Abstract
Purpose
The health of rhizosphere soil microorganisms is an important indicator to evaluate soil quality. Therefore, understanding the response of rhizosphere soil microorganisms to tobacco crop succession is crucial for promoting the sustainable development of agriculture.
Methods
The microbial diversity and community structure of rhizosphere soil in continuous cropping and non-cropped tobacco for 7 years were analyzed by the Illumina platform.
Result
(1) Continuous cropping tobacco cause rhizosphere soil acidification and reduction in alkaline nitrogen (AN) and soil organic matter (SOM). (2) Continuous cropping tobacco reduces the diversity of rhizosphere soil microbial communities, increasing harmful functional microorganisms and declining beneficial ones. (3) The abundance of bacteria that perform nitrification and saprophytic fungi in the rhizosphere soil of continuous cropping areas decreases, inhibiting carbon and nitrogen cycling processes. (4) The composition and diversity of the soil rhizosphere microbial community are affected by the imbalance in the physicochemical property of the rhizosphere.
Conclusion
Continuous cropping tobacco cause rhizosphere soil acidification and nutrient imbalance, and the carbon and nitrogen cycles involved in microorganisms were damaged. Furthermore, the decreased diversity of rhizosphere soil microorganisms and the increased abundance of pathogenic fungi contribute to the continuous cropping obstacles of tobacco.
Similar content being viewed by others
Introduction
Tobacco plays a crucial role in the economic industry of Guizhou Province and is a major contributor to tobacco production in China. Due to the limitations of arable land resources and the market economy, as well as a lack of sustainable planting concepts (Wang et al., 2020), continuous tobacco cropping has become prevalent (He et al. 2011). Several issues have arisen due to long-term continuous cropping, such as decreased soil quality, increased disease-causing pathogens (Rao et al., 2022; Li et al. 2022), and decreased tobacco yields (Liu et al. 2012; Guo et al. 2017; You et al. 2015). These problems negatively impact the sustainability of the tobacco growing industry.
Rhizosphere soil microorganisms are important components of soil ecosystems and play crucial roles in soil nutrient transformation and cycling (Jacoby et al. 2017), plant growth and development, and pathogen defense (Shao et al. 2021; Liu et al. 2021; Wu et al. 2022; Yang et al. 2019; Jacoby et al. 2017). They are also significant indicators of soil health. Continuous cropping cause many issues, for instance, the decline in the diversity of soil microbial populations and the rise of the dominant pathogenic species (Liu et al., 2023; Liu et al. 2022; Zheng et al. 2022). Those conditions are not conducive to the sustainable development of soil ecosystems. Few studies have investigated the alterations in the structure and physicochemical characteristics of rhizosphere soil microbial communities and their interrelationships in the “honey-sweet and fragrant type” of tobacco continuous cropping in Guizhou Province. Thus, comprehending the alterations in the microbial community structure and physicochemical properties of soil in tobacco rhizosphere due to continuous cropping obstacles is crucial to alleviate the problems related to such cropping in tobacco plantation soil. This understanding will contribute to the sustainable utilization of the soil and ensure the superior quality of tobacco production.
In this study, high-throughput sequencing technology were used to investigate the structure and diversity of rhizosphere soil microorganisms in continuous cropping tobacco fields, and Pearson correlation and mantel correlation analysis was adopted to analyze the relationship between rhizosphere soil microorganisms and physicochemical properties. This study aims to provide a reference for mitigating continuous crop barriers.
Materials and methods
Study site and soil sampling
The sampling location for soil was in Longping Village (106° 3′ E, 27° 23′ N), Bijie City, Guizhou Province. The region sits between 1390 and 1420 m above sea level. The local region has an average annual temperature of around 15 °C. This study was set two tobacco cropping systems, including non-continuous tobacco soil, and soil continuous tobacco for 7 years. Both cropping patterns had the same soil characteristics and management practices. Soil samples were collected from the rhizosphere of five tobacco plants in each field during the peak period of tobacco growth (60 days after transplanting) using the 5-point sampling method. A total of six mixed samples (two treatments with three replicates each) were collected. The tobacco plants were carefully extracted and shaken to remove any excess soil, and the rhizosphere soil within 0–4 mm of the roots was collected. The soil samples were immediately transported to the laboratory with dry ice. After removing visible residues, part of the samples was dried naturally to measure soil physical and chemical indices, and the remaining was stored at − 80 °C for measuring microbial indicators.
Soil physicochemical analysis
The soil’s physical and chemical properties were determined according to the “Soil Agricultural Chemical Analysis” (Bao et al., 2000). Soil pH was measured using a pH meter with a suspension ratio (w/v) of 1:2.5 using the potentiometric method. The soil organic matter (SOM) was oxidized using the potassium dichromate heating method in an oil bath under heating conditions. The soil organic carbon was oxidized with potassium dichromate-sulfuric acid and titrated using a 0.2 mol/L ferrous ammonium sulfate standard solution. The alkaline nitrogen (AN) was measured by the alkaline diffusion method, in which nitrate nitrogen (NO − 3-N) in the soil was converted to ammonium nitrogen (NH + 4-N) under alkaline conditions. The absorption was done using 20 g/L boric acid, and titration was performed using a 0.01 mol/L standard sulfuric acid solution. The soil’s available phosphorus (AP) was extracted using a 0.5 mol/L sodium bicarbonate solution and measured by the molybdenum blue colorimetric method. Color development was achieved using an aqueous solution of molybdenum-antimony (a combination of molybdate and antimony potassium tartrate), followed by detection using an ultraviolet spectrophotometer at a wavelength of 700 nm. The soil’s available potassium (AK) was measured via flame photometry, with a neutral 1 mol/L solution of ammonium acetate as the leaching agent. Ammonium ions within the solution were substituted with potassium ions on the soil colloid surface, and potassium ions in the leachate were detected using flame photometry.
