Abstract
Across metazoans, visual systems employ different types of photoreceptor neurons (PRs) to detect light. These include rhabdomeric PRs, which exist in distantly related phyla and possess an evolutionarily conserved phototransduction cascade. While the development of rhabdomeric PRs has been thoroughly studied in the fruit fly Drosophila melanogaster, we still know very little about how they form in other species. To investigate this question, we tested whether the transcription factor Glass, which is crucial for instructing rhabdomeric PR formation in Drosophila, may play a similar role in other metazoans. Glass homologues exist throughout the animal kingdom, indicating that this protein evolved prior to the metazoan radiation. Interestingly, our work indicates that glass is not expressed in rhabdomeric photoreceptors in the planarian Schmidtea mediterranea nor in the annelid Platynereis dumerilii. Combined with a comparative analysis of the Glass DNA-binding domain, our data suggest that the fate of rhabdomeric PRs is controlled by Glass-dependent and Glass-independent mechanisms in different animal clades.
Similar content being viewed by others
Introduction
Most animals can detect visual cues, which provide them with detailed information about their environment. This information may include the shape of nearby objects, colours, movements, or the day–night cycle, and it is relevant for surviving. As a consequence, animals have evolved various types of photoreceptor neurons (PRs) such as ciliary and rhabdomeric PRs [1, 2], which play different roles in different animal species. For instance, rhabdomeric PRs are critical for image-forming vision (e.g. in Drosophila compound eye PRs) and for identifying the direction of a light source (e.g. in the planarian Schmidtea mediterranea and in the annelid Platynereis dumerilii) [3,4,5]. Nevertheless, in the case of most metazoan clades, very little is known about how rhabdomeric PRs develop.
Interestingly, all known rhabdomeric PRs appear to use a similar assortment of phototransduction proteins. These PRs possess rhabdomeric-type opsins that can modify their spatial conformation upon light stimulation, which allows them to activate Gαq. Gαq signals through phospholipase C (PLC) causing the opening of cation channels on the cytoplasmic membrane of PRs and thus leading to the formation of action potentials. This light-sensing machinery is present in distantly related animal phyla [1, 6, 7], including vertebrates (due to the ‘intrinsically photosensitive retinal ganglion cells’, ipRGCs [8]), which poses the question of to what degree the development of rhabdomeric PRs is evolutionarily conserved. Is the acquisition of the rhabdomeric phototransduction cascade regulated by a similar set of transcription factors across metazoans? Using the fruit fly Drosophila melanogaster as a model system, we have recently shown that the zinc finger transcription factor Glass acts as critical cell fate selector by directing the maturation of PR precursors into adult, light-sensing PRs. In Drosophila, Glass is required for the expression of virtually all the phototransduction proteins [9], and it regulates the development of all types of rhabdomeric PRs (including those in the Bolwig organ, the ocelli, and the compound eye) [10,11,12]. Therefore, we investigated whether Glass may also be involved in rhabdomeric PR differentiation in other animal species.
The planarian Schmidtea and the annelid Platynereis are emerging model organisms whose visual systems have been well characterised [3, 5, 13,14,15,16,17,18,19]. Interestingly, by analysing recently published single-cell sequencing data of Schmidtea we found that glass is not expressed in rhabdomeric PRs in this species. Moreover, using in situ hybridisation we could not detect glass expression in rhabdomeric PRs in Platynereis. Thus, while Glass is critical for the specification of rhabdomeric PR identity in Drosophila, the absence of Glass in rhabdomeric PRs in Schmidtea and Platynereis supports that different genetic programmes are required for controlling rhabdomeric PR cell fate in different animal clades. Therefore, while the initial specification of the eye field appears to be controlled by an evolutionarily conserved group of transcription factors (called the retinal determination network, RDN) [17, 20, 21], the subsequent steps that diversify distinct cell types, including rhabdomeric PRs, are likely controlled by diverse developmental programmes.
Methods
Phylogenetic analysis
We searched for protein sequences similar to Drosophila Glass [22] and Platynereis Glass [23] (see sequences in Additional files 1 and 2) by using NCBI BLAST [24] and the Schmidtea mediterranea Genome Database [25]. Redundant sequences were removed from the collection using CD-HIT with an identity cut-off of 90% [26]. To obtain cluster maps based on all-against-all pairwise similarity, we used CLANS2 with the BLOSUM62 matrix and a p value cut-off of 1e−60 [27]. For phylogenetic tree construction, we selected a non-derived set of sequences from the glass cluster and aligned them with MUSCLE [28]. Sequences shorter than 300 amino acids were removed prior to the alignment. We trimmed the alignments with TrimAI in ‘Automated 1’ mode [29]. We identified the JTT + I + G4 model as the best by IQ-TREE [30]. Maximum likelihood trees and bootstrap analysis were performed with IQ-TREE. Trees were visualised with FigTree [31] (for the data corresponding to this analysis, see Additional file 3).
Glass-binding site analysis
We examined a subset of Glass-like protein sequences by aligning them with either BLAST [24] or MUSCLE [28], and analysed them with ‘DNA-binding site predictor for Cys2His2 Zinc Finger Proteins’ [32, 33] (for details on the sequences that we used, see Fig. 3 and Additional file 4). To investigate the DNA-binding specificity of each of these candidates, we copied its full amino acid sequence as input and asked the software to search for Cys2His2 domains [32]. Then, we predicted the binding sites for those regions that best aligned with the fourth and the fifth zinc fingers of Glass, which are responsible for recognising its targets in vivo [34,35,36,37]. We used ‘expanded linear SVM’ as prediction model.
