Abstract
Background
Rhesus monkeys (Macaca mulatta) exhibit pronounced individual differences in social traits as measured by the macaque Social Responsiveness Scale-Revised. The macaque Social Responsiveness Scale was previously adapted from the Social Responsiveness Scale, an instrument designed to assess social and autistic trait variation in humans. To better understand potential biological underpinnings of this behavioral variation, we evaluated the trait-like consistency of several biological measures previously implicated in autism (e.g., arginine vasopressin, oxytocin, and their receptors, as well as ERK1/2, PTEN, and AKT(1–3) from the RAS-MAPK and PI3K-AKT pathways). We also tested which biological measures predicted macaque Social Responsiveness Scale-Revised scores.
Methods
Cerebrospinal fluid and blood samples were collected from N = 76 male monkeys, which, as a sample, showed a continuous distribution on the macaque Social Responsiveness Scale-Revised. In a subset of these subjects (n = 43), samples were collected thrice over a 10-month period. The following statistical tests were used: “Case 2A” intra-class correlation coefficients of consistency, principal component analysis, and general linear modeling.
Results
All biological measures (except AKT) showed significant test–retest reliability within individuals across time points. We next performed principal component analysis on data from monkeys with complete biological measurement sets at the first time point (n = 57), to explore potential correlations between the reliable biological measures and their relationship to macaque Social Responsiveness Scale-Revised score; a three-component solution was found. Follow-up analyses revealed that cerebrospinal fluid arginine vasopressin concentration, but no other biological measure, robustly predicted individual differences in macaque Social Responsiveness Scale-Revised scores, such that monkeys with the lowest cerebrospinal fluid arginine vasopressin concentration exhibited the greatest social impairment. Finally, we confirmed that this result held in the larger study sample (in which cerebrospinal fluid arginine vasopressin values were available from n = 75 of the subjects).
Conclusions
These findings indicate that cerebrospinal fluid arginine vasopressin concentration is a stable trait-like measure and that it is linked to quantitative social trait variation in male rhesus monkeys.
Similar content being viewed by others
Background
All primate species are social for at least some portion of their lifespan [1]. Sociality requires the ability to recognize and remember conspecifics, communicate information effectively, interact in a species-typical manner, and acquire and use knowledge about social relationships that exist between those in one’s social group [2]. Yet, pronounced individual variation in social functioning is evident within non-human primate species [3, 4]. Although this social variation has been well documented, it has not been systematically studied [5].
Introduction of the Social Responsiveness Scale (SRS) facilitated rapid and large-scale assessment of social trait variation in humans [6, 7]. This scale provides a quantitative measure of typical and atypical social functioning in natural social settings [8, 9]. This scale is scored such that higher SRS scores indicate greater social impairment. SRS scores have been shown to be continuously distributed across the general human population [10]. At the extreme of the population distribution, higher SRS scores overlap significantly with a diagnosis of autism spectrum disorder (ASD) [8, 11], a brain disorder characterized by core social cognitive and interaction deficits [12]. This collective evidence has enabled use of the SRS as both a research tool for measuring the presence of autistic traits in members of the general human population [7, 13], and as a clinical screening tool for ASD [8, 9].
The SRS has proven to be a useful measure of social variation in multiple and diverse human societies [14,15,16,17]. Given its broad applicability in humans, the SRS has been adapted for use in chimpanzees [18, 19] and rhesus macaques [20] to enable assessment of the presence of similar social traits in these species. Consistent with the human SRS total score [10], chimpanzee SRS and macaque SRS (mSRS) scores are continuously distributed across their respective general populations [18,19,20,21]. Moreover, rhesus monkeys observed to be 1.5 standard deviations (SD) above the population mean on naturally occurring non-social behavior are also rated as having a greater burden of autistic-like traits [21].
Very little is known at present about the biological underpinnings of quantitative social trait variation in human and non-human primates. This is because prior biological investigations have typically focused on the development of species-typical social behavior (e.g., parental behavior onset; social bond formation) in rodent species [22]. Nevertheless, several cellular and neuroanatomical signalling pathways implicated in prosocial behavior, ASD, and/or ASD-related syndromes may underlie the regulation of social trait variation.
The first class of biological candidates includes the hypothalamic neuropeptides oxytocin (OXT) and arginine vasopressin (AVP) [23], and the main receptors to which they bind to regulate social behavior, OXTR and AVPR1A [24, 25]. It is well established that these neuropeptides are critical for the expression of prosocial behavior (e.g., affiliative behavior, social bond formation, parental care, social learning, and memory) [26, 27]. Moreover, experimental (pharmacological or genetic) disruption of OXT and AVP brain signaling produces a variety of social behavior impairments in rodents [24, 25, 28, 29]. Deficits in these neuropeptide signaling pathways have also been documented in mouse models of neurogenetic syndromes with high penetrance for ASD (e.g., Fragile X syndrome, Prader-Willi syndrome, CNTNAP2) [30, 31].
The second class of biological candidates includes two main kinase signaling pathways, RAS-MAPK and PI3K-AKT, which include ERK1/2, PTEN and AKT(1–3). ASD is a common comorbidity in rare syndromes (e.g., Costello, Noonan, LEOPARD, and cardio-facio-cutaneous syndromes) caused by mutations in signaling molecules immediately upstream of ERK (MEK, RAF, SPRED1, and RAS) [32, 33]. Similarly, ERK is hyperphosphorylated in Fragile X syndrome patients [34] and implicated in biochemical dysregulation of tuberous sclerosis, another childhood syndrome associated with ASD [35]. In tuberous sclerosis, the TSC1 and TSC2 heterodimeric complex regulates the signaling pathway from the kinase AKT to mTOR and downstream protein synthesis, and this pathway has been implicated in ASD [36]. Similarly, PTEN and PI3K are upstream of these signaling molecules and there is increasing evidence linking both to ASD [37, 38].