DNA extraction and sequencing
The DNA was extracted with the TGuide S96 Magnetic Soil /Stool DNA Kit (Tiangen Biotech (Beijing) Co., Ltd.) according to manufacturer instructions. The DNA concentration of the samples was measured with the Qubit dsDNA HS Assay Kit and Qubit 4.0 Fluorometer (Invitrogen, Thermo Fisher Scientific, Oregon, USA). The 338F: 5′-ACTCCTACGGGAGGCAGCA-3′ and 806R: 5′-GGACTACHVGGGTWTCTAAT-3′ universal primer set was used to amplify the V3-V4 region of 16S rRNA gene from the genomic DNA extracted from each sample. The internal transcribed spacer 1 between 18S and 5.8S rRNA genes (ITS1 rRNA) was amplified by PCR using primers 5′-CTTGGTCATTTAGAGGAAGTAA-3′ and 5′-GCTGCGTTCTTCATCGATGC-3′. The PCR was performed in a total reaction volume of 10 μl: DNA template 5–50 ng, Vn F (10 μM) 0.3 μl, Vn R (10 μM) 0.3 μl, KOD FX Neo Buffer 5 μl, dNTP (2 mM each) 2 μl, KOD FX Neo 0.2 μl, ddH2O up to 10 μl. Vn F and Vn R were selected according to the amplification area. After with initial denaturation at 95 °C for 5 min, followed by 25 cycles of denaturation at 95 °C for 30 s, annealing at 50 °C for 30 s, extension at 72 °C for 40 s, and a final step at 72 °C for 7 min. The PCR amplicons were purified with Agencourt AMPure XP Beads (Beckman Coulter, Indianapolis, IN) and quantified using the Qubit dsDNA HS Assay Kit and Qubit 4.0 Fluorometer (Invitrogen, Thermo Fisher Scientific, Oregon, USA). After the individual quantification step, amplicons were pooled in equal amounts. Illumina NovaSeq 6000 (Illumina, Santiago CA, USA) was used for sequencing for the constructed library.
Bioinformatics
The bioinformatics analysis of this study was performed with the aid of the BMK Cloud (Biomarker Technologies Co., Ltd., Beijing, China). According to the quality of a single nucleotide, raw data was primarily filtered by Trimmomatic v0.33 (Edgar et al., 2013). Identification and removal of primer sequences were achieved by Cutadapt v1.9.1 (Callahan et al. 2016). PE reads previously obtained were assembled by USEARCH v10 (Segata et al., 2011), followed by chimera removal using UCHIME v8.1 (Quast et al. 2013). The obtained high-quality reads were used in the bioinformatics statistical analysis.
Statistical analysis
Sequences with similarity ≥ 97% were clustered into the same operational taxonomic unit (OTU) by USEARCH v10.0 (Edgar et al., 2013), and the OTUs with abundance < 0.005% were filtered. Taxonomy annotation of the OTUs was performed based on the naive Bayes classifier in QIIME2 (Bolyen et al. 2019) using the SILVA database (Quast et al. 2013) (release 132) with a confidence threshold of 70%. Sequences with similarity ≥ 97% were clustered into the same operational taxonomic unit (OTU) by USEARCH v10.0 (Edgar et al., 2013), and the OTUs with abundance < 0.005% were filtered. Taxonomy annotation of the OTUs was performed based on the Naive Bayes classifier in QIIME2 (Bolyen et al. 2019) using the SILVA database (Quast et al. 2013) (release 132) with a confidence threshold of 70%. The Alpha diversity was calculated using the QIIME2 and R software. Beta diversity was calculated to measure the similarity of microbial communities from different samples using QIIME. Principal component analysis (PCA) was used to analyze the beta diversity. Linear discriminant analysis (LDA) effect size (LEfSe) was adopted to test the significant taxonomic difference among groups. A logarithmic LDA score 4.0 was set as the discriminative feature threshold to identify significant variation. To explore the dissimilarities of the microbiome among different factors, the correlation between microbial communities and soil properties was determined using the Mantel test and the Pearson test. The fungal functions were analyzed using FUNGuild v1.0 (Nguyen et al. 2016). The bacterial functions were analyzed using FAPROTA v1.2.6 (Louca et al. 2016). Soil physicochemical parameters, microbial alpha diversity indices, and functional prediction data were compared, and a normal distribution was ensured. Statistical analyses were conducted using SPSS Statistics v26. The statistical significance of the two treatments was determined using an analysis of variance (ANOVA) with post hoc multiple comparisons of variances (Amenu et al., 2023).
Results
Physicochemical properties of rhizosphere soils
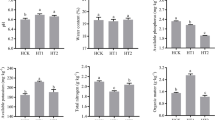
The physical and chemical properties of continuous and non-continuous rhizosphere tobacco soils are shown in Table 1. There has a significant difference in physicochemical properties between the CK and CM groups. All physicochemical properties (pH, SOM, AP, AK, and AN) of CM are lower than those of the CK group, indicating that the nutrient status of the soil with continuous cropping diseases is lower than that of the soil with normal non-cropped conditions. According to the classification of soil nutrient levels for tobacco cultivation (Wu et al. 2014; Yang et al. 2022), the rhizosphere soil in the CM group presents a nutrient imbalance with acidic pH and AN deficiency. However, the effective nutrients in the rhizosphere soils of the CK group were generally harmonized and showed partial enrichment levels, and the soil pH was neutral. It can be inferred that continuous tobacco cultivation over multiple years has led to soil acidification, resulting in an imbalance and deficiency of soil nutrients.
Diversity of bacterial and fungal communities in rhizosphere soil
Table 2 presents the results of the Alpha diversity indices ANOVA for bacteria and fungi obtained through amplicon sequencing. The diversity and richness indices (Simpson and Shannon indices, ACE, and Chao1 indices) of soil bacterial communities in the CM group were significantly lower than those of the CK group. This finding suggests that successive years of cultivating tobacco have significantly reduced the diversity of soil bacterial communities. Within the fungi group, the Simpson and Shannon indices of the CM community were considerably lower than those of the CK group. The ACE and Chao1 indices were higher in the CM group than in the CK group, but the differences were not statistically significant. These findings reveal that the continuous cultivation of tobacco increases the richness of rhizosphere soil fungi. However, a significant decrease is observed in both the abundance and evenness of these fungi.
The structure and composition of bacterial and fungal communities in the rhizosphere soil
OTU clustering was performed on each set of soil samples to conduct a comparative study of the bacterial and fungal community composition in rhizosphere soils of continuous cropping areas and non-continuous normal soils. This analysis has yielded 25 phyla, 77 classes, 170 orders, 282 families, 493 genera, and 554 species of bacteria. Additionally, 8 phyla, 27 classes, 65 orders, 135 families, 262 genera, and 307 species were analyzed for fungi. In the bacterial phylum-level analysis (Fig. 1a), the top 10 bacteria in relative abundance are Proteobacteria, Acidobacteria, Actinobacteria, Chloroflexi, Gemmatimonadetes, Bacteroidetes, Verrucomicrobia, Patescibacteria, Nitrospirae, Firmicutes, and Proteobacteria is the most dominant bacterium in this group. The relative abundance shares of Proteobacteria, Actinobacteria, Verrucomicrobia, and Firmicutes increased in the CM group compared with the CK group. Conversely, the relative abundance percentage of Acidobacteria, Gemmatimonadetes, Chloroflexi, Bacteroidetes, and Nitrospirae decreases. At the genus level of bacteria (Fig. 1c), the ten most abundant bacteria in terms of relative abundance are Sphingobium, Rhodanobacter, Bradyrhizobium, and Gemmatimonas, plus one additional “Candidatus Udaeobacter” and five “uncultured bacterium” bacteria. Sphingobium is the dominant bacterium. Compared with the CK group, the relative abundance percentage of Sphingobium decreases in the CM group, while the relative abundance ratio of Rhodanobacter and Bradyrhizobium increases. These findings suggest that continuous cropping alters the community structure of rhizosphere soil bacteria.