Animal caretaking
Drosophila melanogaster stocks were cultured at 25 °C in a 12:12 h light–dark cycle, and we fed them with cornmeal medium (which was supplemented with molasses, fructose, and yeast). We used Canton-S as a wild-type strain (courtesy of R. Stocker), glass-Gal4 (courtesy of S. Kim) [38] and UAS-mCD8::RFP (Bloomington Stock Center, no. 32219).
Our wild-type P. dumerilii were a mixed population of worms, originally captured in the sea in Naples (Italy) and Arcachon (France). We also used r-opsin1-GFP worms (courtesy of F. Raible) [14]. These animals were kept in sea water at 22 °C, in a 16:8 h light–dark cycle. We maintained them synchronised to an artificial moon cycle, induced by slightly increasing the light intensity at night for 1 week per month (using a 10-W light bulb, to simulate the full moon). Platynereis had a varied diet that included Artemia salina, Tetraselmis marina, fish food, and spinach leaves. For our experiments (i.e. in situ hybridisation and microinjections), we crossed males and females and collected the fertilised eggs, as previously described [39]. The larvae that hatched from these eggs were kept at 18 °C.
Immunohistochemistry and in situ hybridisation
In the case of Drosophila antibody staining, these were performed on cryosections of glass > mCD8::RFP flies, as previously described [9, 40]. We dissected heads (removing the proboscis to improve the penetration of our reagents) and fixed them for 20 min with 3.7% formaldehyde in 0.01 M phosphate buffer (PB; pH 7.4). Then, we washed our samples with PBT (Triton X-100 0.3% in PB) and incubated them with a cryoprotectant solution (sucrose 25% in PB) overnight at 4 °C. The following day, we embedded the fly heads in OCT, froze them with liquid nitrogen, and cut 14-μm cryosections in the transverse plane. Once the samples were dry, we proceeded to immunostain them. For this, we washed the slides with PBT (this buffer was also used in subsequent washing steps) and incubated them in primary antibody (rabbit anti-DsRed, 1:100, Clontech, no. 632496) at 4 °C overnight. Then, we washed the cryosections and incubated them in secondary antibody (goat anti-rabbit conjugated to Alexa Fluor 568, 1:200, Molecular Probes, no. A-11011) at 4 °C overnight, and washed again the next day. We mounted our samples by using Vectashield that contained DAPI (Vector, H-1200) and took images with a Leica SP5 confocal microscope.
To detect the glass transcript in Drosophila, we used the ViewRNA in situ hybridisation kit of Affimetrix (no. QVT0012)—which is a proprietary method—and proceeded according to the instructions of the company. Briefly, we took head cryosections (as described above for antibody staining) and ordered a mix of labelled RNA probes against glass from Affimetrix. Then, we processed the samples by digesting them with protease QF, and washed with PB and with various commercial solutions included in the kit. We incubated our cryosections with the glass probes for 2 h, at 40 °C. After this, we continued with a series of washing and signal amplification steps, followed by a colour reaction. (We used Fast Red as a fluorophore.) We finished by washing the samples with PB, and used Vectashield containing DAPI (Vector, H-1200) to cover the slides. Imaging was done with a Leica SP5 confocal microscope.
To perform double in situ hybridisation in Platynereis, we followed—with few modifications—a protocol that has been previously used for characterising the expression pattern of r-opsin1 [16, 41]. For the present work, we also produced an RNA probe against the glass transcript (for details on the glass probe, see Additional file 1). We fixed 3–5-day-old larvae in 4% formaldehyde, and we subjected them to a mild proteinase K digestion to improve the penetration of our reagents. These larvae were prehybridised at 65 °C by using a hybridisation mix (Hyb-Mix), containing 50% formamide, 5× saline-sodium citrate buffer (SSC), 50 µg/ml heparin, 0.1% Tween 20, and 5 mg/ml torula RNA. Then, we dissolved the probes against r-opsin1 and glass (labelled with either fluorescein-UTP or digoxigenin-UTP) in Hyb-Mix, denatured them at 80 °C for 10 min, and added this solution to our samples. We hybridised both probes simultaneously by incubating at 65 °C overnight. Then, we washed the samples at 65 °C with a series of solutions that initially contained 50% formamide and 2× SSCT (obtained from a stock solution with Tween 20 0.1% in 4× SSC), and we progressively decreased the concentration of both formamide and SSCT throughout successive washes. After washing, we placed the larvae at room temperature and proceeded to immunostain them. We detected the two probes sequentially, by using peroxidase-conjugated primary antibodies against fluorescein (1:250, Roche) and digoxigenin (1:50, Roche). To detect the first probe, we incubated our samples overnight at 4 °C in one of these antibodies, washed them with Tris NaCl Tween 20 buffer (TNT; 0.1 M Tris-HCl, 0.15 M NaCl, 0.1% Tween 20; pH 7.5), and started the colour reaction by adding a solution that contained fluorescent tyramide (conjugated to either Cy3 or fluorescein). We controlled the development of the signal by using a fluorescence microscope, and, when it was ready, we washed in TNT and stopped the peroxidase activity with H2O2. To detect the second probe, we repeated these immunostaining steps similarly. We mounted our samples with 90% glycerol and scanned them in a confocal microscope (example confocal stacks can be found within the Additional file 5).