To better understand the biological basis of social trait variation in primates, here, we measured mSRS scores in a sample of socially housed rhesus monkeys using a refined and highly reliable version of the mSRS [21], as described below. We next evaluated the trait-like consistency of various biological “readouts” of the AVP, OXT, RAS-MAPK, and PI3K-AKT signaling pathways. Finally, we tested whether any of the reliable biological measures could robustly predict quantitative social trait variation in this species.
Methods
Study design
A detailed study timeline is provided in Table 1. Briefly, behavioral observations were conducted over a 15-month period on N = 76 rhesus monkeys. Subjects were studied in two cohorts to accommodate project workload, and to ensure that all behavioral observations were conducted during the non-breeding season. (Restricting behavioral observations to the non-breeding season minimizes the potential impact of seasonal changes in macaque social behavior on mSRS score ascertainment.) Cerebrospinal fluid (CSF) and blood samples were collected from all N = 76 subjects. In a subset of subjects (n = 43), biological samples were collected thrice over a 10-month period. This enabled us to assess the trait-like consistency of our CSF and blood biological measures across multiple time points in a manner not readily achievable in human participants. Collecting biological samples over much of the year also enabled us to examine the stability of these biological measures across the breeding and non-breeding seasons. Finally, experimenters were blinded to monkeys’ mSRS scores during biological sample collection and quantification.
Subjects and study site
Subjects were N = 76 male rhesus monkeys (Macaca mulatta), born and reared at the California National Primate Research Center (CNPRC). All subjects lived in mixed age and sex groups of 58 to 145 individuals, with 98.22 ± 24.14 (Mean ± SD) animals per group. Groups comprised 2–18 matrilines (Median: 14 matrilines per group). Each group was housed in a large, outdoor, half-acre (0.19 ha) field corral (30.5 m wide X 61 m deep X 9 m high). Subjects were housed among 16 of the 24 field corrals.
Subjects were tattooed as infants and dye-marked periodically to facilitate easy identification for husbandry- and research-related procedures. Monkeys had ad libitum access to Lixit-dispensed water. Primate laboratory chow was provided twice daily, and fruit and vegetable supplements were provided once weekly. Various toys, swinging perches, and other forms of enrichment in each corral, along with outdoor and social housing, provided a stimulating environment.
Subjects were 3.73 ± 1.17 (Mean ± SD) years old and ranged in age from 1.25 to 6.27 years at the time of study enrollment. Subjects underwent quantitative behavioral data collection as part of a larger research program focused on the biology of macaque social functioning [21]. Eligibility criteria for the parent investigation included: male, 1–7 years of age, socially housed in any of the outdoor field corrals (i.e., not housed indoors in individual cages), medically healthy, and not simultaneously enrolled in another CNPRC project. Preference for study inclusion was given to animals that had been tested as infants in the CNPRC BioBehavioral Assessment Program and which were housed in a field corral with a minimum of five other eligible animals.
Rank was assessed in each corral by behavioral management personnel who recorded aggressive and submissive interactions following food provisioning. Because each corral contained a different number of males, using the raw rank was ineffective as a direct measure that could be compared across all subjects. Thus, rank was calculated as the proportion of males in the group that the focal individual outranked, such that the highest-ranked individual had a value of 1 and the lowest-ranked individual had a value of 0 [39]. Ranks are assessed monthly at CNPRC; thus, we used monkeys’ ranks collected contemporaneously with their mSRS ratings for the purpose of statistical analysis.
Behavioral data collection
Subjects were observed unobtrusively in their home field corrals. Inter-observer reliabilities of > 85% agreement were established prior to commencing experimental data collection. Each animal was observed for two 10-min focal samples per day (0800–1030 and 1045–1300) over a 2-week period (called a “biweek”). A maximum of eight subjects (range: six to eight), residing in one or two corrals, were observed per biweek per observer. Behavior was recorded at 15-s intervals using instantaneous sampling and time-ruled check-sheets [40]. At the end of a biweek (at least 1 h after the final observation was concluded and no more than 24 h after the last observation), observers rated each subject on the 36-item original mSRS [20], which we modified from a four-point to a seven-point Likert scale (1 = total absence of the trait, 7 = extreme manifestation of the trait) for each item [21]. Prior to final summary, questions written in the infrequent direction were reverse scored such that higher scores always indicated greater impairment. Since only 17 of the original 36 mSRS items exhibit inter-rater and test-rest reliability [21], here we extracted and tabulated ratings for the 17 reliable items, which form the basis of the mSRS-Revised (mSRS-R) [21]. Final summed mSRS-R total scores can range between 17 and 119. Please see Additional file 1 for the mSRS-R instrument.
Biological sample collection and processing procedures
Subjects in the present study underwent biological sample collection as part of the larger research program. CSF and blood samples were first collected 79.37 ± 33.42 days (range: 21 to 157 days) after mSRS-R score ascertainment in all N = 76 monkeys. A complete biological measurement set was available from n = 57 of these subjects. A further subset of these monkeys (n = 43) underwent sample collection at two additional time points to enable determination of within-individual consistency in the biological measures for the present study. Sampling procedures were the same as those employed in our prior research [41], and identical for each sample collection time point.
Samples were collected between 0800 and 1100 to minimize any potential circadian effects on the biological measurements. Collection of both CSF and blood samples was accomplished within 10–15 min of initial cage entry; only one monkey per day was sampled from the same corral. Briefly, each subject was captured from his home corral, rapidly immobilized with telazol (5–8 mg/kg), and moved to an indoor procedure room. Supplementary ketamine (5–8 mg/kg) was used as needed to facilitate complete immobilization. Immediately following relocation, CSF (2 mL) was drawn from the cisterna magna using standard sterile procedure. CSF samples were immediately aliquoted into 1.5-mL siliconized polypropylene tubes and flash-frozen on dry ice.