Regarding the fungal phylum level (Fig. 1b), the most abundant fungi, in descending order, are Ascomycota, Basidiomycota, Mortierellomycota, Chytridiomycota, Rozellomycota, Olpidiomycota, Glomeromycota, and Kickxellomycota. Ascomycota is the most prevalent fungus. Compared with the CK group, the relative abundance of Ascomycota and Mortierellomycota decreases in the CM group, whereas the proportion of Basidiomycota and Chytridiomycota increases. At the genus level of fungi (Fig. 1d), the most abundant genera among the top 10 are Purpureocillium, Fusarium, Mortierella, Penicillium, Aspergillus, Gliocladiopsis, Cladosporium, Ilyonectria, Epicoccum, and Conocybe, with Purpureocillium being the most dominant. Compared with the CK group, the proportion of Purpureocillium, Cladosporium, Epicoccum, and Conocybe increases, while the proportion of Fusarium, Mortierella, Penicillium, Aspergillus, Gliocladiopsis, and Ilyonectria decreases. These results suggest that the community structure of rhizosphere soil fungi has undergone a significant alteration due to continuous cropping.
Differences in the distribution of bacterial and fungal communities in the rhizosphere soil
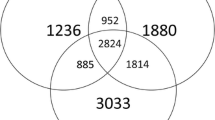
The differences in bacterial community structure between non-continuous cropping normal rhizosphere soil and continuous cropping diseased rhizosphere soil were compared using PCA based on the Bary Curtis algorithm. The variation in microbial composition can be explained by the total sum of the first and second principal component axes being over 50%. As shown in Fig. 2, the overall explanatory reliability of bacteria and fungi PC1 and PC2 is high, while the CK and CM groups are distributed independently with a significant distance between them. The result indicates a significant difference in bacterial and fungal community structure between normal rhizosphere soils with continuous and non-continuous cropping practices. To further determine the differential distribution of species in soil between the CM and CK groups, LEfSe analysis was performed on the results (Fig. 3). There are six species of bacteria at the level of the phylum and three at the level of the cultivable genus of bacteria. These species are classified as Proteobacteria, Patescibacteria, Acidobacteria, Actinobacteria, Chloroflexi, and Bacteroidetes at the phylum level and Sphingobium, Bradyrhizobium, and Rhodanobacter at the cultivable genus level, as shown in Fig. 3a. There are numerous distinct species of fungi, with two phylum-level and seven genus-level species. The phylum-level species included Ascomycota and Chytridiomycota, while the genus-level species were Purpureocillium, Fusarium, Aspergillus, Gliocladiopsis, Cladosporium, Epicoccum, and Conocybe, as shown in Fig. 3b.
The Cladogram showing that LEfSe analysis of soil bacteria and fungi. a Bacteria. b Fungi. CK: soil from non-continuous crop farmland. CM: soil from planting tobacco for 7 years. Taxa with significant differences in abundance between groups are indicated by colored dots, yellow indicates no differences, and the branching map circles represent phylogenetic taxa from phylum to genus
The relationship between rhizosphere microorganisms and physicochemical properties
To investigate the relationship among rhizosphere soil microbiota, physicochemical properties, and species diversity, we employed Pearson correlation analysis and the Mantel test to perform a correlation analysis of species diversity, physicochemical properties, and alpha diversity. The results indicate (Fig. 4) positive correlations among the physicochemical properties of the rhizosphere soil each other. Within the bacterial community (Fig. 4a), the bacterial genera showed a significantly positive correlation with soil SOM and AP. Chao1 and ACE, the indicators for bacterial abundance, were also significantly positively correlated with soil pH, SOM, and AK. In addition, the diversity indices (Shannon and Simpson) were significantly positively correlated with soil SOM, AN, and AP. These observations indicate that the species composition and diversity of bacterial rhizosphere soils are affected by soil physicochemical properties. The impacts of soil SOM and AN on the bacterial community were particularly significant. Similarly, the fungal genera showed a significantly positive correlation with soil SOM and AK (Fig. 4b). Shannon and Simpson exhibited a significantly positive correlation with soil SOM, AN, and AK. In contrast, Chao1 and ACE were not significantly correlated with soil physicochemical properties. These findings suggest that soil physicochemical properties primarily affect the rhizosphere soil community composition and species diversity of fungi. Additionally, AN was observed to have a highly significant impact on fungal diversity.
A correlation heatmap and network map of environmental factors with alpha diversity index and bacterial and fungal communities at the genus level. a Bacteria. b Fungi. Mantel’s p is the p-value of correlation of soil physicochemical properties with genus and Alpha index. Mantel’s r is the r value of Mantel analysis of soil physicochemical properties with genus and Alpha index. Pearson’s r is the p-value for correlation of soil physicochemical properties with genus and Alpha index
Pearson correlation analyses were conducted to analyze the top 10 rhizosphere soil microorganism communities to explore the effects of soil physicochemical properties on rhizosphere soil species. Concerning the bacterial community (Fig. 5a), the amount of Bradyrhizobium was significantly negatively correlated with SOM and AN. Furthermore, the quantity of Rhodanobacter was significantly negatively correlated with pH, SOM, and AN. In the fungal community (Fig. 5b), the abundance of Purpureocillium was significantly negatively correlated with SOM and AK. The concentration of Fusarium was significantly positively correlated with AK. Additionally, the content of Penicillium demonstrated a significantly positive correlation with AK, AN, pH, and SOM. The abundance of Aspergillus indicates significant positive correlations with SOM, AP, AN, and pH. A significantly positive correlation was observed between the quantity of Ilyonectria in soil and AK, AN, and pH. The abundance of Epicoccum was significantly negatively correlated with the pH of SOM, AP, and AN.