Microinjection of glass-Tomato
We used an unpublished assembly of the Platynereis genome (courtesy of D. Arendt, EMBL Heidelberg) for making a glass-Tomato reporter (see Additional files 1 and 2 for details). We PCR-amplified a fragment of the Platynereis glass promoter and cloned it into a plasmid in frame with the tandem dimer version of Tomato (courtesy of L. A. Bezares-Calderón) by using sticky end ligation with ApaI and SgsI [42]. The fragment that we cloned included a 5789 bp long upstream sequence and also the beginning of the Glass coding sequence: the first ATG codon was predicted both by aligning the Platynereis version of Glass to the Glass homologues of other species and by using the ATGpr software [43, 44]. For details on the plasmid that we injected, see its annotated sequence in Additional file 6.
For microinjections, we collected freshly fertilised Platynereis eggs and proceeded as previously described [14]. Briefly, we removed the jelly of the eggs by digesting with proteinase K and washing with abundant sea water, using a sieve. We diluted the glass-Tomato plasmid to a final concentration of about 200 ng/μl, and delivered it into 1-cell embryos with a microinjection set-up, by using Femtotip II microcapillaries (Eppendorf). Larvae were kept at 18 °C, and we imaged them with a Leica SP8 confocal microscope to study the expression of the reporter (representative confocal stacks are available in the Additional file 5). The expression of this reporter showed some degree of mosaicism, given that it was not integrated into the genome, which allowed us to investigate the morphology of the individual neurons that expressed it. We investigated over 100 surviving, Tomato-positive Platynereis larvae.
Results
Glass homologues are present throughout metazoans
Glass plays a fundamental role for the differentiation of rhabdomeric PRs in the fruit fly [9, 11, 45, 46]. To investigate whether it provides a similar function across metazoans, we first decided to search for Glass homologues in other species.
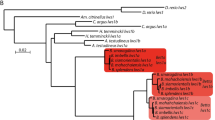
To do this, we obtained Glass-like sequences by using NCBI BLAST [24] and the Schmidtea mediterranea Genome Database [25]. We analysed these sequences with the CLANS2 software (using the BLOSUM62 matrix and a p value cut-off of 1e−60) to produce a cluster map (Fig. 1a) [27]. In this type of graph, closely related sequences (represented as points) are clustered together and connected by dark lines. Based on their similarities, we were able to identify multiple Glass homologues across distantly related species. Some more derived sequences (e.g. from Strongylocentrotus and Saccoglossus) were also clearly supported as Glass homologues in our analysis. Using these data, we constructed a maximum likelihood phylogenetic tree for Glass, which was visualised with FigTree (Fig. 1b) [31] (for more details on our analysis, see the “Methods” section and the Additional file 3). Importantly, our data reveal that Glass homologues are widely present throughout the animal kingdom.
Glass phylogeny. To identify Glass homologues, we searched for Glass-like sequences with BLAST and obtained a cluster map by using all-against-all pairwise similarity. In this graph, those sequences that are most similar appear clustered together, and connected by a darker line (a). Based on these data, we built a maximum likelihood tree for Glass (b) (for further details, see the “Methods” section, the tree file and the sequences that we used for it are included in the Additional file 3)
Neither vertebrates nor choanoflagellates have clear Glass homologues
Based on the distribution of Glass homologues, it seems this protein was present in the common ancestor of all metazoans, but not in choanoflagellates (the sister group of metazoans). Intriguingly, we could also not find any Glass homologue in vertebrates (Fig. 2). Since we identified Glass across multiple animal clades, we wondered why we could not find its vertebrate homologue. Several species have fully sequenced, well-annotated genomes, such as zebrafish, mice, or humans [47,48,49,50,51]. For this reason, we decided to further investigate the evolutionary conservation of Glass by scrutinising its sequence.
Glass homologues exist in most animal groups. Based on sequence comparison (Additional file 4, also see Fig. 3), we infer that glass appeared in the common ancestor of all metazoans, and that it has been transmitted to most present-day animals (shown in green on the phylogenetic tree [74]). However, we were not able to identify glass in vertebrates
Glass homologues share a distinctive cluster of five Cys2His2 zinc fingers in most species. (One exception is Caenorhabditis, in which it only has four zinc fingers because the first one is missing.) Particularly, the fourth and the fifth zinc fingers are especially important because they are responsible for guiding Glass towards its targets, given that they recognise its DNA-binding motif in vivo, GAARCC [34,35,36,37]. Therefore, we modified our bait by using the consensus sequence of either the full cluster of five zinc fingers, or only the fourth and fifth zinc fingers, and we repeated our BLAST search against vertebrates and choanoflagellates. By doing this, we obtained results such as, for example, ZSCAN22, ZNF253, or KOX 26 in humans, which still showed less similarity to Glass than any of those homologues that we identified in other species (Fig. 3, sequences available in Additional file 4). We also considered the human candidates that appeared annotated as putative Glass orthologues in Flybase via DIOPT [22, 52], including ZNF764, ZNF683, or ZNF500, but, likewise, they aligned poorly with the consensus sequence of the Glass zinc fingers (Fig. 3, sequences available in Additional file 4). Next, we analysed whether any of these proteins would be able to functionally substitute Glass by recognising its DNA-binding motif, the GAARCC sequence [34, 35, 37]. For this, we used the online tool ‘DNA-binding site predictor for Cys2His2 Zinc Finger Proteins’, which predicts the DNA-binding behaviour of zinc finger proteins [32, 33]. This software indicates that those Glass-like proteins that exist in vertebrates and choanoflagellates cannot recognise the GAARCC motif, in contrast to the clear Glass homologues that we found in other animals (i.e. in Amphimedon, Schmidtea, Platynereis, Aplysia, Caenorhabditis, Drosophila, Strongylocentrotus, and Branchiostoma) (Fig. 3). Consequently, it remains unclear what happened to the glass gene during the evolution of vertebrates: it could be that they lost Glass, or that it severely changed its amino acid sequence and its DNA-binding motif. Intriguingly, similar to Drosophila, some cells in the vertebrate retina also use the rhabdomeric phototransduction cascade—the ipRGCs, which detect irradiance [8]—and, based on our data, it seems highly probable that these cells develop through different mechanisms in Drosophila and in vertebrates.
