevelopmental disorder. To generate this list, we used curated lists of CNV from ClinGen and DECIPHER. We accessed the ClinGen ftp site [27] and downloaded the region curation list for hg19. We merged this list with the list from ClinGen Dosage Sensitivity Curation Page [28]. We chose the regions with haploinsufficiency and/or triplosensitivity scores of 3 (sufficient evidence) and treated deletions and duplications separately wherever indicated. For DECIPHER, we downloaded the list of CNV syndromes from [29]. We excluded those with a grade of 3 (susceptibility locus) and treated deletions and duplications separately wherever indicated. We then merged these lists (see Additional file 3: Table S2 for the final list). For a few regions with discrepant classifications in the two databases, we used the ClinGen classification.
We called CNV potentially damaging if it satisfied one or more of the following three conditions: (1) if the CNV occurred within a locus associated with known genomic disorders curated by ClinGen and/or DECIPHER (as noted above); (2) if the CNV was larger than 3 Mb; or, (3) if the CNV was larger than 1 Mb and included one or more coding exons from at least one brain-expressed gene (as determined from the UCSC Genome Browser). For chromosome X, we included only known loci associated with genomic disorders due to potentially lower quality of CNV calls from the sex chromosomes [30]. Five individuals with evidence for three or more large CNV (> 1 Mb) were removed due to concerns about the quality of the sample. In addition, we removed any sample with a called CNV > 45 Mb, eliminating one individual with a CNV of 75 Mb. The 45 Mb threshold was derived from an ongoing analysis by the GATK Team at Broad Institute to generate CNV calls for ASD samples (including PAGES) from WES data using gCNV [31]. In the GATK calls, pdCNV status for 77% of the variants associated with known genomic disorders from this study was confirmed, and no CNV in autosomes was larger than 45 Mb. We retained 996 high-quality samples for further analyses.
Note that the American College of Medical Genetics and Genomics (ACMG) guidelines were not used in this study to classify damaging variants (CNV or SNV) [32, 33]. Some of the variants that we classified as potentially damaging could be variants of uncertain significance based on ACMG guidelines. In addition, we did not have de novo information to aid in classification.
Phenotypic information
We extracted information for the following variables from the Swedish National Patient Register and the Swedish National Birth Register: ID (IQ < 70), attention-deficit/hyperactivity disorder (ADHD), psychotic disorders (schizophrenia, schizotypal, delusional, and other non-mood psychotic disorders), obsessive–compulsive disorder (OCD), anxiety disorder, speech and language disorders, scholastic skill disorders, motor function disorders, epilepsy, sleeping disorders, hypotonia, birth defects, prenatal growth rate, gestational age in weeks, weight, height and head circumference at birth, and Apgar scores (Additional file 2: Table S3). Thirteen individuals with a diagnosis of Down syndrome and one with a diagnosis of Turner’s syndrome were not included in downstream analyses. (More broadly, individuals with sex chromosome aneuploidies were excluded from PAGES at the time of recruitment.)
The average head circumference of healthy newborns is 33–35 cm [34]. While the range depends on the length of the newborn, among other attributes, we used head circumference without adjustment, defining "HC-small" if the circumference was smaller than 32 cm and "HC-large" if it was larger than 38. Small for gestational age was defined as birth weight less than two standard deviations below the mean using Swedish growth charts [35], while large for gestational age was defined as birth weight more than two standard deviations above the mean.
Statistical analysis
To identify comorbidities and birth characteristics associated with ASD probands who carry damaging mutations, we used a logit model in which carrier status of the damaging variant type was the dependent variable (carrier of pdCNV or pdSNV or not) and predictors were sex, used as a covariate, and potential comorbidity or characteristic. Thus, a series of models were fit, one for each potentially associated feature. We reported the resulting odds ratio (OR), p values, and 95% confidence intervals (CIs) for the OR after adjusting for the sex variable.
Results
Demographic data
After quality control, whole-exome sequencing (WES) data were available for 808 probands, and genotype (chromosomal microarray or CMA) data were available for 996 probands (Table 1). Of these individuals, 70% were male (Table 1).
Of the comorbidities and birth characteristics for the population of ASD probands (Table 2), ID was most common (48%), and epilepsy was second (31%). Individuals with congenital anomalies had the lowest age of ASD diagnosis, while individuals with psychotic disorders had the highest age of ASD diagnosis. Comorbidities and birth characteristics of the probands which were not genotyped or sequenced are presented in Table S4 (Additional file 2).