Next, whole blood samples (up to 25 mL) were drawn from the femoral vein. Blood was collected at room temperature into EDTA-treated vacutainer tubes for kinase quantification. Samples were spun over a Ficoll-hypaque gradient and mononuclear cells collected from the interface were washed in PBS 2x, pelleted, and solubilized (in 50 mM Tris pH 7.4, 10 mM EGTA, 0.5% NP-40 and protease and phosphatase inhibitor cocktails). Samples were spun to remove insoluable material and then aliquoted. Whole blood was also collected into PAXgene tubes for neuropeptide receptor gene expression. Samples were left at room temperature for 2 h, subsequently transferred to − 20 °C for 24 h, and then transferred to − 80 °C per manufacturer’s guidelines. All biological samples were stored at − 80 °C until quantification.
After sample collection, each subject was prophylactically administered metoclopramide and ketoprofen. Additionally, replacement fluids were given if needed per veterinary guidelines. Subjects were placed in a standard laboratory cage located in a hospital/transition room for recovery overnight, and then returned to their home corrals the next day.
CSF neuropeptide quantification
CSF OXT and AVP concentrations were quantified using commercially available enzyme immunoassay kits (Enzo Life Sciences, Farmingdale, NY) [41,42,43,44]. These kits have been validated for use in rhesus monkeys and are highly specific and exclusively recognize OXT and AVP, respectively, and not related peptides (i.e., the OXT cross-reactivity with AVP is < 0.02% and the minimum assay sensitivity is 15 pg/mL; and the AVP cross-reactivity with OXT is < 0.001% and the minimum assay sensitivity is 2.84 pg/mL). A research scientist performed sample preparation and OXT and AVP quantification following established procedures. CSF samples were directly assayed (without prior extraction) for OXT and AVP. All CSF samples were assayed in duplicate (100 µL per well) with a tunable microplate reader for 96-well format per manufacturer’s instructions. The intra-assay and inter-assay coefficients of variation were 7.75% and 11.78%, respectively, for OXT. The intra-assay and inter-assay coefficients of variation were 7.70% and 14.86%, respectively, for AVP.
Blood neuropeptide receptor quantification
Measurement of OXTR and AVPR1A mRNA levels was conducted using protocols developed for rhesus monkeys [41]. Total RNA was isolated and purified using a PAXgene blood RNA kit from blood stabilized in PAXgene RNA tubes (Qiagen, CA). The first strand cDNA synthesis reaction was carried out with iScript Reverse Transcription Supermix (Bio-Rad, CA) with a starting RNA quantity of 1 µg in a 20 µl final volume. qPCR was performed to determine gene expression levels of OXTR and AVPR1A using RT2 qPCR Primer Assays for Rhesus Macaque OXTR and AVPR1A (Qiagen, CA), and endogenous control (GAPDH, Life Technologies, CA) was used for normalization. qPCR was performed on the QuantStudio 3 Real-Time PCR System (Applied Biosystems, CA) with SYBR Green (Qiagen, CA). cDNA was PCR amplified in triplicate, and Ct values from each sample were obtained using QuantStudio 3 qPCR software. Analyses were conducted using the comparative Ct method (2−ΔΔCt) [45].
Blood kinase signaling quantification
Blood kinase activation levels were determined using previously published protocols [46], which had been optimized for use in rhesus monkeys [41]. Denatured protein (20 µg per lane) from the previously generated soluble cytoplasmic fraction was electrophoresed through an 8–12% polyacrylamide gradient gel for 2 h at 100 V at room temperature. The gel was electroblotted onto a PVDF membrane for 1 h at 50 V at 4 °C. After blocking, the blot was incubated with primary antibody (Ras, MEK1/2, Phospho-MEK1/2, Erk1/2, Phospho-Erk1/2, Pan AKT and phospho-AKT, PTEN and phospho-PTEN, and GAPDH, Cell Signaling, MA) overnight at 4 °C, washed and followed by fluorescent secondary antibody (Anti-Rabbit IgG, Cell Signaling, MA) incubation for 1 h at room temperature. The blots were visualized using the LI-COR Odyssey Imager (LI-COR Biosciences, NE).
Statistical analyses
Data were analyzed using JMP Pro 14 (SAS Institute Inc., Cary, NC). We first calculated the test–retest reliability of the neuropeptide and kinase signaling pathway data as a “Case 2A” intra-class correlation coefficient of consistency estimated by a Restricted Maximum Likelihood Mixed Model following [47]. This yields an effect size equivalent to a correlation coefficient [47]. As noted above, three samples were collected from a subset of subjects for this purpose. Subjects were included in this analysis as long as they had at least 2 out of 3 useable time points per biological measure represented in the data; all n = 43 monkeys met this inclusion threshold.
We next performed principal component analysis (PCA) to better understand potential collinearities between the reliable biological measures and their relationship to mSRS-R score. This is critical to prevent collinearities from producing false negatives (or less likely, false positives) in our final General Linear Model (GLM) analysis (see below). This approach is an example of best-practice “sensitivity analysis” [48]. Since almost all of the biological measures exhibited high test–retest reliability, we were able to assess n = 57 subjects which had a complete set of biological measurements at the first collection time point. Eigenvalues and scree plots suggested a 3-component solution, accounting for 69.1% of the total variance. This was extracted by varimax rotation for principal components. PCA loadings are effect sizes equivalent to the correlation coefficient between the component and the variable [49].
This PCA suggested that CSF AVP concentration and differential neuropeptide receptor gene expression might predict mSRS-R score, whereas the other biological measures appeared unrelated (as assessed by variable loadings). In order to test this hypothesis, we performed a GLM, predicting mSRS-R score, given the reliable biological measures. The analysis controlled for corral, cohort, age, and rank. Blood OXTR and AVPR1A gene expression were highly correlated, and collinear with total and differential neuropeptide receptor gene expression. We therefore included the total neuropeptide receptor gene expression as the sum of the OXTR and AVPR1A gene expression to capture correlated expression of the two genes, and differential neuropeptide receptor gene expression as the difference between OXTR and AVPR1A gene expression to capture relative up or down regulation of these receptors. All of the other reliable biological measures were included in the analysis. This approach conservatively tests each biological measure controlling for all others, and thus identifies variables driving a relationship while excluding variables that are only secondarily correlated. The assumptions of GLM (normality of error, homogeneity of variance, and linearity) were tested graphically. No transformations were required. As noted above, n = 57 subjects had complete biological sample sets available at the first collection time point for analysis. However, CSF AVP concentration had been quantified on all N = 76 subjects, and an AVP value was available from n = 75 of them. This enabled us to run a follow-up analysis in the entire study sample excluding all biological measures except CSF AVP concentration, but including all of the aforementioned blocking factors, in the model.