Analysis of the Glass zinc fingers. Generally, Glass homologues possess a cluster of five Cys2His2 zinc fingers, each of them containing the following motif: Cys-X2,4-Cys-X12-His-X3,4,5-His. Of these, we compared the sequences of the fourth and the fifth zinc fingers, which are responsible for recognising the DNA Glass-binding motif in PRs in vivo [34,35,36,37], from the following species: Amphimedon (Porifera), Schmidtea (Platyhelminthes), Platynereis (Annelida), Aplysia (Mollusca), Caenorhabditis (Nematoda), Drosophila (Arthropoda), Strongylocentrotus (Echinodermata) and Branchiostoma (Cephalochordata). In the table, those amino acids that match the Glass consensus sequence (deduced by aligning the homologues of different species, in the first column) appear on black background. The 3D structure of the DNA-bound Cys2His2 domain has been resolved [75], and it is expected that four amino acids per zinc finger directly recognise three base pairs. These amino acids are well evolutionarily conserved across different Glass homologues and, in the sequences that we show, they are no. 10 (D), 12 (S), 13 (T), and 16 (K) in the fourth zinc finger, and no. 38 (Q), 40 (G), 41 (N), and 44 (R) in the fifth zinc finger. Other residues and neighbouring zinc fingers are also expected to contribute to the DNA-binding specificity of Glass [76]. Similarly, we aligned Glass-like proteins from vertebrates (e.g. human) and choanoflagellates (e.g. Salpingoeca) with BLAST [24] and MUSCLE [28], but they showed little similarity to the Glass consensus sequence (shown in the second column). Furthermore, a ‘DNA-binding site predictor for Cys2His2 Zinc Finger Proteins’ has been developed and is available online [32, 33]. This software predicts that, based on their amino acid sequence, all Glass homologues (in the first column) can bind to the same DNA motif: GAAGCC, which was expected from experimental works in Drosophila and Caenorhabditis [34, 35]. By contrast, it appears that the Glass-like proteins of vertebrates and choanoflagellates (in the second column) would not be able to recognise this motif. All sequences are available in the Additional file 4
glass is not expressed in rhabdomeric PRs in the Schmidtea eye
Given that Glass is an essential transcription factor for activating the expression of phototransduction proteins in all Drosophila PRs [9, 10], we investigated whether Glass has a similar function in other organisms. For this, we tested whether it is expressed in PRs in the eye of the planarian Schmidtea mediterranea. Planarians typically possess one pair of eyes, located in the head, that mediate light avoidance [5, 17, 53]. Importantly, their eyes contain rhabdomeric PRs, which are evolutionarily homologous to Drosophila PRs [1, 17].
Recently, a single-cell transcriptome atlas has been published for Schmidtea, and it is available online [18, 19, 54]. Using this database, rhabdomeric PRs can be identified because they form a cluster of non-ciliated neurons that express phototransduction proteins, including the opsin gene (Fig. 4a) [19]. Surprisingly, these cells do not co-express Glass (Fig. 4b), suggesting that, in contrast to Drosophila, Glass is not important for the function of rhabdomeric PRs in the Schmidtea eye.
glass is not expressed in rhabdomeric PRs in Schmidtea. These graphs were obtained from the Planarian Digiworm atlas, a single-cell transcriptome database for Schmidtea mediterranea [19, 25]. Each point corresponds to one single cell, and they are clustered according to the similarity of their transcriptome. One of the clusters shown, corresponding to non-ciliated neurons, is formed by 14 rhabdomeric PRs which can be identified because of the expression of the opsin gene (dd_Smed_v4_15036_0_1, a). However, these PRs do not appear to express the Schmidtea glass homologue (annotated as dd_Smed_v4_75162_0_1 in this website [19, 54], b)
glass is not expressed in rhabdomeric PRs in the Platynereis eye
We next tested whether Glass is expressed in rhabdomeric PRs in the marine ragworm P. dumerilii. The visual system of Platynereis has been well studied, both from a molecular and a functional point of view. Platynereis possesses two types of bilateral eyes containing rhabdomeric PRs, called the dorsal and ventral eyes (also known as adult and larval eyes, respectively). These two eye types are able to detect the direction of light, thus mediating phototaxis [3, 13,14,15,16].