Genetic findings
Of the 808 individuals for whom WES was performed, 69 (9%) had pdSNV in an ASC102 gene, and no individuals had more than one (Additional file 4: Table S5). Of the pdSNV, 34 were predicted protein-truncating variants (frameshift, nonsense, splice acceptor or donor), and the remaining 35 were missense variants predicted to be deleterious (MPC score > 2). Genes with the highest frequency of pdSNV were GRIN2B (n = 6), POGZ (n = 5), SATB1 (n = 4), DYNC1H1 (n = 4), and CREBBP (n = 3). Two individuals had pdSNV in each of the following genes: CACNA1E, CHD8, DIP2A, FOXP1, RORB, SETD5, STXBP1, SUV420H1 (now referred to as KMT5B), and SYNGAP1. Combining the ASC102 genes with developmental delay/ID genes from additional curated sources (ASC102 + DGR) led to the identification of 157 pdSNV in 144 probands (18%), and 12 individuals had more than one pdSNV. Using the combined list, two or more individuals had pdSNV in the following genes, not already noted above: BRFA, CACNA1C, EHMT1, HK1, IQSEC2, KMT2A, LRP2, MTOR, PIK3CA SCN8A, and SMARCA4.
Of the 996 probands who were genotyped, 105 (11%) carried one or more pdCNV (Additional file 5: Table S6). Twelve individuals had two pdCNV, for a total of 117 pdCNV overall: 66 of these were heterozygous deletions, and 51 were heterozygous duplications, ranging in size from 218 kb to 44 Mb (median 3.6 Mb). There were 59 pdCNV that were considered to be known genomic disorders (Table 3).
In the 674 probands for which there was both WES and CMA data, 123 (18%) had at least one PDV using the ASC102 gene list and the CNV list, and 182 (27%) had at least one PDV using ASC102 + DGR gene list and the CNV list.
In the PAGES data, seven individuals had a diagnosis of fragile X syndrome. Five individuals with fragile X syndrome were included in the CNV analysis (CMA probands), where one had a recurrent pdCNV (22q11.2 duplication syndrome). Three individuals with fragile X syndrome were included in the SNV analysis (WES probands), none of which had an additional pdSNV.
Comorbidities and birth characteristics of the probands
Evaluating medical and psychiatric comorbidities among individuals with ASD (Tables 4, 5), the pdCNV and pdSNV groups showed slightly different average ages of ASD diagnosis by group and by sex, although none of these differences were significant (p value > 0.05). Of the phenotypes of individuals with pdCNV and pdSNV (Table 5), ID was the most common disorder.
For probands carrying pdSNV, versus those who did not, ID and epilepsy showed a significant positive association (Table 6), regardless of curated gene list (ASC102 vs. ASC102 + DGR). Similar patterns were observed when only considering pdSNV from the DGR list (Additional file 2: Table S7). Congenital anomalies and scholastic skill disorders were associated with carrying pdCNV (Table 7). When carriers of either pdSNV or pdCNV were assessed, ID and epilepsy showed consistent associations with PDV (Table 7).
We next compared the effect of pdCNV status for ASD subjects who do or do not manifest ID for the largest group of genetically characterized subjects, i.e., those who were genotyped (Table 8). We compared rates of pdCNV and phenotypes of ASD individuals with and without ID; rates were not significantly different between groups (p value > 0.05 for all tests). For instance, the risk for congenital abnormalities is similar for potentially damaging CNV carriers whether or not they meet criteria for ID, 2.46 versus 3.71 (Table 8), thus ID status is not driving this association..
Data for sleeping disorders, hypotonia, birth defects, prenatal growth rate, gestational age in weeks, weight and height at birth, and Apgar scores were underpowered due to a high number of missing values.
Over the course of the review, we conducted a more conservative analysis using additional criteria, in order to address questions raised during review. This resulted in removing seven individuals and reassigning 27 pdCNV as not potentially damaging, impacting 31 individuals [28 individuals in the accompanying manuscript by Klei et al. [19]] (Additional file 5: Table S6). Specifically, seven individuals were removed due to concerns about complex or recurrent PDV: One individual had very large duplications on two different chromosomes; three individuals had a terminal duplication and a terminal deletion in the same chromosome; and three individuals had an almost identical pericentromeric duplication of 13 Mb on chromosome 8. While data for all these seven individuals passed our quality control steps, we removed them in this conservative additional analysis.