The advanced methods employed here greatly increase power and reduce sample size (often by 5- or 10-fold) over analyses such as T-Tests or simple regression, while simultaneously also reducing false positives [50,51,52,53,54]. However, no formal a priori power calculations similar to those used for simpler analyses exist for the analyses employed here. As we and others have argued [50, 55, 56], the best approach to a priori power calculations for advanced analyses is Mead’s Resource Equation [57]. Mead’s Resource Equation provides a sample size above which additional subjects will have little impact on power; this size was smaller than the number of subjects we had available for each analysis. We therefore opted to use all subjects for which we had data, noting that these analyses are all suitably powered. Finally, effect sizes for these GLM analyses are reported as partial eta squared (ηp2), which is the square of the partial correlation coefficient rp for continuous predictors, following best practice for GLMs [50].
Results
Autism-associated biological measures demonstrate robust test–retest reliability
All neuropeptide and kinase signaling measures (except phosphorylated AKT/total AKT) showed significant test–retest reliability within individuals across multiple collection time points (Table 2). The test–retest reliability intra-class correlation coefficients ranged from 42.0 to 88.6% for the reliable biological measures. These measurements spanned a 10-month collection period (inclusive of the breeding and non-breeding seasons) and indicate that circannual changes in breeding behavior do not significantly impact the consistency of these measures.
Continuous distribution of mSRS-R scores in the general monkey population
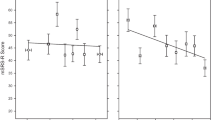
Scores on the mSRS-R in this sample ranged from 26 to 88. As observed in past research studies of SRS score distribution in the general human population [10, 58], mSRS-R scores in this sample of rhesus monkeys were likewise continuously distributed across the general population. As also observed in human populations [9, 59], mSRS-R scores were skewed from a normal distribution, indicative of more severe impairment being relatively rare in this monkey species (Fig. 1a). (We note that these mSRS-R data reflect a subset of animals from a larger behavioral study [21], and thus, do not constitute an independent replication sample.)
macaque Social Responsiveness Scale-Revised (mSRS-R) scores and cerebrospinal fluid (CSF) arginine vasopressin (AVP) concentration in male rhesus macaques from the general population. a mSRS-R scores are plotted as a histogram (N = 76), with higher scores indicating greater social impairment. Total mSRS-R scores could range from 17 to 119. A corresponding box-and-whisker plot is shown above the histogram. The box shows the inter-quartile range, the line the median, the whiskers the 10th and 90th percentiles, and the black circles any data out of this range. b CSF AVP concentration negatively predicts mSRS-R score (n = 57), such that individuals with the lowest CSF AVP concentration demonstrate the greatest social impairment. The observed mSRS-R score is plotted against the partialled CSF AVP concentration. Thus, CSF AVP concentration is corrected (partialled) for the other variables in the analysis and scaled to the range of the original CSF AVP concentration data
A principal component loads mSRS-R score and a subset of autism-associated biological measures
PCA extracted a three-component solution. Only one component loaded mSRS-R score and did so along with two biological measures: CSF AVP concentration and blood differential neuropeptide receptor gene expression. The other biological measures loaded onto two separate components (Table 3).
CSF AVP concentration predicts quantitative social trait variation in the general monkey population
We next used GLM to test the contribution of the biological measures to mSRS-R score. Only CSF AVP concentration significantly predicted mSRS-R score, with lower CSF AVP concentrations predicting higher mSRS-R scores (F1,34 = 6.629; P = 0.0146; ηp2 = 16.3%; rp = − 40.4%; Fig. 1b). We then confirmed this result by re-running the analysis, but excluding the other biological measures, thereby enabling us to include a greater number of subjects with available CSF AVP values. The result held in the larger study sample (F1,55 = 4.555; P = 0.0373; ηp2 = 7.65%; rp = − 27.7%).
Discussion
This study investigated potential biomarkers of quantitative social trait variation in male rhesus monkeys from the general CNPRC population. All biological measures (except AKT) showed significant test–retest reliability within individuals across time points. PCA revealed a potential association between CSF AVP concentration and mSRS-R score; this relationship was subsequently confirmed by GLM (which also indicated that none of the covariates including age and rank, affected mSRS-R scores). Specifically, CSF AVP concentration, but no other biological measure that we tested, robustly predicted individual differences in mSRS-R score, such that monkeys with the lowest CSF AVP concentration exhibited the greatest social impairment. These findings buttress those of a prior study reporting stable within-individual consistency of CSF AVP concentration in a modest sample of N = 10 monkeys [41], and add to growing evidence linking individual differences in CSF AVP concentration to individual differences in primate social functioning, including grooming behavior in macaques [41] and Autism Diagnostic Observation Schedule Calibrated Severity Scores in people with ASD [42].
It has been known for nearly three decades that AVP signaling plays a critical role in mammalian prosocial behavior. AVP was first implicated in social bond formation and paternal care in voles [28, 60, 61]. Subsequently, experimental dysregulation of the brain AVP signaling pathway was shown to produce social deficits in mice and voles [24, 28, 29]. Although far less is known about the biology of social functioning in primates, we have shown that naturally low-social adult monkeys, which initiate fewer affiliative interactions, spend less time in physical contact and grooming with conspecifics, and show appreciable social information processing abnormalities (e.g., face recognition deficits and impaired species-typical gaze aversion in response to conspecific aggression) [41, 62,63,64], exhibit lower CSF AVP concentrations compared to their socially competent peers [41]. Natural silencing of AVP gene activity from birth is also associated with robust social developmental deficits in Brattleboro rats [65, 66], suggesting the intriguing possibility that early impairment in brain AVP signaling may similarly contribute to the pathogenesis of social deficits in young humans.