In Drosophila, glass is expressed in all rhabdomeric PRs [12, 55]. We could detect glass expression in the compound eye of adult flies both with in situ hybridisation and with a glass-Gal4 line crossed to UAS-mCD8::RFP (Fig. 5a–b′), which confirms previous data [12, 55]. By contrast, in the case of Platynereis, in situ hybridisations performed in 3–5-day-old larvae did not show co-expression of the glass transcript with rhabdomeric opsin 1 (r-opsin1), which is a marker for rhabdomeric PRs in both the dorsal and the ventral eyes [14, 16], indicating that glass is not present in these cells (Fig. 5c–c′′′′, also see confocal stacks in Additional file 5). In addition, we also generated a Platynereis glass reporter by cloning 5.7 kb of its upstream sequence into a plasmid, where the glass start codon was in frame with Tomato (a red fluorescent protein). We used this plasmid for transient transgenesis by injecting it in 1-cell embryos containing a stable r-opsin1-GFP insertion [14]. r-opsin1-GFP animals consistently showed strong GFP signal in their dorsal eye PRs, and this signal was weaker in the ventral eye PRs. In the case of the dorsal eyes, all PRs project their rhabdomeres into a pigment cup, and their axons form four nerves that innervate the optic neuropil in the brain [3, 14, 16]. After microinjections, we tested 3–8-day-old larvae (slightly older than those that we used for in situ, to guarantee that positive cells had enough fluorescence to distinguish them) but we did not observe co-expression of GFP and Tomato. glass-Tomato-expressing neurons were consistently located in the head of Platynereis, distant from the ventral eyes. The expression of glass-Tomato showed some degree of mosaicism due to this reporter not being integrated into the genome, which allowed us to observe the morphology of individually labelled cells in detail. Some of these Tomato-positive cells appeared close to the dorsal eyes, but they did not project any rhabdomere-like extension into the pigment cup, and their axons did not innervate the optic neuropil (Fig. 5d–e′′, confocal stacks are available in Additional file 5), indicating that they were not part of the eye rhabdomeric PRs. We conclude that, while Glass is expressed in all types of rhabdomeric PRs in Drosophila, it is not present in known rhabdomeric PRs in Platynereis.
glass is not expressed in rhabdomeric PRs in Platynereis. a, b glass is present in all Drosophila rhabdomeric PRs, including those in the compound eye [12, 55]. This can be observed in head cryosections, either by using in situ hybridisation (magenta in a and greyscale in a′) or with glass > mCD8::RFP flies (magenta in b and greyscale in b′). In both cases, samples were counterstained with DAPI (green). c–e In contrast to Drosophila, double in situ hybridisation against the glass (red) and r-opsin1 (green) transcripts shows that glass is not present in Platynereis rhabdomeric PRs. Samples were counterstained with antibodies against acetylated Tubulin (ac-Tub, blue), which is a neuropil marker (c, transversal view of a whole-mounted, 5-day-old larva). To the right, close-ups of the dorsal (arrow in c; c′, c′′) and ventral eyes (arrowhead in c; c′′′, c′′′′) show that glass (in magenta/greyscale) is not expressed in either of these visual organs. Similarly, we found that a microinjected glass-Tomato reporter (magenta/greyscale) was not co-expressed with a stable r-opsin1-GFP insertion (green). Brightfield (BF, greyscale) was imaged as a reference (d–d′′, dorsal view of a whole-mounted, 8-day-old larva). The positions of the dorsal and ventral eyes are shown with an arrow and an arrowhead, respectively. Close-ups to the right show how the axons of Tomato, and GFP-positive neurons project into two different areas in the brain (d′, d′′; orthogonal views taken along the Z segment are shown below). As a control, we also imaged an 8-day-old, wild-type, uninjected larva to test its autofluorescence (using two excitation laser wavelengths: 552 nm, same as for Tomato; and 488 nm, same as for GFP). Scale bars: 10 μm in c′, c′′′; 20 μm in d–e; and 50 μm in a, b. Axes: D, dorsal; M, medial; P, posterior; V, ventral
Glass is expressed in Platynereis sensory neurons
Since glass is predominantly expressed in PRs in Drosophila, we wondered what types of cells express glass in Platynereis. We observed that most of the neurons that were labelled with the glass-Tomato reporter innervated the neurosecretory neuropil (which is ventral to the optic neuropil, Fig. 5d–d′′) [56], and, interestingly, many of them were bipolar neurons (Fig. 6). These two features are relevant because an ongoing electron microscopy (EM) connectome reconstruction shows that, in Platynereis larvae, most neurons are either unipolar or pseudounipolar [3, 56,57,58,59]. Based on their position and on their morphology, all bipolar neurons in this EM reconstruction are considered sensory neurons because they possess distinctive membranous specialisations (called sensory dendrites) that project towards the surface [3, 56,57,58,59]. Therefore, it is very likely that a subset of glass-expressing cells in Platynereis are sensory neurons.
Glass-expressing cells in Platynereis include sensory neurons. When we injected our glass-Tomato reporter, we observed that many of the neurons that were appeared labelled in the Platynereis head were bipolar, located close to the surface, and they often possessed membranous specialisations resembling sensory dendrites (arrows) (a–d). Scale bars: 5 μm
The neurosecretory neuropile of Platynereis contains multiple sensory neurons, and it has been characterised both from an anatomical and a molecular point of view [56]. However, it is still unknown whether this region is homologous to any structure of the Drosophila brain. Given that glass is also required for the development of the corpora cardiaca in Drosophila [60], it could be possible that Glass has an evolutionarily conserved function in neurosecretory cells. In addition, it could also be that Glass regulates the formation of other sensory neurons. Notably, the Caenorhabditis homologue of Glass (called CHE-1) is expressed in ASE chemosensory neurons, and it regulates their development [34, 61].