Furthermore, for this additional analysis, following discussion with the Editor, the following pdCNV were reclassified as not being pdCNV: (1) large CNV in pericentromeric regions (n = 11); (2) for pdCNV > 1 Mb and < 3 Mb, we included only deletions with one or more coding exons from at least one brain-expressed gene that was also constrained for truncating variants (probability of loss-of-function intolerant (pLI) ≥ 0.9 in gnomAD), reclassifying 12 pdCNV as not potentially damaging; (3) CNV reported in Decipher, but with lesser evidence reported in ClinGen, specifically, two 16p13.11 duplications and two 16p12.1 were reclassified as not potentially damaging (see Table 3); (4) one large duplication in the 15q13.3 microdeletion syndrome region which met our criteria for large CNV was reclassified as not potentially damaging since duplications in this region are not considered risk loci according to DECIPHER and ClinGen. In this more conservative, supplemental analysis, two individuals had two pdCNV, for a total of 76 pdCNV across the cohort (Additional file 5: Table 6). In addition to congenital anomalies and scholastic skill disorders previously shown to be associated with individuals carrying pdCNV (Table 6), we observed associations for ID, HC-small, and small for gestational (Additional file 2: Table S8). When carriers of pdSNV and/or pdCNV were assessed, ID and epilepsy were associated with carrying PDV (Additional file 2: Table S8), similar to the previous results (Table 7).
Discussion
Frequently, large-scale gene discovery studies are carried out on convenience samples, often with limited clinical data. Hence, the prevalence of PDV in the population cannot be readily estimated. Furthermore, while the spectrum of comorbid medical, neurological and psychiatric phenotypes for CNV has been studied extensively, less is known about comorbidities associated with pdSNV. In this study, we investigated pdSNV and pdCNV in a population sample of individuals from Sweden identified with autistic disorder. In this population sample, 27% of individuals had pdSNV (ASC102 + DGR gene list) and/or pdCNV. Carriers of pdCNV made up 11% of the individuals with autistic disorder in the Swedish population, similar to that reported for European ancestry in other studies [8, 36], while 18% of the individuals were carriers of pdSNV (ASC102 + DGR gene list). Of the 674 probands for which both WES and CMA data were available, 16 individuals had two or more PDV. Twelve individuals had two pdCNV, and five individuals had a pdCNV and pdSNV (one individual had one pdSNV and two pdCNV). One might be tempted to attribute oligogenic mechanisms for ASD on the basis of these 16 carriers; however, if PDVs occur at either a Poisson rate of 0.182 (ASC102 gene list) or 0.270 (ASC102 + DGR gene list), this number of carriers of two or more PDVs is consistent with random chance. Hence, while individuals with more than one PDV exist, and have been shown in some instances to have more severe phenotypes, our epidemiological analyses do not support what has been termed an oligogenic model in autism, i.e., where there is a nonrandom occurrence of 2 or more high-risk variants in individuals [37,38,39].
Consistent with prior reports [40, 41], CNV in the 15q11q13 Prader–Willi syndrome/Angelman syndrome region were most common (n = 10), including eight duplications and two deletions. Because we don't have access to detailed phenotype information and we could not determine the parent of origin of the deletions, we don't know if they are associated with Prader–Willi syndrome (loss of paternal allele) or Angelman syndrome (loss of maternal allele). Two other common regions in the cohort were 2q37 deletion syndrome (n = 4) and 22q11.2 deletion syndrome (velo‐cardio‐facial syndrome/DiGeorge syndrome) (n = 4), both of which are known risk factors for ASD and ID. Genes most commonly impacted by pdSNV included GRIN2B (n = 6), which is reported in individuals with ID, epilepsy, and ASD [42]. POGZ is emerging as a major gene in ASD [13], similar to what is observed here. Other genes with several pdSNV were SATB1 (n = 4), DYNC1H1 (n = 4), SCN8A (n = 3), and CREBBP (n = 3).