These findings implicating AVP in mammalian prosocial functioning led us recently to investigate whether CSF AVP concentration is lower in ASD cases vs. controls. This was indeed the case, as in two independent ASD cohorts, CSF AVP concentration accurately differentiated pediatric ASD cases and controls (aged 1.5–19 years). Specifically, across the range of CSF AVP concentrations, the likelihood of ASD increases over 1,000-fold, corresponding to nearly a 500-fold increase in risk with each 10-fold decrease in CSF AVP concentration [41, 42]. We also recently tested the predictive value of AVP by mining over 900 banked CSF samples collected during standard of care from 0 to 3-month-old human newborns. We found that individuals diagnosed with ASD later in childhood have significantly lower neonatal CSF AVP concentrations compared to those who do not later receive an ASD diagnosis [43]. These collective findings suggest that a neurochemical marker of impaired social functioning may be present very early in life, many months or even years, before behavioral symptoms first emerge.
It is well established that ASD is a principally polygenic inherited brain disorder [67], and approximately 100 ASD susceptibility genes have now been identified [68]. Interestingly, AVP itself has not been identified as a high confidence ASD risk gene by SFARI gene, a comprehensive database that catalogues and scores all known human genes for ASD susceptibility [69]. How then might we reconcile findings on AVP’s role in mammalian prosocial functioning [28, 60, 70] with well documented evidence that endophenotypic autistic traits are common, highly heritable, and continuously distributed across the general human population [58, 71]? We suggest that brain AVP signaling may be a downstream pathway critical for the expression of ASD symptoms, and a point of convergence for multiple and diverse ASD susceptibility genes. This would explain our findings linking variation in CSF AVP concentration to variation in grooming behavior in monkeys [41] and clinical symptom severity in ASD patients [42]. This would also explain why intranasal AVP treatment improves social abilities in idiopathic ASD patients, as we recently reported in a double-blind, randomized, placebo-controlled phase 2a pilot trial [72].
Limitations
This study had several limitations that warrant comment. First, although we used a sophisticated primate model that enabled reverse-translation of a scale that measures social and autistic trait variation in humans, we acknowledge that animal models nevertheless are approximations for examining human neurodevelopmental disease. Second, the relationship between CSF AVP concentration and mSRS-R score in this study was correlational, not causal. Moreover, measurement of AVP in cisternal CSF precluded a more mechanistic understanding of AVP’s role in primate social functioning. Future research using viral vector, optogenetic, gene editing, and/or positron emission tomography radiotracer tools will be required to address this issue in a more tractable manner. Third, unlike the biological measures in this study which were assessed three times, mSRS-R scores were only assessed once per subject, using a relatively new instrument. However, we previously assessed the psychometric properties of the 36-item mSRS, and in the process omitted all items that did not show robust test–retest and inter-rater reliability, resulting in the highly reliable 17-item mSRS-R. We also note that mSRS-R scores are closely related to multiple other measures of social functioning [21]. Fourth, although we assessed biological measures that had been previously implicated in prosocial behavior, ASD, and/or ASD-related syndromes, we did not observe relationships between CSF OXT concentration or blood kinase signaling and social trait variation here. This is in contrast to prior research in humans that has linked neonatal CSF OXT concentration to later social engagement [73], and blood kinase signaling to ASD and its symptom severity [74,75,76]. However, absence of evidence implicating these biological measures in monkey social functioning is not necessarily evidence of absence. This may be particularly true for the blood kinase signaling measures, as the present study did not capture the theoretical extreme of mSRS-R scores (i.e., the highest mSRS-R score here was an 88 out of a possible 119), and the distribution of scores was statistically skewed toward animals with less social impairment. It therefore remains possible that the relationship between blood kinase signaling and social behavior variation is only evident when studied in a sample of the most socially impaired monkeys. Finally, this study was restricted to male subjects, due to our broader research interest in better understanding the biological underpinnings of ASD, which impacts four times as many males as females [77]. Nevertheless, ASD does affect girls, and it remains to be determined whether the relationship between CSF AVP concentration and social trait variation holds in female subjects.
Conclusions
In conclusion, this study marked a first step toward better understanding the biological basis of quantitative social trait variation in primates. Findings from this study established the trait-like consistency of nearly all of the biological “readouts” from the AVP, OXT, RAS-MAPK, and PI3K-AKT signaling pathways in rhesus monkeys. Finally, CSF AVP concentration robustly predicted quantitative social trait variation in this species. Research is now required to better understand the role of the AVP signaling pathway in species typical and atypical primate social functioning, and in human primates, AVP’s involvement at the extreme of the social trait distribution, particularly in the pathogenesis of ASD.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AAALAC International:
-
Association for Assessment and Accreditation of Laboratory Animal Care International
- ASD:
-
Autism spectrum disorder
- AVP:
-
Arginine vasopressin
- CNPRC:
-
California National Primate Research Center
- CSF:
-
Cerebrospinal fluid
- GLM:
-
General linear model
- IACUCs:
-
Institutional Animal Care and Use Committees
- ICC:
-
Intra-class correlation coefficient
- mSRS:
-
Macaque Social Responsiveness Scale
- mSRS-R:
-
Macaque Social Responsiveness Scale-Revised
- OXT:
-
Oxytocin
- PC:
-
Principal component
- PCA:
-
Principal component analysis
- SD:
-
Standard deviations
- SE:
-
Standard error
- SRS:
-
Social Responsiveness Scale
- UCSF:
-
University of California, San Francisco
References
Sussman RW. The origins and nature of sociality. London: Routledge; 2017. p. 353.
Talbot CF. Ability to recognize individuals. In: Weekes-Shackelford V, Shackelford TK, Weekes-Shackelford VA, editors. Encyclopedia of evolutionary psychological science. New York: Springer; 2016. p. 1–9. https://doi.org/10.1007/978-3-319-16999-6_1253-1.