Microbial gene function prediction in rhizosphere soil of different groups
The bacterial function was predicted in the two treatment groups using the functional annotation of prokaryotic taxa (FAPROTAX), and bacteria were annotated into 45 functional groups. Ten functional groups with the highest relative abundance were chemoheterotrophy, aerobic chemoheterotrophy, nitrification, ureolysis, nitrate reduction, aerobic ammonia oxidation, nitrogen fixation, fermentation, aerobic nitrite oxidation, and aromatic compound degradation (Fig. 6a).
FUNGuild was employed to predict the trophic functions of fungal communities subjected to different treatments. They are dominated by Pathogens and Saprotrophs and were further categorized into 23 functional groups. According to Fig. 6b, the top ten fungal functional groups were ranked by relative abundance, including undefined saprotroph, fungal parasite, plant pathogen, animal pathogen, wood saprotroph, algal parasite, dung saprotroph, plant saprotroph, endophyte, and ectomycorrhizal.
We conducted a variance analysis on the predicted functions in bacteria and fungi to examine the variations in functional genes within the rhizosphere soil microbiome under different treatments. The results show that the bacterial functional genes involved in the nitrogen cycle change significantly in the continuous cropping soils (Fig. 7a), with a significant decrease in the relative abundance of nitrification-related functional genes and an increase in the relative abundance of nitrogen-fixing functional and ureolytic genes. The study suggests a decline in the population of nitrification-related bacteria in soils undergoing continuous cropping, and the proportion of bacteria capable of converting external nitrogen sources is increased. In addition, the relative abundance of aromatic compound degradation is significantly upgraded, indicating that continuous cropping increases the organic contaminants in the soils.
Notably, alterations in the function of fungi were apparent in continuous cropping soils (Fig. 7b). The relative abundance of fungal parasites significantly increases, and that of plant pathogens decreases noticeably, indicating that the parasitic pathogenic fungi gradually increase in successional cropping soils. In contrast, the relative abundance of wood, dung, and plant saprotrophs declines significantly, along with a reduction in the relative abundance of symbiotrophs. This decrease implies that the beneficial functional fungi gradually decrease in the continuous cropping soil.
Discussion
Soil microorganisms are crucial in agroecosystems since they determine soil fertility, crop productivity, and resistance reversal (Hartmann and Six 2023). This study compares the diversity, structural characteristics, physicochemical properties, and correlations between the fungal and bacterial communities in the rhizosphere soil without continuous cropping and that of a tobacco plant affected by disease after 7 years of continuous cropping. The results indicate that continuous cropping leads to soil acidification in roots, nutrient imbalances, and reduced diversity within the microbial community and alters community structure. Alterations in soil nutrient levels in roots influence the rhizosphere microbial community composition. Such changes in microbial community characteristics cause tobacco morbidity.
Continuous cropping leads to nutrient imbalances and soil acidification in the rhizosphere soil
Crops are characterized by selective uptake or enrichment of nutrients (Lu et al. 2020). Prolonged and continuous monoculture may disturb the nutrient equilibrium in the soil (Liu et al. 2002; Jin et al. 2002; Zhang et al. 2010). This study has shown that continuous cropping leads to a decline in soil nutrient status and an imbalance in the rhizosphere compared to the balanced and high-nutrient status found in non-continuous cropping soils. Effective nutrients in the continuous cropping soils exhibit significantly lower pH, SOM, AN, AP, and AK than those in non-continuous cropping soils. Prolonged continuous cultivation causes progressive soil acidification (Zhang et al. 2010; Tian et al. 2011). In this study, the pH of the soils between the roots of continuous cultivation decreased to 4.83, much lower than the pH range of high-quality tobacco (5.8–6.5) (Yang et al. 2022). The low pH could potentially affect the uptake and use of effective soil nutrients by the tobacco root system (Fig. 4). Soil acidification may be attributed to the prolonged continuous cultivation of tobacco, a result of the accumulation of phenolic acid-secreted substances in the root system (Zhang et al. 2018; Bai et al. 2019), and the imbalanced application of fertilizers (Yan et al. 2018, 2020). The SOM stability is critical for maintaining soil fertility and crop yields (Githongo et al. 2023; Wood et al. 2018; Oldfield et al. 2019). The present study show that prolonged continuous cropping severely reduces SOM, consistent with the studies of Fujii et al. (2022) and Xie et al. (2023). Nitrogen is a crucial element for tobacco plant growth. Nitrogen deficiency can significantly affect physiological and biochemical processes essential for plant growth, such as plant cell synthesis, enzyme activity, and chlorophyll synthesis (Bai et al. 2020), ultimately influencing the intrinsic chemical qualities of tobacco (Lisuma et al. 2020). The AN content in soil subjected to continuous cropping is reduced below the threshold required for optimal growth of top-quality tobacco (Wu et al. 2014), aligning with the findings reported by Li et al. (2016). According to the above analysis, we speculate that soil alkaline nitrogen deficiency may be a triggering factor for tobacco plant susceptibility, consistent with the research results of Tian et al. (2011). Phosphorus is a limiting nutrient for plant growth in agricultural ecosystems (Hartmann and Six 2023). Our results indicate that continuous cropping can maintain the normal growth and development of tobacco despite the induced reduction in soil AP.
In summary, continuous cropping tends to acidify the soil and cause nutrient imbalances, depriving the roots of effective nutrients. Organic fertilizer and appropriate quicklime can improve the soil pH during tobacco cultivation. Growing application of organic fertilizers can also enhance the soil environment (Zhu et al. 2022; Zhang et al., 2022) and maintain fertility.
The characteristics of microbial communities in rhizosphere soil were significantly altered due to continuous cropping
Rhizosphere soil microorganisms can reflect the arable ecosystem health and are considered a valuable indicator of soil quality (Yang et al. 2021). Rhizosphere microorganisms, known as the second genome of plants (Berendsen et al. 2012), contribute to the conversion of substances in the soil. This process is crucial for plant nutrient acquisition, development, and stress resistance (Zhang 2020; Raaijmakers and Mazzola 2016; Berendsen et al. 2012). With growing years of continuous cropping, soil microbial communities shift from “bacterial” to “fungal” soils, as observed in the works of Li et al. (2012) and Liu et al. (2016). This study found that the biodiversity of microorganisms (bacteria and fungi) in the rhizosphere with replant disease decreased, consistent with the findings reported by Liu et al. (2022) and Hou et al. (2016). When the abundance of fungi increases, the microbiota within the rhizosphere shifts towards a fungal-dominated soil.