Some genes were impacted by either pdSNV or pdCNV. For example, SHANK3 was disrupted in one individual by pdSNV (nonsense variant) and three individuals by pdCNV (22q13.3 deletion); SHANK3 encodes a scaffold protein of the postsynaptic density that is essential for proper functioning of the synapse and loss of one functional copy of this gene, leading to Phelan-McDermid syndrome, has been estimated to account for ~0.5% of ASD [43]. Other ASD genes impacted by pdCNV or pdSNV include SCN2A (missense variant; the same variant is reported in ClinVar as de novo and likely pathogenic, variation ID: 207016) and ASLX3 (frameshift variant).
Among individuals with autistic disorder, we observed a significant association between PDV and ID, scholastic skills disorders, and epilepsy. This association was not observed in studies with smaller sample sizes, likely due, in part, to lack of information for the comorbid conditions [44]. Individuals with pdCNV had an elevated risk for congenital anomalies, a relevant risk factor for autism. Because of the near-universal health care and national health registers in Sweden, the findings of comorbid neurological and developmental conditions were not likely to be due to ascertainment bias.
We compared the effect of pdCNV status for ASD subjects who do or do not manifest ID. ID (IQ < 70) was the most common comorbidity (47% had ID) and had sufficient sample size to make such an exploration meaningful. Although ASD subjects who had pdCNV were more likely to have ID, ASD subjects with and without ID showed no significant differences in the association of pdCNV status with other potentially associated phenotypes. Thus, conditioning on ID status does not appear to explain much of the variation for other CNV-related associations.
Research suggests epilepsy and ASD have shared etiological mechanisms [45]. A large study of 5815 children with ASD found that 12.5% had epilepsy among children aged 2–17 years, and 26% among children aged 13 years and older [46]. In the PAGES cohort, 31% of individuals with autistic disorder had epilepsy. There were multiple findings of pdSNV and pdCNV in known epilepsy genes in our study.
In the PAGES cohort, thirteen individuals had a diagnosis of Down syndrome and were not included in the current analyses. The prevalence of Down syndrome is reported to be higher for those with ASD than in the general population [47] and represent an additional genetic diagnosis for ASD in PAGES.
Limitations
The results of this study should be interpreted in the context of some limitations. First, not all variants were validated by a second method; therefore, some could be artifacts. Nonetheless, a substantial portion of the CNV were independently validated by calling CNV from the whole exome data [31], and the validation rate of SNV is similarly high, as documented by variant calls from whole-genome versus whole-exome sequencing [48]. To further limit potentially miscalled or misclassified CNV, we went so far as to run an additional analysis, removing seven individuals with presumed pdCNV and reassigning pdCNV status for 27 pdCNV. We observed significant associations of ID, HC-small, and small for gestational with carrying pdCNV, in addition to the previously observed associations. Second, judgment calls and empirically defined thresholds were used to identify PDV. It is also important to note that this study focused on autistic disorder, and future studies on individuals with less profound ASD are warranted in order to draw a more comprehensive picture of the genetic architecture of the autism spectrum. Third, head circumference at birth, and indeed, most birth-related variables are dependent and should be interpreted with caution. Fourth, phenotypes are defined from medical registers, which may lead to under-ascertainment of comorbid diagnoses, particularly of milder findings, since the Swedish National Patient Register would not include comorbid diagnoses for those who do not seek clinical services for the relevant condition or only seek help at a primary care facility.
Conclusions
This population survey, with its characterization of developmental impact and frequency of rare PDV, provides greater insight into the genetic architecture of ASD and associated comorbidities. pdSNV were frequent, even more frequent than pdCNV. This indicates that high-throughput sequencing is an important part of the genetic characterization of ASD. Reliable methods for calling genic CNV from sequencing data have been established [49,50,51,52]; hence, there is good reason to use sequencing as a first-tier clinical approach, especially when one considers the co-occurrence of pdSNV and pdCNV in some subjects. The high rates of genetic findings in this epidemiological cohort provide a very strong rationale for developing precision medicine approaches in ASD, with treatment tailored to differences in underlying etiology and biology.
Rare pdCNV and pdSNV had a statistically higher occurrence in ASD subjects with ID, scholastic skill disorders, congenital anomalies, and epilepsy. These findings are consistent with prior reports, and given the nature of our sample, we can exclude ascertainment bias as the cause of this association.