Phillips KA, Bales KL, Capitanio JP, Conley A, Czoty PW, Hart BA, et al. Why primate models matter. Am J Primatol. 2014;76(9):801–27.
Clark AB, Ehlinger TJ. Pattern and adaptation in individual behavioral differences. In: Bateson PPG, Klopfer PH, editors. Perspectives in ethology: volume 7 alternatives. New York: Springer; 1987. p. 1–47. https://doi.org/10.1007/978-1-4613-1815-6_1.
Talbot CF, Maness AC, Capitanio JP, Parker KJ. The factor structure of the macaque social responsiveness scale-revised predicts social behavior and personality dimensions. Am J Primatol. 2021;e23234.
Constantino J, Gruber C. Social responsive scale (SRS) manual. Los Angeles: Western Psychological Services; 2005.
Constantino JN, Gruber CP. Social responsiveness scale: SRS-2. Los Angeles: Western Psychological Services; 2012.
Constantino JN, Davis SA, Todd RD, Schindler MK, Gross MM, Brophy SL, et al. Validation of a brief quantitative measure of autistic traits: comparison of the social responsiveness scale with the autism diagnostic interview-revised. J Autism Dev Disord. 2003;33(4):427–33.
Constantino JN, Lavesser PD, Zhang Y, Abbacchi AM, Gray T, Todd RD. Rapid quantitative assessment of autistic social impairment by classroom teachers. J Am Acad Child Adolesc Psychiatry. 2007;46(12):1668–76.
Constantino JN. The quantitative nature of autistic social impairment. Pediatr Res. 2011;69(5 Pt 2):55R-62R.
Pine E, Luby J, Abbacchi A, Constantino JN. Quantitative assessment of autistic symptomatology in preschoolers. Autism. 2006;10(4):344–52.
American Psychiatric Association. Diagnostic and statistical manual of mental disorders: diagnostic criteria for autistic disorder, 5th edition. 2013. https://doi.org/10.5555/appi.books.9780890425596.x00pre.
Chan W, Smith LE, Hong J, Greenberg JS, Mailick MR. Validating the social responsiveness scale for adults with autism. Autism Res. 2017;10(10):1663–71.
Bölte S, Poustka F, Constantino JN. Assessing autistic traits: cross-cultural validation of the social responsiveness scale (SRS). Autism Res. 2008;1(6):354–63.
Stickley A, Tachibana Y, Hashimoto K, Haraguchi H, Miyake A, Morokuma S, et al. Assessment of autistic traits in children aged 2 to 4 1/2 years with the preschool version of the social responsiveness scale (SRS-P): findings from Japan. Autism Res. 2017;10(5):852–65.
Wigham S, McConachie H, Tandos J, Le Couteur AS. Gateshead Millennium Study core team. The reliability and validity of the Social Responsiveness Scale in a UK general child population. Res Dev Disabil. 2012;33(3):944–50.
Jussila K, Kuusikko-Gauffin S, Mattila M-L, Loukusa S, Pollock-Wurman R, Joskitt L, et al. Cross-cultural differences in the Parent Rated Social Responsiveness Scale (SRS)? Evaluation of the Finnish version among high-functioning school aged males with and without autism spectrum disorder. Res Autism Spectrum Disorders. 2015;9:38–44.
Faughn C, Marrus N, Shuman J, Ross SR, Constantino JN, Pruett JR, et al. Brief report: chimpanzee social responsiveness scale (CSRS) detects individual variation in social responsiveness for captive chimpanzees. J Autism Dev Disord. 2015;45(5):1483–8.
Marrus N, Faughn C, Shuman J, Petersen SE, Constantino JN, Povinelli DJ, et al. Initial description of a quantitative, cross-species (chimpanzee-human) social responsiveness measure. J Am Acad Child Adolesc Psychiatry. 2011;50(5):508–18.
Feczko EJ, Bliss-Moreau E, Walum H, Pruett JR, Parr LA. The Macaque Social Responsiveness Scale (mSRS): a rapid screening tool for assessing variability in the social responsiveness of rhesus monkeys (Macaca mulatta). PLoS ONE. 2016;11(1):e0145956.
Talbot CF, Garner JP, Maness AC, McCowan B, Capitanio JP, Parker KJ. A psychometrically robust screening tool to rapidly identify socially impaired monkeys in the general population. Autism Res. 2020;13(9):1465–75.
Rogers FD, Bales KL. Mothers, fathers, and others: neural substrates of parental care. Trends Neurosci. 2019;42(8):552–62.
Baribeau DA, Anagnostou E. Oxytocin and vasopressin: linking pituitary neuropeptides and their receptors to social neurocircuits. Front Neurosci. 2015;9:335.
Bielsky IF, Hu S-B, Szegda KL, Westphal H, Young LJ. Profound impairment in social recognition and reduction in anxiety-like behavior in vasopressin V1a receptor knockout mice. Neuropsychopharmacology. 2004;29(3):483–93.
Takayanagi Y, Yoshida M, Bielsky IF, Ross HE, Kawamata M, Onaka T, et al. Pervasive social deficits, but normal parturition, in oxytocin receptor-deficient mice. Proc Natl Acad Sci USA. 2005;102(44):16096–101.
Insel TR. The challenge of translation in social neuroscience: a review of oxytocin, vasopressin, and affiliative behavior. Neuron. 2010;65(6):768–79.
Donaldson ZR, Young LJ. Oxytocin, vasopressin, and the neurogenetics of sociality. Science. 2008;322(5903):900–4.
Parker KJ, Lee TM. Central vasopressin administration regulates the onset of facultative paternal behavior in microtus pennsylvanicus (meadow voles). Horm Behav. 2001;39(4):285–94.
Donaldson ZR, Spiegel L, Young LJ. Central vasopressin V1a receptor activation is independently necessary for both partner preference formation and expression in socially monogamous male prairie voles. Behav Neurosci. 2010;124(1):159–63.