In continuous cropping soils, the relative abundance of Fusarium and Aspergillus decreases significantly, while that of Cladosporium increases significantly. These substances are typically considered plant pathogens (Ge et al. 2022; Li et al. 2017; Wu et al. 2018; Rao et al., 2022; Heuchert et al., 2005; Prasannath et al., 2021). Furthermore, Ilyonectria and Penicillium are harmful fungi inducing plant diseases (Shen et al. 2018; Bhatta 2022; Xu et al. 2021; Stošić et al. 2021), and their relative abundance decreases slightly. The relative abundance of Purpureocillium, Epicocum, Rhodanobacter, and Bradyrhizobium elevates; by contrast, Sphingomonas decreases in relative abundance; these species are commonly considered beneficial colonies for plants (Zhang et al. 2021a, b; Taguiam et al. 2021; Huang et al. 2021b; Huo et al. 2018; Wu et al. 2022; Hartmann and Six 2023). In summary, long-term continuous cropping disrupted the balance of soil microbial communities, consistent with the study by Wu et al. (2018). The relative abundance of beneficial microbial communities increased and that of harmful pathogen communities decreased, contrary to the research results of Hou et al. (2016), Li (2022), and Wang et al. (2020). We proposed two possible explanations for this divergence: firstly, a reduction in effective soil nutrients results from prolonged continuous cropping (as shown in Table 1), leading to rapid proliferation of beneficial bacteria to transform the soil nutrients, meeting crop growth requirements. For example, Rhodanobacter can adapt to low-nutrient and acidic conditions and participate in the nitrogen nutrition cycle (Avijit Ghosh et al. 2022; Heuvel et al. 2010), Bradyrhizobium, nitrogen-fixing bacteria, participates in soil nitrogen transformation (Wu et al. 2022; Hartmann and Six 2023). It is hypothesized that the causal agent of root rot (Fusarium) prompts the rapid proliferation of its antagonistic bacteria and fungi to uphold the balance of soil microecology. Notably, Purpureocillium effectively suppresses Fusarium-induced root rot (Zhang et al. 2021), and Rhodanobacter reportedly delivers antagonistic effects against root rot pathogens (Huo et al. 2018), and both flora pronouncedly elevates in successive cropping soils, as illustrated in Fig. 1 (c and d). In conclusion, continuous cropping cultivation disturbs the equilibrium among microbial communities in rhizosphere soils and alters the microbiome configuration.
The physicochemical properties impact the characteristics of rhizosphere soil microbial communities
The soil microbial community structure and plant growth are closely related to the physicochemical properties of soils (Liu et al. 2021a, b). Soil physicochemical properties can impact the structure and function of rhizosphere microorganisms (Song et al. 2019). The study reveals that soil acidification considerably decreases bacterial abundance and diversity, with minor changes observed in soil fungal communities. The changes reported here are consistent with the findings of the study conducted by Li et al. (2023). Organic matter and nitrogen are important components of microorganisms, providing nutrients for microorganisms (Obalum et al. 2017; Kuypers et al. 2018). In this study, it is found that SOM and AN significantly impact seven species (Bradyrhizobium, Rhodanobacter, Purpureocillium, Aspergillus, Cladosporium, Epicoccum, and Conocybe) between the two treatment groups, and Fusarium was only significantly positively correlated with AK. We hypothesize that decreased organic matter and nitrogen in continuous soils (refer to Table 1) enhances the dependence on parasitized tobacco for nutrients. Additionally, we propose that excessive Fusarium parasitism on tobacco can lead to diseases (i.e., root rot). It is demonstrated that Sphingomonas can degrade pesticide residues and organic contaminants in soils (Bhatia et al. 2022; Guo et al. 2010; Kant Bhatia et al., 2021; Zhang et al. 2021a), and soil physicochemical properties does not significantly impact the relative abundance of Sphingobium, which decreases in continuous cropping soils. Combining this observation with FAPROTAX analysis (Fig. 5), it is hypothesized that this reduction is due to the toxic effects of hazardous substance accumulation resulting from long-term continuous cropping. In addition, Penicillium and Aspergillus can exist in the soil as phosphate-solubilizing fungi (Li et al. 2023), requiring phosphorus as a nutrient. They decrease with the reduced AP, indicating a decrease in Phosphate-solubilizing content in the soil. In summary, the imbalance of soil physicochemical properties inhibits the proliferation of most rhizosphere soil microorganisms, as evidenced by the studies of Huang et al. (2021a), Yuan et al. (2019), and Zhong et al. (2020). Overall, changes in the physicochemical properties of the rhizosphere soils in continuous cropping are important factors contributing to rhizosphere microbial differences.
Significant changes were observed in the functional genes of rhizosphere soil bacteria and fungi
To elucidate the impact of continuous cropping on microbial activity, applying FAPROTAX functional prediction demonstrated a marked alteration in the relative abundance of genes implicated in the nitrogen cycle in the continuous cropping soils. The significant increase in the relative abundance of genes for nitrogen fixation and ureolysis suggests that bacteria actively contribute to nitrification. However, a pronounced reduction in the relative abundance of genes linked to nitrification is observed, specifically, aerobic ammonia, aerobic nitrite, and nitrite oxidation. This may be due to soil acidification arising from continuous cropping (Table 1) and the death of acid-intolerant bacteria. The notable rise in the proportion of aromatic compound degradation indicates a surge in soil organic pollutants, possibly attributed to pesticide residue and plastic remnants generated by the prolonged cultivation of tobacco crops for disease control.
Saprotrophic fungi represent the most significant fungal group within the fungal communities. They can degrade indissoluble residues of other organisms, including lignocellulose and chitin. This indicates that saprotrophic fungi are critical components of the carbon cycle’s metabolic process (Várnai A et al. 2014). This study found a significant reduction in the relative abundance of saprophytic fungi in the continuous soil. Consequently, we hypothesized that the carbon cycle of the soil’s metabolic processes had been negatively affected. Endophytic microorganisms are essential components of plants, residing within their tissues without inducing any disease (Porras-Alfaro et al., 2011). Importantly, fungal parasites are capable of transmitting viruses by contacting with the host plant through free spores and entering the plant by disrupting its cell walls (Gallet et al. 2018). Furthermore, they can attack all organisms utilizing a polysaccharide catabolic mechanism (Várnai A et al., 2014). The study found a significant increase in the relative abundance of fungal parasites in continuous cropping soils, which may be the cause of reduced levels of plant pathogens, animal pathogens and other pathogenic fungi, wood saprotrophs, dung saprotrophs, plant saprotrophs and other saprophytic fungi, and endophytes. Overall, continuous cropping has caused a shift in the rhizosphere soil fungi functions from saprotroph to pathotroph, resulting in a disturbance to the metabolic processes of the soil carbon cycle and an elevated risk of disease susceptibility in tobacco.