Importantly, because many of the same subjects have been characterized for genotypes from common variants, we can explore the genetic architecture of ASD in even greater detail, relating common and rare variant risk. Indeed, in an accompanying manuscript by Klei et al. [19], we explore the joint contributions of rare and common variation to liability for ASD, finding that they work together approximately additively.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Abbreviations
- ACMG:
-
American College of Medical Genetics and Genomics
- ADHD:
-
Attention-deficit/hyperactivity disorder
- ASD:
-
Autism spectrum disorder
- ASC102:
-
102 Genes identified by the Autism Sequencing Consortium (ASC)
- CI:
-
Confidence interval
- CMA:
-
Chromosomal microarray
- CNV:
-
Copy number variation
- DGR:
-
Gene2phenotype and the Radboud University Medical Center intellectual disability gene lists
- DSM:
-
Diagnostic and Statistical Manual of Mental Disorders
- gnomAD:
-
Genome aggregation database
- ICD:
-
International classification of diseases
- ID:
-
Intellectual disability
- MPC:
-
"Missense badness, Polyphen-2, Constraint" pathogenicity score
- OCD:
-
Obsessive–compulsive disorder
- OR:
-
Odds ratio
- PAGES:
-
Population-based autism genetics and environmental study
- pdCVN:
-
Potentially damaging copy number variation
- pdSNV:
-
Potentially damaging single nucleotide variation
- PDV:
-
Potentially damaging variation
- pLI:
-
Probability of loss-of-function intolerant
- SNP:
-
Single nucleotide polymorphisms
- SNV:
-
Single nucleotide variation
- WES:
-
Whole-exome sequencing
References
Levy SE, Mandell DS, Schultz RT. Autism. Lancet. 2009;374:1627–38.
American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 4th ed. Washington, DC: American Psychiatric Association; 1994.
American Psychiatric Association. Diagnostic and statistical manual of mental disorders (5th edn.). 2013.
Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, et al. Most genetic risk for autism resides with common variation. Nat Genet. 2014;46:881–5.
Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 2015;87:1215–33.
Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L, et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet. 2014;94:677–94.
Kushima I, Aleksic B, Nakatochi M, Shimamura T, Okada T, Uno Y, et al. Comparative analyses of copy-number variation in autism spectrum disorder and schizophrenia reveal etiological overlap and biological insights. Cell Rep. 2018;24:2838–56.
Shen Y, Dies KA, Holm IA, Bridgemohan C, Sobeih MM, Caronna EB, et al. Clinical genetic testing for patients with autism spectrum disorders. Pediatrics. 2010;125:e727–35.
Poultney CS, Goldberg AP, Drapeau E, Kou Y, Harony-Nicolas H, Kajiwara Y, et al. Identification of small exonic CNV from whole-exome sequence data and application to autism spectrum disorder. Am J Hum Genet. 2013;93:607–19.
Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–72.
Gudmundsson OO, Walters GB, Ingason A, Johansson S, Zayats T, Athanasiu L, et al. Attention-deficit hyperactivity disorder shares copy number variant risk with schizophrenia and autism spectrum disorder. Transl Psychiatry. 2019;9:258.
Satterstrom FK, Walters RK, Singh T, Wigdor EM, Lescai F, Demontis D, et al. Autism spectrum disorder and attention deficit hyperactivity disorder have a similar burden of rare protein-truncating variants. Nat Neurosci. 2019;22:1961–5.
Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An J-Y, et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180:568-84.e23.
Chawner SJRA, Doherty JL, Anney RJL, Antshel KM, Bearden CE, Bernier R, et al. A genetics-first approach to dissecting the heterogeneity of autism: phenotypic comparison of autism risk copy number variants. Am J Psychiatry. 2021;178:77–86.
Coe BP, Witherspoon K, Rosenfeld JA, van Bon BWM, Vulto-van Silfhout AT, Bosco P, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet. 2014;46:1063–71.
Moreno-De-Luca D, Sanders SJ, Willsey AJ, Mulle JG, Lowe JK, Geschwind DH, et al. Using large clinical data sets to infer pathogenicity for rare copy number variants in autism cohorts. Mol Psychiatry. 2013;18:1090–5.
Tammimies K, Marshall CR, Walker S, Kaur G, Thiruvahindrapuram B, Lionel AC, et al. Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. JAMA. 2015;314:895–903.
PAGES [Internet]. [cited 2021 Apr 15]. https://ki.se/en/meb/pages.
Klei L, McClain LL, Mahjani B, Panayidou K, de Rubeis S, Grahnat ACS, et al. How rare and common risk variation jointly affect liability for autism spectrum disorder. Mol Autism. 2021. https://doi.org/10.1101/2020.10.27.20220095.