Peñagarikano O, Lázaro MT, Lu X-H, Gordon A, Dong H, Lam HA, et al. Exogenous and evoked oxytocin restores social behavior in the Cntnap2 mouse model of autism. Sci Transl Med. 2015;7(271):271ra8.
Francis SM, Sagar A, Levin-Decanini T, Liu W, Carter CS, Jacob S. Oxytocin and vasopressin systems in genetic syndromes and neurodevelopmental disorders. Brain Res. 2014;1580:199–218.
Samuels IS, Saitta SC, Landreth GE. MAP’ing CNS development and cognition: an ERKsome process. Neuron. 2009;61(2):160–7.
Vithayathil J, Pucilowska J, Landreth GE. ERK/MAPK signaling and autism spectrum disorders. Prog Brain Res. 2018;241:63–112.
Michalon A, Sidorov M, Ballard TM, Ozmen L, Spooren W, Wettstein JG, et al. Chronic pharmacological mGlu5 inhibition corrects fragile X in adult mice. Neuron. 2012;74(1):49–56.
Winter JN, Jefferson LS, Kimball SR. ERK and Akt signaling pathways function through parallel mechanisms to promote mTORC1 signaling. Am J Physiol Cell Physiol. 2011;300(5):C1172-1180.
Numis AL, Major P, Montenegro MA, Muzykewicz DA, Pulsifer MB, Thiele EA. Identification of risk factors for autism spectrum disorders in tuberous sclerosis complex. Neurology. 2011;76(11):981–7.
Levitt P, Campbell DB. The genetic and neurobiologic compass points toward common signaling dysfunctions in autism spectrum disorders. J Clin Invest. 2009;119(4):747–54.
Rademacher S, Eickholt BJ. PTEN in autism and neurodevelopmental disorders. Cold Spring Harb Perspect Med. 2019;9(11).
Linden JB, McCowan B, Capitanio JP, Isbell LA. Male-inflicted wounds have opposite effects on hair cortisol for captive male and female rhesus macaques (Macaca mulatta) following new group formation. Primates. 2019;60(1):51–62.
Altmann J. Observational study of behavior: sampling methods. Behaviour. 1974;49(3):227–67.
Parker KJ, Garner JP, Oztan O, Tarara ER, Li J, Sclafani V, et al. Arginine vasopressin in cerebrospinal fluid is a marker of sociality in nonhuman primates. Sci Transl Med. 2018;10(439).
Oztan O, Garner JP, Partap S, Sherr EH, Hardan AY, Farmer C, et al. Cerebrospinal fluid vasopressin and symptom severity in children with autism. Ann Neurol. 2018;84(4):611–5.
Oztan O, Garner JP, Constantino JN, Parker KJ. Neonatal CSF vasopressin concentration predicts later medical record diagnoses of autism spectrum disorder. Proc Natl Acad Sci USA. 2020;117(19):10609–13.
Carson DS, Garner JP, Hyde SA, Libove RA, Berquist SW, Hornbeak KB, et al. Arginine vasopressin is a blood-based biomarker of social functioning in children with autism. PLoS ONE. 2015;10(7):e0132224.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–8.
Faridar A, Jones-Davis D, Rider E, Li J, Gobius I, Morcom L, et al. Mapk/Erk activation in an animal model of social deficits shows a possible link to autism. Mol Autism. 2014;5:57.
McGraw KO, Wong SP. Forming inferences about some intraclass correlation coefficients. Psychol Methods. 1996;1(1):30–46.
Thabane L, Mbuagbaw L, Zhang S, Samaan Z, Marcucci M, Ye C, et al. A tutorial on sensitivity analyses in clinical trials: the what, why, when and how. BMC Med Res Methodol. 2013;13:92.
Howitt D, Cramer D. Introduction to statistics in psychology. London: Pearson Education; 2007.
Gaskill BN, Garner JP. Power to the people: power, negative results and sample size. J Am Assoc Lab Anim Sci. 2020;59(1):9–16.
Grafen A, Hails R. Modern statistics for the life sciences. Oxford: Oxford University Press; 2002.
Garner JP. The significance of meaning: why do over 90% of behavioral neuroscience results fail to translate to humans, and what can we do to fix it? ILAR J. 2014;55(3):438–56.
Richter SH, Garner JP, Auer C, Kunert J, Würbel H. Systematic variation improves reproducibility of animal experiments. Nat Methods. 2010;7(3):167–8.
Richter SH, Garner JP, Würbel H. Environmental standardization: cure or cause of poor reproducibility in animal experiments? Nat Methods. 2009;6(4):257–61.
Festing MFW. On determining sample size in experiments involving laboratory animals. Lab Anim. 2018;52(4):341–50.
Festing MFW. Randomized block experimental designs can increase the power and reproducibility of laboratory animal experiments. ILAR J. 2014;55(3):472–6.
Mead R. The design of experiments: statistical principles for practical applications. New York, NY, US: Cambridge University Press; 1988. xiv, 620 p. (The design of experiments: Statistical principles for practical applications).
Constantino JN, Todd RD. Autistic traits in the general population: a twin study. Arch Gen Psychiatry. 2003;60(5):524–30.
Kamio Y, Inada N, Moriwaki A, Kuroda M, Koyama T, Tsujii H, et al. Quantitative autistic traits ascertained in a national survey of 22 529 Japanese schoolchildren. Acta Psychiatr Scand. 2013;128(1):45–53.
Winslow JT, Hastings N, Carter CS, Harbaugh CR, Insel TR. A role for central vasopressin in pair bonding in monogamous prairie voles. Nature. 1993;365(6446):545–8.
Wang Z, Young LJ, De Vries GJ, Insel TR. Voles and vasopressin: a review of molecular, cellular, and behavioral studies of pair bonding and paternal behaviors. Prog Brain Res. 1998;119:483–99.
Capitanio JP. Personality dimensions in adult male rhesus macaques: prediction of behaviors across time and situation. Am J Primatol. 1999;47(4):299–320.