Conclusions
Remarkably, the earliest steps of eye development are controlled by a group of transcription factors, called the ‘retinal determination network’ (RDN), which is both required and sufficient for eye formation in distantly related species [20, 62,63,64,65,66,67,68]. RDN members, such as Eyeless, Sine oculis, or Eyes absent are important for inducing eye field specification. To achieve this, they establish complex epistatic interactions with each other. These interactions occur similarly across model organisms, suggesting that this is an evolutionarily conserved process [20, 69]. In contrast to the early steps of eye field specification, subsequent mechanisms that specify the cell fate of PRs are not well understood. Here, we provide evidence that, during the late stages of eye development, rhabdomeric PRs mature through different mechanisms in different species.
In Drosophila, we have recently shown that Sine oculis (a core component of the RDN) directly activates the expression of the transcription factor glass, which is crucial for activating the expression of virtually all the phototransduction proteins in all types of Drosophila PRs [9, 10, 70]. Based on similarities in their light-sensing machinery, Drosophila PRs are considered homologous to the ipRGCs of vertebrates, and also to the rhabdomeric PRs that exist in Schmidtea and Platynereis [1, 6, 7, 15, 17, 19]. Intriguingly, while we did identify Glass homologues in most metazoans, we could not find a clear Glass homologue in vertebrates. Moreover, our data indicate that glass is not expressed in the rhabdomeric PRs of Schmidtea or Platynereis. This suggests that metazoans have evolved alternative transcriptional pathways to direct the formation of rhabdomeric PRs. One of these pathways requires Glass (e.g. in Drosophila), while others do not (e.g. in vertebrates, Schmidtea, or Platynereis larvae).
It could be possible that Glass started being expressed in rhabdomeric PRs at some point during the evolution of ecdysozoans and that it became specialised in regulating the differentiation of these cells. Therefore, comparing the differentiation of Glass-expressing and non-Glass-expressing PRs provides a valuable entry point to dissect shared and dissimilar aspects of the developmental programme. Additionally, it would also be interesting to know the identity of Glass-expressing cells for understanding the ancestral function of Glass. The glass transcript is rare and lowly expressed in the Schmidtea single-cell transcriptome data that we have currently available [18, 19], and it was also lowly expressed in the single-cell transcriptome datasets of Platynereis, to the point of being removed from the analyses of the two papers in which the sequencing was published [71, 72], which makes it impossible to compare the function of glass-expressing cells between different species at this moment. It could be possible that this is because only a few cells in the brain express Glass, and these may not have been included in the samples that were sequenced. Therefore, we expect that, in the near future, increasing both the number and the quality of such single-cell transcriptomes for these and other species will be useful to address several questions about the evolution of specific cell fates. For example, some opsins may have other functions apart from light sensing [73], and it would be relevant to know whether glass regulates the expression of any such opsin outside the Platynereis eye (for example), at any stage.
The absence of Glass in rhabdomeric PRs in the eye of some species argues for other transcription factors being capable of activating the expression of phototransduction proteins; however, the underlying mechanism remains unknown. Our data support a rather complex scenario for the evolution of rhabdomeric PRs, but future works on the targets of the RDN may help to better understand how rhabdomeric PR identity is regulated.
Abbreviations
- ac-Tub:
-
acetylated Tubulin
- EM:
-
electron microscopy
- PB:
-
phosphate buffer
- PBT:
-
phosphate buffer with Triton X-100
- PR:
-
photoreceptor neuron
- RDN:
-
retinal determination network
- r-opsin1:
-
rhabdomeric opsin 1
- SSC:
-
saline-sodium citrate buffer
- SSCT:
-
saline-sodium citrate buffer with Tween 20
References
Fain GL, Hardie R, Laughlin SB. Phototransduction and the evolution of photoreceptors. Curr Biol. 2010;20:R114–24.
Nilsson D-E. Photoreceptor evolution: ancient siblings serve different tasks. Curr Biol. 2005;15:R94–6.
Randel N, Asadulina A, Bezares-Calderon LA, Veraszto C, Williams EA, Conzelmann M, Shahidi R, Jekely G. Neuronal connectome of a sensory-motor circuit for visual navigation. Elife. 2014;3:e02730.
Borst A. Drosophila’s view on insect vision. Curr Biol. 2009;19:R36–47.
Paskin TR, Jellies J, Bacher J, Beane WS. Planarian phototactic assay reveals differential behavioral responses based on wavelength. PLoS ONE. 2014;9:e114708.
Provencio I, Warthen DM. Melanopsin, the photopigment of intrinsically photosensitive retinal ganglion cells. WIREs Membr Transp Signal. 2012;1:228–37.
Montell C. Drosophila visual transduction. Trends Neurosci. 2012;35:356–63.
Hankins MW, Hughes S. Vision: melanopsin as a novel irradiance detector at the heart of vision. Curr Biol. 2014;24:R1055–7.
Bernardo-Garcia FJ, Fritsch C, Sprecher SG. The transcription factor Glass links eye field specification with photoreceptor differentiation in Drosophila. Development. 2016;143:1413–23.
Bernardo-Garcia FJ, Humberg T-H, Fritsch C, Sprecher SG. Successive requirement of Glass and Hazy for photoreceptor specification and maintenance in Drosophila. Fly. 2017;11:112–20.
Moses K, Ellis MC, Rubin GM. The glass gene encodes a zinc-finger protein required by Drosophila photoreceptor cells. Nature. 1989;340:531–6.
Ellis MC, O’Neill EM, Rubin GM. Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development. 1993;119:855–65.
Jékely G, Colombelli J, Hausen H, Guy K, Stelzer E, Nédélec F, Arendt D. Mechanism of phototaxis in marine zooplankton. Nature. 2008;456:395–9.