Mahjani B, Dellenvall K, Grahnat A-CS, Karlsson G, Tuuliainen A, Reichert J, et al. Cohort profile: Epidemiology and Genetics of Obsessive–compulsive disorder and chronic tic disorders in Sweden (EGOS). Soc Psychiatry Psychiatr Epidemiol. 2020;55:1383–93.
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinform. 2013;43:11.10.1–33.
Samocha KE, Kosmicki JA, Karczewski KJ, O’Donnell-Luria AH, Pierce-Hoffman E, MacArthur DG, et al. Regional missense constraint improves variant deleteriousness prediction [Internet]. bioRxiv. 2017 [cited 2021 Apr 17]. p. 148353. https://doi.org/10.1101/148353v1.abstract.
Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–8.
Thormann A, Halachev M, McLaren W, Moore DJ, Svinti V, Campbell A, et al. Flexible and scalable diagnostic filtering of genomic variants using G2P with Ensembl VEP. Nat Commun. 2019;10.
gene2phenotype [Internet]. [cited 2021 Jul 8]. https://www.ebi.ac.uk/gene2phenotype/downloads.
Intellectual disability [Internet]. [cited 2021 Jul 8]. https://www.radboudumc.nl/en/patientenzorg/onderzoeken/exome-sequencing-diagnostics/exomepanelspreviousversions/exomepanelspreviousversions/intellectual-disability.
ClinGen [Internet]. [cited 2021 Jul 23]. https://ftp.clinicalgenome.org/.
ClinGen Genome Dosage Map [Internet]. [cited 2021 May 13]. https://dosage.clinicalgenome.org/recurrent_region.shtml.
DECIPHER v11.3: Mapping the clinical genome [Internet]. [cited 2021 May 13]. https://decipher.sanger.ac.uk/disorders/syndromes/list.
Pinto D, Darvishi K, Shi X, Rajan D, Rigler D, Fitzgerald T, et al. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotechnol. 2011;29:512–20.
(How to) Call common and rare germline copy number variants [Internet]. [cited 2021 Apr 13]. https://gatk.broadinstitute.org/hc/en-us/articles/360035531152-How-to-Call-common-and-rare-germline-copy-number-variants.
Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med. 2020;22:245–57.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–23.
Ricci SS, Kyle T. Maternity and pediatric nursing. Philadelphia: Lippincott Williams & Wilkins; 2009.
Marsál K, Persson PH, Larsen T, Lilja H, Selbing A, Sultan B. Intrauterine growth curves based on ultrasonically estimated foetal weights. Acta Paediatr. 1996;85:843–8.
Schaefer GB, Mendelsohn NJ, Professional Practice and Guidelines Committee. Clinical genetics evaluation in identifying the etiology of autism spectrum disorders: 2013 guideline revisions. Genet Med. 2013;15:399–407
Du Y, Li Z, Liu Z, Zhang N, Wang R, Li F, et al. Nonrandom occurrence of multiple de novo coding variants in a proband indicates the existence of an oligogenic model in autism. Genet Med. 2020;22:170–180.
Turner TN, Coe BP, Dickel DE, Hoekzema K, Nelson BJ, Zody MC, et al. Genomic Patterns of De Novo Mutation in Simplex Autism. Cell. 2017;171:710-722.e12.
Schaaf CP, Sabo A, Sakai Y, Crosby J, Muzny D, Hawes A, et al. Oligogenic heterozygosity in individuals with high-functioning autism spectrum disorders. Hum Mol Genet. 2011;20:3366–75.
Reiter LT. Chapter 9—Developmental disabilities, autism, and schizophrenia at a single locus: complex gene regulation and genomic instability of 15q11-q13 cause a range of neurodevelopmental disorders. In: Rubenstein J, Rakic P, Chen B, Kwan KY, editors. Neurodevelopmental Disorders. Academic Press; 2020. pp. 201–21.
Lu Y, Liang Y, Ning S, Deng G, Xie Y, Song J, et al. Rare partial trisomy and tetrasomy of 15q11-q13 associated with developmental delay and autism spectrum disorder. Mol Cytogenet. 2020;13:21.
Platzer K, Lemke JR. GRIN2B-Related Neurodevelopmental Disorder. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Mirzaa G, et al., editors. GeneReviews®. Seattle (WA): University of Washington, Seattle; 2018.