Conclusions
This study indicates that continuous cropping leads to rhizosphere soil acidification, nutrient imbalance, and carbon and nitrogen cycling involving microorganisms. It also reduces rhizosphere microbial diversity and increases pathogenic fungi abundance, major causes of continuous cropping disorder in tobacco. These findings complement the investigation of continuous cropping disorder in the Bijie city, Guizhou Province, providing an effective approach for managing continuous cropping in tobacco soils.
Availability of data and materials
All data generated and analyzed during this study are included in this article.
References
Amenu D, Bacha K (2023) Probiotic potential and safety analysis of lactic acid bacteria isolated from ethiopian traditional fermented foods and beverages. Ann Microbiol 73:37. https://doi.org/10.1186/s13213-023-01740-9
Bai YX, Shi PX, Yang CC, Jia M, Wang G, Yang HW, Xu ZL (2019) Analysis of the accumulation characteristics of phenolic acids in soil from continuous tobacco cultivation and their interaction relationships, Jiangsu Agricultural Sciences (3): 22–8. https://doi.org/10.11838/sfsc.1673-6257.18277.
Bai WQ, Hu MY, Wang CP, Jiang XY, Lei KR, Wu H (2020) A review of nitrogen utilisation in plants. Jiangsu Agricultural Science, 48:1–11. https://doi.org/10.15889/j.issn.1002-1302.2020.04.001.
Bao SD (2000) Soil Agricultural Chemical Analysis, 3rd edn. China Agriculture Press, Beijing
Berendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17(8): 478–86. https://doi.org/10.1016/j.tplants.2012.04.001.
Bhatia SK, Gurav R, Kim B, Kim S, Cho DH, Jung H, Kim YG, Kim JS, Yang YH (2022) Coproduction of exopolysaccharide and polyhydroxyalkanoates from Sphingobium yanoikuyae BBL01 using biochar pretreated plant biomass hydrolysate. Bioresour Technol 361:127753. https://doi.org/10.1016/j.biortech.2022.127753
Bhatta UK (2022) Alternative management approaches of citrus diseases caused by penicillium digitatum (green mold) and penicillium italicum (blue mold). Front Plant Sci 12. https://doi.org/10.3389/fpls.2021.833328
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu Y, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, Mcdonald D, Mciver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using qiime 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Callahan BJ, Mcmurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) Dada2: high-resolution sample inference from illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Edgar RC (2013) Uparse: highly accurate otu sequences from microbial amplicon reads. Nat Methods 10:996–998. https://doi.org/10.1038/nmeth.2604
Fujii K, Mitani R, Inagaki Y, Hayakawa C, Shibata M, Kosaki T, Ueda MU (2022) Continuous maize cropping accelerates loss of soil organic matter in northern thailand as revealed by natural 13c abundance. Plant Soil 474:251–262. https://doi.org/10.1007/s11104-022-05333-4
Gallet R, Michalakis Y, Blanc S (2018) Vector-transmission of plant viruses and constraints imposed by virus–vector interactions. Curr Opin Virol 33:144–150. https://doi.org/10.1016/j.coviro.2018.08.005
Ge XT, Dai K, Hu YX, Wang JM, Jiang N, Lu CH, Xia ZY (2022) Analysis of genetic diversity of Fusarium root rot fungi of tobacco in Yunnan Province. Acta Tabacaria Sinica 28(05):113–120. https://doi.org/10.16472/j.chinatobacco.2022.017.
Ghosh A, Manna MC, Jha S, Singh AK, Misra S, Srivastava RC, Srivastava PP, Laik R, Bhattacharyya R, Prasad SS, Singh SP, Singh SK, Kumar V, Tiwari S, Singh AK (2022) Chapter six - impact of soil-water contaminants on tropical agriculture, animal and societal environment. In: Sparks DL (ed) Advances in Agronomy. Academic Press, p. 209–274 https://doi.org/10.1016/bs.agron.2022.07.006.
Githongo M, Ngatia L, Kiboi M, Muriuki A, Fliessbach A, Musafiri C, Fu R, Ngetich F (2023) The structural quality of soil organic matter under selected soil fertility management practices in the central highlands of Kenya. Sustainability 15:6500. https://doi.org/10.3390/su15086500
Guo P, Wang B, Hang B, Li S, He J (2010) Sphingobium faniae sp. Nov., A pyrethroid- degrading bacterium isolated from activated sludge treating wastewater from pyrethroid manufacture. Int J Syst Evol Microbiol 60:408–412. https://doi.org/10.1099/ijs.0.009795-0
Guo FY, Wu ZH, Yan XM, Li JH, Han HG, Wang ZS, Cao XT, Liu QZ (2017) Effects of different crop rotation patterns on soil nutrients, root vigour and tobacco leaf quality of flue-cured tobacco forecrop. J Henan Agricult Sci 46(5):45–50. https://doi.org/10.15933/j.cnki.1004-3268.2017.05.008.
Hartmann M, Six J (2023) Soil structure and microbiome functions in agroecosystems. Nat Rev Earth Environ 4:4–18. https://doi.org/10.1038/s43017-022-00366-w
He C, Liu GS, Li ZL, Qiao BM, Dong NY, Jiang SJ (2011) Effects of continuous cropping on soil organic carbon and enzyme activities of tobacco planting and their relationship with soil-borne diseases. J Henan Agricult Univ 45:701–705. https://doi.org/10.16445/j.cnki.1000-2340.2011.06.011.
Heuchert B, Braun U, Schubert K (2005) Morphotaxonomic revision of fungicolous Cladosporium species (hyphomycetes)
Hou H, Dong K, Yang ZX, Dong Y, Tang L, Zheng Y. (2016). Progress of research on the mechanism of continuous crop disorder. Soils 48(06):1068–1076.https://doi.org/10.13758/j.cnki.tr.2016.06.002.
Huang Y, Wang Q, Zhang W, Zhu P, Xiao Q, Wang C, Wang WuL, Tian Y, Xu M, Gunina A (2021a) Stoichiometric imbalance of soil carbon and nutrients drives microbial community structure under long-term fertilization. Appl Soil Ecol 42:3269–3277. https://doi.org/10.1016/j.apsoil.2021.104119
Huang YL, Yin QX, Jiang SL, Bao XT, Wu X, Wang DL, Zhu C (2021b) Identification and biological characterisation of Epicoccum sorghinum, the causal agent of tea leaf spot disease. Chin J Trop Crops 42:3269–3277
Huo Y, Kang J, Park J, Li J, Chen L, Yang D (2018) Rhodanobacter ginsengiterrae sp. Nov., An antagonistic bacterium against root rot fungal pathogen fusarium solani, isolated from ginseng rhizospheric soil. Arch Microbiol 200:1457–1463. https://doi.org/10.1007/s00203-018-1560-9
Jacoby R, Peukert M, Succurro A, Koprivova A, Kopriva S (2017) The role of soil microorganisms in plant mineral nutrition—current knowledge and future directions. Front Plant Sci 8. https://doi.org/10.3389/fpls.2017.01617.