Backfisch B, Veedin Rajan VB, Fischer RM, Lohs C, Arboleda E, Tessmar-Raible K, Raible F. Stable transgenesis in the marine annelid Platynereis dumerilii sheds new light on photoreceptor evolution. Proc Natl Acad Sci USA. 2013;110:193–8.
Arendt D, Tessmar-Raible K, Snyman H, Dorresteijn AW, Wittbrodt J. Ciliary photoreceptors with a vertebrate-type opsin in an invertebrate brain. Science. 2004;306:869–71.
Randel N, Bezares-Calderón LA, Gühmann M, Shahidi R, Jékely G. Expression dynamics and protein localization of rhabdomeric opsins in Platynereis larvae. Integr Comp Biol. 2013;53:7–16.
Lapan SW, Reddien PW. Transcriptome analysis of the planarian eye identifies ovo as a specific regulator of eye regeneration. Cell Rep. 2012;2:294–307.
Plass M, Solana J, Wolf FA, Ayoub S, Misios A, Glazar P, Obermayer B, Theis FJ, Kocks C, Rajewsky N. Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science. 2018;360:eaaq1723.
Fincher CT, Wurtzel O, de Hoog T, Kravarik KM, Reddien PW. Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science. 2018;360:eaaq1736.
Silver SJ, Rebay I. Signaling circuitries in development: insights from the retinal determination gene network. Development. 2005;132:3–13.
Arendt D, Tessmar K, Medeiros de Campos-Baptista M-I, Dorresteijn A, Wittbrodt J. Development of pigment-cup eyes in the polychaete Platynereis dumerilii and evolutionary conservation of larval eyes in Bilateria. Development. 2002;129:1143–54.
Flybase. http://flybase.org/. Accessed 26 Feb 2019.
Max Planck Institute for Developmental Biology: Platynereis dumerilii transcriptome. http://jekely-lab.tuebingen.mpg.de/blast/. Accessed 26 Feb 2019.
NCBI BLAST service. http://blast.ncbi.nlm.nih.gov/. Accessed 26 Feb 2019.
Schmidtea mediterranea Genome Database. http://smedgdkc03.stowers.org. Accessed 26 Feb 2019.
Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–9.
Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–4.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7.
Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3.
IQ-TREE software for phylogenomic inference. http://www.iqtree.org/. Accessed 26 Feb 2019.
FigTree viewer of phylogenetic trees. http://tree.bio.ed.ac.uk/software/figtree/. Accessed 26 Feb 2019.
DNA-binding site predictor for Cys2His2 zinc finger proteins. http://zf.princeton.edu/. Accessed 26 Feb 2019.
Persikov AV, Singh M. De novo prediction of DNA-binding specificities for Cys2His2 zinc finger proteins. Nucleic Acids Res. 2014;42:97–108.
Etchberger JF, Lorch A, Sleumer MC, Zapf R, Jones SJ, Marra MA, Holt RA, Moerman DG, Hobert O. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 2007;21:1653–74.
Naval-Sánchez M, Potier D, Haagen L, Sánchez M, Munck S, Van de Sande B, Casares F, Christiaens V, Aerts S. Comparative motif discovery combined with comparative transcriptomics yields accurate targetome and enhancer predictions. Genome Res. 2013;23:74–88.
O’Neill EM, Ellis MC, Rubin GM, Tjian R. Functional domain analysis of glass, a zinc-finger-containing transcription factor in Drosophila. Proc Natl Acad Sci USA. 1995;92:6557–61.
Enuameh MS, Asriyan Y, Richards A, Christensen RG, Hall VL, Kazemian M, Zhu C, Pham H, Cheng Q, Blatti C, et al. Global analysis of Drosophila Cys2-His2 zinc finger proteins reveals a multitude of novel recognition motifs and binding determinants. Genome Res. 2013;23:928–40.
Park S, Bustamante EL, Antonova J, McLean GW, Kim SK. Specification of Drosophila corpora cardiaca neuroendocrine cells from mesoderm is regulated by Notch signaling. PLoS Genet. 2011;7:e1002241.
Hauenschild C, Fischer A. Platynereis dumerilii: Mikroskopische Anatomie, Fortpflanzung, Entwicklung. Stuttgart: Gustav Fischer Verlag; 1969.
Wolff T. Histological techniques for the Drosophila eye. Part II: Adult. In: Sullivan W, Ashburner M, Hawley RS, editors. Drosophila protocols. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2000.
Tessmar-Raible K, Steinmetz PR, Snyman H, Hassel M, Arendt D. Fluorescent two-color whole mount in situ hybridization in Platynereis dumerilii (Polychaeta, Annelida), an emerging marine molecular model for evolution and development. Biotechniques. 2005;39:460–2.
Day RN, Davidson MW. The fluorescent protein palette: tools for cellular imaging. Chem Soc Rev. 2009;38:2887–921.
ATGpr service. http://atgpr.dbcls.jp/. Accessed 26 Feb 2019.
Nishikawa T, Ota T, Isogai T. Prediction whether a human cDNA sequence contains initiation codon by combining statistical information and similarity with protein sequences. Bioinformatics. 2000;16:960–7.
Liang X, Mahato S, Hemmerich C, Zelhof AC. Two temporal functions of Glass: ommatidium patterning and photoreceptor differentiation. Dev Biol. 2016;414:4–20.
Morrison CA, Chen H, Cook T, Brown S, Treisman JE. Glass promotes the differentiation of neuronal and non-neuronal cell types in the Drosophila eye. PLoS Genet. 2018;14:e1007173.
Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–51.
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921.
Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62.
Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503.