Betancur C, Buxbaum JD. SHANK3 haploinsufficiency: a “common” but underdiagnosed highly penetrant monogenic cause of autism spectrum disorders. Mol Autism. 2013;4:17.
Barone R, Gulisano M, Amore R, Domini C, Milana MC, Giglio S, et al. Clinical correlates in children with autism spectrum disorder and CNVs: Systematic investigation in a clinical setting. Int J Dev Neurosci. 2020;80:276–86.
Richard AE, Scheffer IE, Wilson SJ. Features of the broader autism phenotype in people with epilepsy support shared mechanisms between epilepsy and autism spectrum disorder. Neurosci Biobehav Rev. 2017;75:203–33.
Viscidi EW, Triche EW, Pescosolido MF, McLean RL, Joseph RM, Spence SJ, et al. Clinical characteristics of children with autism spectrum disorder and co-occurring epilepsy. PLoS ONE. 2013;8:e67797.
DiGuiseppi C, Hepburn S, Davis JM, Fidler DJ, Hartway S, Lee NR, et al. Screening for autism spectrum disorders in children with Down syndrome: population prevalence and screening test characteristics. J Dev Behav Pediatr. 2010;31:181–91.
An J-Y, Lin K, Zhu L, Werling DM, Dong S, Brand H, et al. Genome-wide de novo risk score implicates promoter variation in autism spectrum disorder. Science. 2018;362.
Ruderfer DM, Hamamsy T, Lek M, Karczewski KJ, Kavanagh D, Samocha KE, et al. Patterns of genic intolerance of rare copy number variation in 59,898 human exomes. Nat Genet. 2016;48:1107–11.
Minoche AE, Lundie B, Peters GB, Ohnesorg T, Pinese M, Thomas DM, et al. ClinSV: clinical grade structural and copy number variant detection from whole genome sequencing data. Genome Med. 2021;13:32.
Trost B, Walker S, Wang Z, Thiruvahindrapuram B, MacDonald JR, Sung WWL, et al. A comprehensive workflow for read depth-based identification of copy-number variation from whole-genome sequence data. Am J Hum Genet. 2018;102:142–55.
Chanwigoon S, Piwluang S, Wichadakul D. inCNV: an integrated analysis tool for copy number variation on whole exome sequencing. Evol Bioinform Online. 2020;16:1176934320956577.
Acknowledgements
We thank the PAGES families and clinicians for their participation.
Funding
This study was supported by the National Institute of Mental Health (NIMH) Grants R01MH097849, R01MH097849-S1, U01MH111661, U01MH111658, U01MH111660, and U01MH111662, and the Beatrice and Samuel A. Seaver Foundation.
Author information
Authors and Affiliations
Contributions
BM had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. JDB, BD, KR, SDR, BM contributed to study concept and design. JDB, BD, SDR, BM, CGM, MM contributed to acquisition, analysis, or interpretation of data. JDB, BD, SDR, BM, CGM, MM contributed to drafting of the manuscript. All authors contributed to critical revision of the manuscript for important intellectual content. BD, BM contributed to statistical analysis. JDB, BD, KR obtained funding. JDB, BD, KR contributed to study supervision. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Regional Ethical Review Board in Stockholm, Sweden, and the Institutional Review Board at the Icahn School of Medicine at Mount Sinai, New York, USA.
Consent for publication
Not relevant.
Competing interests
The last author is Editor-in-Chief of Molecular Autism. The other authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
: Table S1. List of genes used for the analysis of potentially damaging SNV
Additional file 2
: Table S3. ICD codes used in this study. Table S4. Comorbidities and birth characteristics of the probands that were not genotyped or sequenced. Table S7. Odds ratios for comorbidities and birth characteristics probands with potentially damaging SNV, DGR list. Table S8. Odds ratios for comorbidities and birth characteristics of probands with potentially damaging CNV or SNV as defined for the additional analysis (see text).
Additional file 3
: Table S2. List of genomic disorders derived from ClinGen and DECIPHER
Additional file 4
: Table S5. List of potentially damaging SNV
Additional file 5
: Table S6. List of potentially damaging CNV
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Mahjani, B., De Rubeis, S., Gustavsson Mahjani, C. et al. Prevalence and phenotypic impact of rare potentially damaging variants in autism spectrum disorder. Molecular Autism 12, 65 (2021). https://doi.org/10.1186/s13229-021-00465-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13229-021-00465-3