Capitanio JP. Sociability and responses to video playbacks in adult male rhesus monkeys (Macaca mulatta). Primates. 2002;43(3):169–77.
Sclafani V, Del Rosso LA, Seil SK, Calonder LA, Madrid JE, Bone KJ, et al. Early predictors of impaired social functioning in male rhesus macaques (Macaca mulatta). PLoS ONE. 2016;11(10):e0165401.
Paul MJ, Peters NV, Holder MK, Kim AM, Whylings J, Terranova JI, et al. Atypical social development in vasopressin-deficient brattleboro rats. eNeuro. 2016;3(2).
Schank JC. Early locomotor and social effects in vasopressin deficient neonatal rats. Behav Brain Res. 2009;197(1):166–77.
Iakoucheva LM, Muotri AR, Sebat J. Getting to the cores of autism. Cell. 2019;178(6):1287–98.
Wang T, Hoekzema K, Vecchio D, Wu H, Sulovari A, Coe BP, et al. Large-scale targeted sequencing identifies risk genes for neurodevelopmental disorders. Nat Commun. 2020;11(1):4932.
SFARI Gene Archive AVP gene. https://gene-archive.sfari.org/database/human-gene/AVP. Accessed 22 Jan 2021.
Albers HE. The regulation of social recognition, social communication and aggression: vasopressin in the social behavior neural network. Horm Behav. 2012;61(3):283–92.
Baron-Cohen S, Wheelwright S, Skinner R, Martin J, Clubley E. The autism-spectrum quotient (AQ): evidence from Asperger syndrome/high-functioning autism, males and females, scientists and mathematicians. J Autism Dev Disord. 2001;31(1):5–17.
Parker KJ, Oztan O, Libove RA, Mohsin N, Karhson DS, Sumiyoshi RD, et al. A randomized placebo-controlled pilot trial shows that intranasal vasopressin improves social deficits in children with autism. Sci Transl Med. 2019;11(491).
Clark CL, St John N, Pasca AM, Hyde SA, Hornbeak K, Abramova M, et al. Neonatal CSF oxytocin levels are associated with parent report of infant soothability and sociability. Psychoneuroendocrinology. 2013;38(7):1208–12.
Argilli E, Berson T, Owen J, Thomas S, Sherr E. Toward the development of an ASD biomarker: altered activity of kinase signaling pathways in blood. International Meeting for Autism Research. 2017. https://insar.confex.com/insar/2017/webprogram/Paper25718.html.
Rosina E, Battan B, Siracusano M, Di Criscio L, Hollis F, Pacini L, et al. Disruption of mTOR and MAPK pathways correlates with severity in idiopathic autism. Transl Psychiatry. 2019;9(1):50.
Erickson CA, Ray B, Wink LK, Bayon BL, Pedapati EV, Shaffer R, et al. Initial analysis of peripheral lymphocytic extracellular signal related kinase activation in autism. J Psychiatr Res. 2017;84:153–60.
Maenner MJ. Prevalence of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 Sites, United States, 2016. MMWR Surveill Summ. 2020;69.
Acknowledgements
We thank JoAnn Yee, Josh Herrington, Alicia Bulleri, Kylee Beck, Jiang Li, Adam Myers, Jesus Madrid, Natalie Lange, and Sophie Rose for helping with various aspects of this research, and the CNPRC staff for maintaining the health and wellbeing of the animals.
Funding
This research was supported by the Simons Foundation (SFARI 342873), the National Institutes of Health (R01HD087048; P51OD011107), and the Stanford Department of Psychiatry and Behavioral Sciences. The funders had no role in study design, data collection, analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
OO, CFT, JPG, EHS, JPC, and KJP designed the study. KJP, JPC, JPG, and EHS secured funding for this research. KJP oversaw execution of the overall study. JPC oversaw execution of behavioral data and biological sample collection at CNPRC. LADR conducted behavioral reliabilities with personnel for outdoor field corral observations. LADR trained personnel on biological sample collection. OO trained personnel on biological sample processing. CFT, ACM, and SMS performed behavioral data collection. CFT, ACM, SMS, and LADR performed biological sample collection. KJP and EHS oversaw biological sample quantification at Stanford University and UCSF, respectively. OO, EA, and NM performed the biological sample preparation and/or quantification. OO, CFT, EA, ACM, SMS, LADR, and JPG performed data aggregation, data entry, and/or data cleaning. JPG performed the statistical analyses and created the tables and figure. KJP, OO, and JPG wrote the first draft of the manuscript. All authors read and approved the final manuscript.
Authors’ information
Our research team includes a biostatistician (JPG). JPG has long-standing expertise in advanced statistical models and power. He is also a leading authority on the reproducibility and translation of animal models.
Corresponding author
Ethics declarations
Ethical approval
All procedures were reviewed and approved by the Institutional Animal Care and Use Committees (IACUCs) at the University of California, Davis and Stanford University. These procedures complied with the Guide for the Care and Use of Laboratory Animals and National Institutes of Health policies on the care and use of animals. The CNPRC and University of California, Davis, are accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC International).
Consent for publication
Not applicable.
Competing interests
The Board of Trustees of the Leland Stanford Junior University and the regents of the University of California, San Francisco (UCSF) have filed patent applications related to biological measures studied herein (Stanford University: PCT/US2019/019029 “Methods for diagnosing and for determining severity of an autism spectrum disorder”; UCSF: PCT/US2016/014623 “Methods of diagnosing and treating autism spectrum disorders”). These patents have not been granted or licensed, and no study author is receiving any financial compensation at this time. EHS serves on the advisory board for Retrophin Inc. All other authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
. macaque Social Responsiveness Scale-Revised Instrument.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Oztan, O., Talbot, C.F., Argilli, E. et al. Autism-associated biomarkers: test–retest reliability and relationship to quantitative social trait variation in rhesus monkeys. Molecular Autism 12, 50 (2021). https://doi.org/10.1186/s13229-021-00442-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13229-021-00442-w