Ensembl genome browser. http://www.ensembl.org/. Accessed 26 Feb 2019.
Hu Y, Flockhart I, Vinayagam A, Bergwitz C, Berger B, Perrimon N, Mohr SE. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinform. 2011;12:357.
Arees EA. Absence of light response in eyeless planaria. Physiol Behav. 1986;36:445–9.
Planarian Digiworm atlas. https://digiworm.wi.mit.edu. Accessed 26 Feb 2019.
Moses K, Rubin GM. Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev. 1991;5:583–93.
Williams EA, Veraszto C, Jasek S, Conzelmann M, Shahidi R, Bauknecht P, Mirabeau O, Jekely G. Synaptic and peptidergic connectome of a neurosecretory center in the annelid brain. Elife. 2017;6:e26349.
Bezares-Calderon LA, Berger J, Jasek S, Veraszto C, Mendes S, Guhmann M, Almeda R, Shahidi R, Jekely G. Neural circuitry of a polycystin-mediated hydrodynamic startle response for predator avoidance. Elife. 2018;7:e363262.
Randel N, Shahidi R, Veraszto C, Bezares-Calderon LA, Schmidt S, Jekely G. Inter-individual stereotypy of the Platynereis larval visual connectome. Elife. 2015;4:e08069.
Veraszto C, Ueda N, Bezares-Calderon LA, Panzera A, Williams EA, Shahidi R, Jekely G. Ciliomotor circuitry underlying whole-body coordination of ciliary activity in the Platynereis larva. Elife. 2017;6:e26000.
De Velasco B, Shen J, Go S, Hartenstein V. Embryonic development of the Drosophila corpus cardiacum, a neuroendocrine gland with similarity to the vertebrate pituitary, is controlled by sine oculis and glass. Dev Biol. 2004;274:280–94.
Uchida O, Nakano H, Koga M, Ohshima Y. The C. elegans che-1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development. 2003;130:1215–24.
Hoge MA. Another gene in the fourth chromosome of Drosophila. Am Nat. 1915;49:47–9.
Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994;265:785–9.
Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, Gehring WJ. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998;125:2181–91.
Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–92.
Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997;91:881–91.
Chow RL, Altmann CR, Lang RA, Hemmati-Brivanlou A. Pax6 induces ectopic eyes in a vertebrate. Development. 1999;126:4213–22.
Loosli F, Winkler S, Wittbrodt J. Six3 overexpression initiates the formation of ectopic retina. Genes Dev. 1999;13:649–54.
Donner AL, Maas RL. Conservation and non-conservation of genetic pathways in eye specification. Int J Dev Biol. 2004;48:743–53.
Jusiak B, Karandikar UC, Kwak S-J, Wang F, Wang H, Chen R, Mardon G. Regulation of Drosophila eye development by the transcription factor Sine oculis. PLoS ONE. 2014;9:e89695.
Achim K, Eling N, Vergara HM, Bertucci PY, Musser J, Vopalensky P, Brunet T, Collier P, Benes V, Marioni JC, et al. Whole-body single-cell sequencing reveals transcriptional domains in the annelid larval body. Mol Biol Evol. 2018;35:1047–62.
Achim K, Pettit JB, Saraiva LR, Gavriouchkina D, Larsson T, Arendt D, Marioni JC. High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin. Nat Biotechnol. 2015;33:503–9.
Leung NY, Montell C. Unconventional roles of opsins. Annu Rev Cell Dev Biol. 2017;33:241–64.
Dunn CW, Giribet G, Edgecombe GD, Hejnol A. Animal phylogeny and its evolutionary implications. Annu Rev Ecol Evol S. 2014;45:371.
Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–17.
Garton M, Najafabadi HS, Schmitges FW, Radovani E, Hughes TR, Kim PM. A structural approach reveals how neighbouring C2H2 zinc fingers influence DNA binding specificity. Nucleic Acids Res. 2015;43:9147–57.
Authors’ contributions
FJB-G, GJ, and SGS conceived the study. FJB-G, GJ, and MS performed the experiments. FJB-G, GJ, and SGS wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank the Bloomington Stock Center, R. Stocker, and S. Kim for fly stocks; F. Raible for the r-opsin1-GFP Platynereis strain; D. Arendt for providing access to the draft Platynereis genome; and L. A. Bezares-Calderón for plasmids. We are also grateful to C. Adler, J. Cleland, and to our colleagues of the Sprecher and Jékely labs for valuable discussions.
Competing interests
The authors do not declare competing or financial interests.
Availability of data and materials
All data presented in this article are accessible online, as indicated in either the main text or in the Additional file section.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was funded by the Swiss National Science Foundation (31003A_149499 to S.G.S.) and the European Research Council (ERC-2012-StG 309832-PhotoNaviNet to S.G.S.).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding authors
Additional files
Additional file 1.
Supplementary methods.
Additional file 2.
Platynereis glass supplementary nucleotide sequences (both genomic and transcriptomic).
Additional file 3.
Glass phylogenetic tree and sequences data.
Additional file 4.
Subset of glass-like sequences for which the DNA-binding affinity was investigated.
Additional file 5.
Example confocal stacks.
Additional file 6.
Annotated sequence of the glass-tomato plasmid that was used for Platynereis microinjections.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Bernardo-Garcia, F.J., Syed, M., Jékely, G. et al. Glass confers rhabdomeric photoreceptor identity in Drosophila, but not across all metazoans. EvoDevo 10, 4 (2019). https://doi.org/10.1186/s13227-019-0117-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13227-019-0117-6