Jin Y, Yang YH, Duan YQ, Long YH, Ye CB. (2002). A preliminary study on the effect of continuous cropping of flue-cured tobacco on tobacco yield and quality. Tobac Sci Technol 41–45. https://doi.org/10.3969/j.issn.1002-0861.2002.01.016.
Kant BS, Gurav R, Choi YK, Choi TR, Kim HJ, Song HS, Mi LS, Lee PS, Soo LH, Kim YG, Ahn J, Yang YH (2021) Bioprospecting of exopolysaccharide from marine Sphingobium yanoikuyae BBL01: production, characterization, and metal chelation activity. Bioresour Technol 324:124674. https://doi.org/10.1016/j.biortech.2021.124674
Kuypers MMM, Marchant HK, Kartal B (2018) The microbial nitrogen-cycling network. Nat Rev Microbiol 16:263–276. https://doi.org/10.1038/nrmicro.2018.9
Li Z, Zu C, Wang C, Yang J, Yu H, Wu H (2016) Different responses of rhizosphere and non-rhizosphere soil microbial communities to consecutive piper nigrum l. Monoculture Sci Rep 6:35825. https://doi.org/10.1038/srep35825
Li HJ, Wang ZP, Song XL, Lin J, Liu YC, Zhao FX (2017) Investigation on the incidence of tobacco root rot disease in Pingdingshan tobacco area of Henan Province and research on the control effect of EM fungicide. J Agriculture 7:25–30
Li H, Li C, Song X, Liu Y, Gao Q, Zheng R, Li J, Zhang P, Liu X (2022) Impacts of continuous and rotational cropping practices on soil chemical properties and microbial communities during peanut cultivation. Sci Rep 12:2758. https://doi.org/10.1038/s41598-022-06789-1
Li X, Chen D, Carrión VJ, Revillini D, Yin S, Dong Y, Zhang T, Wang X, Delgado-Baquerizo M (2023) Acidification suppresses the natural capacity of soil microbiome to fight pathogenic fusarium infections. Nat Commun 14:5090. https://doi.org/10.1038/s41467-023-40810-z
Li X, Zhang XL, Sun BY, Yue BB, Zhang HH, Xu N, Zhu WX, Sun GY (2012) Effects of continuous cropping of flue-cured tobacco on enzyme activities and microbiota of ploughed soil. Soils 44:456–460. https://doi.org/10.13758/j.cnki.tr.2012.03.015.
Lisuma J, Mbega E, Ndakidemi P (2020) Influence of tobacco plant on macronutrient levels in sandy soils. Agronomy 10:418. https://doi.org/10.3390/agronomy10030418
Liu QZ, Guo FY, Wu ZH, Li BJ, Chen GQ, Zhu JW (2012) Soil barrier factors and control measures for continuous cropping of tobacco. Chin Agric Sci Bull 28:87–90
Liu Y, Jiang E, Wang GW, Zhang Y, Yang YQ, Yue MC, Liu SL, Wang Q (2016) Effects of different years of continuous cropping on physicochemical properties and microbiota of tobacco-planting soil. Chin Agric Sci Bull 32:136–140
Liu Q, Wang S, Li K, Qiao J, Guo Y, Liu Z, Guo X (2021a) Responses of soil bacterial and fungal communities to the long-term monoculture of grapevine. Appl Microbiol Biotechnol 105:7035–7050. https://doi.org/10.1007/s00253-021-11542-1
Liu SR, Wang HL, Sun H, Yang P, Jiang SY, Jiang GH (2022) Study on the effect of continuous cultivation of Pinellia ternata on rhizosphere soil microbial communities. Chin Trad Herbal Drugs 53:1148–1155
Liu F, He TB, Liu YS, Luo HB, Shu YG (2002) Nutrient changes in long-term continuous cropping yellow loam tobacco land and analysis of their fertiliser effects. Tobac Sci Technol :30–33.
Liu WS, Li HQ, He Yi, Huang YY, Qiu KY, Xie YZ (2021) Progress of rhizosphere microbial regulation of plant-soil interactions. Soil Fertil Sci China :318–327
Liu FT, Li MS, Yan CC, Peng ZL, Jiang YL, Huang JH, Zhang YF, Ren TB, Liu GS (2023) Analysis of soil microbial community structural changes and driving factors under continuous cropping conditions of flue-cured tobacco. J Huazhong Agricult Univ 42:139–146. https://doi.org/10.13300/j.cnki.hnlkxb.2023.02.018.
Louca S, Parfrey LW, Doebeli M (2016) Decoupling function and taxonomy in the global ocean microbiome. Science 353:1272–1277. https://doi.org/10.1126/science.aaf4507
Lu WH, Zhang NM, Bao L, Zhang L, Qin TF (2020) Progress of research on the characteristics and causes of continuous crop barriers and control measures in China's facility cultivation. Soils 52:651–658. https://doi.org/10.13758/j.cnki.tr.2020.04.001.
Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG (2016) Funguild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20:241–248. https://doi.org/10.1016/j.funeco.2015.06.006
Obalum SE, Chibuike GU, Peth S, Ouyang Y (2017) Soil organic matter as sole indicator of soil degradation. Environ Monit Assess 189:176. https://doi.org/10.1007/s10661-017-5881-y
Oldfield EE, Bradford MA, Wood ASA (2019) Global meta-analysis of the relationship between soil organic matter and crop yields. Soil 5:15–32. https://doi.org/10.5194/soil-5-15-2019
Porras-Alfaro A, Bayman P (2011) Hidden fungi, emergent properties: endophytes and microbiomes. Annu Rev Phytopathol 49:291–315. https://doi.org/10.1146/annurev-phyto-080508-081831
Prasannath K, Shivas RG, Galea VJ, Akinsanmi OA (2021) Novel Botrytis and Bladosporium species associated with flower diseases of macadamia in Australia. J Fungi 7:898. https://doi.org/10.3390/jof7110898
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
Raaijmakers JM, Mazzola M (2016) Soil immune responses. Science 352:1391–1392. https://doi.org/10.1126/science.aag1039
Rao D, Liu P, Zou L, Teng Y, Yu H, Yin G (2022) Effects of long-term continuous cropping and reductive soil disinfestation on fungal community in flue-cured tobacco rhizosphere. Soil Fertilizer Sci China 4:47–56
Shao QY, Dong HB, Han YF, Liang ZQ (2021) Progress in the study of rhizosphere microbiome of plants. J Plant Nutr Fertilizers 27:144–152
Shen GH, Xue QH, Zhao J (2018) Identification and biological characteristics of root rot pathogen of strawberry soil red husk fungus. Acta Agriculturae Boreali-Occidentalis Sinica 27:1032–1040
Song J, Han Y, Bai B, Jin S, He Q, Ren J (2019) Diversity of arbuscular mycorrhizal fungi in rhizosphere soils of the Chinese medicinal herb Sophora flavescens Ait. Soil Tillage Res 195:104423. https://doi.org/10.1016/j.still.2019.104423
Stošić S, Ristić D, Savković Ž, Grbić ML, Vukojević J, živković S, (2021) Penicillium and talaromyces species as postharvest pathogens of pear fruit (pyrus communis) in serbia. Plant Dis 105(11):3510–3521. https://doi.org/10.1094/PDIS-01-21-0037-RE
Taguiam JD, Evallo E, Balendres MA (2021) Epicoccum species: ubiquitous plant pathogens and effective biological control agents. Eur J Plant Pathol 159:713–725. https://doi.org/10.1007/s10658-021-02207-w
Tian L, Yin LC, Hu RZ, Luo LF, Peng Y, Zhou SG, Luo JX (2011) Analysis of soil nutrient status of tobacco plantation with different years of continuous cropping. Hunan Agricult Sci :49–51. https://doi.org/10.16498/j.cnki.hnnykx.2011.03.018.
Van Den Heuvel RN, Van Der Biezen E, Jetten MS, Hefting MM, Kartal B (2010) Denitrification at pH 4 by a soil-derived Rhodanobacter-dominated community. Environ Microbiol 12:3264–3271. https://doi.org/10.1111/j.1462-2920.2010.02301.x
Várnai A, Mäkelä MR, Djajadi DT, Rahikainen J, Hatakka A, Viikari L (2014) Chapter Four - carbohydrate-binding modules of fungal cellulases: occurrence in nature, function, and relevance in industrial biomass conversion. In: Advances in Applied Microbiology. Sariaslani S, Gadd GM (eds). Academic Press. pp 103–165. https://doi.org/10.1016/B978-0-12-800260-5.00004-8.
Wang S, Cheng J, Li T, Liao Y (2020) Response of soil fungal communities to continuous cropping of flue-cured tobacco. Sci Rep 10:19911. https://doi.org/10.1038/s41598-020-77044-8
Wood SA, Tirfessa D, Baudron F (2018) Soil organic matter underlies crop nutritional quality and productivity in smallholder agriculture. Agr Ecosyst Environ 266:100–108. https://doi.org/10.1016/j.agee.2018.07.025
Wu Y, Sui XH, Dai LX, Zheng YM, Zhang ZM, Tian YY, Yu TY, Sun XW, Sun QQ, Wu ZF (2022) Progress in the study of slow-growing rhizobacteria and their symbiotic mechanism with peanut. Chinese Agricult Sci 55:1518–1528
Wu CH, Liu JZ (2022) Progress in the study of rhizosphere microbial influences and their interactions with plants. J Hebei Norm Univ (Natural Science) 46:603–613. https://doi.org/10.13763/j.cnki.jhebnu.nse.202204014
Wu DZ, Luo HX, Song ZM, Guo GD, Chen YA, Li YX, Jiang YP, Li ZH (2014) Spatial variability and management zoning of major nutrients in soils of flue-cured tobacco in Qiannan City, Guizhou Province. Chin J Appl Ecol ;25:1701–1707. https://doi.org/10.13287/j.1001-9332.20140415.006.
Wu AZ, Cheng YZ, Wu SX, Liu GK, Zhang SX, Xiao SH (2018) Pathogen identification of Fusarium root rot of tobacco. Acta Tabacaria Sin 24:135–140. https://doi.org/10.16472/j.chinatobacco.2017.318
Wu L, Wang J, Wu H, Chen J, Xiao Z, Qin X, Zhang Z, Lin W (2018) Comparative metagenomic analysis of rhizosphere microbial community composition and functional potentials under Rehmannia glutinosa consecutive monoculture. Int J Mol Sci 19. https://doi.org/10.3390/ijms19082394
Xie D, Yuan W, Xie J, Shao L, Wang W, Yang T, Cai Y, Yan M, Jiang J, Wang W (2023) Influences of different proportions of organic fertilizer and nitrogen application rate on yield, quality and main chemical components of flue-cured tobacco. Jiangsu Agricult Sci 51:87–97. https://doi.org/10.15889/j.issn.1002-1302.2023.19.014
Xu Y, Wei J, Wei Y, Han P, Dai K, Zou X, Jiang S, Xu F, Wang H, Sun J, Shao X (2021) Tea tree oil controls brown rot in peaches by damaging the cell membrane of monilinia fructicola. Postharvest Biol Technol 175:111474. https://doi.org/10.1016/j.postharvbio.2021.111474
Yan P, Shen C, Fan L, Li X, Zhang L, Zhang L, Han W (2018) Tea planting affects soil acidification and nitrogen and phosphorus distribution in soil. Agr Ecosyst Environ 254:20–25. https://doi.org/10.1016/j.agee.2017.11.015
Yan P, Wu L, Wang D, Fu J, Shen C, Li X, Zhang L, Zhang L, Fan L, Wenyan H (2020) Soil acidification in Chinese tea plantations. Sci Total Environ 715:136963. https://doi.org/10.1016/j.scitotenv.2020.136963
Yang Z, Dai CC, Wang XX, Li XG (2019) Advances in rhizosphere microbial mechanisms of soil-borne fungal disease development in crops. Acta Pedol Sin 56:12–22
Yang JY, Zhao YW, Dong NY, Li JY, Liu XY, Li HC, Chang JB, Chen YC, Yang JJ (2021) Causes and control methods of tobacco continuous crop disorder. Modern Agricult Sci Technol :101–103.
Yang C, Yang WS, Lu YL, Pan YH, Ma A G, Zhang XL, Zhang XL Zhou FF (2022) Characterisation of soil nutrient characteristics of tobacco plantation at different altitudes in Chuxiong City, Yunnan Province. Hunan Agricult Sci :34–39. https://doi.org/10.16498/j.cnki.hnnykx.2022.006.009.
You C, Gao F, Wang Fj, Tang SK, Gu L, Zhang T, Xu ZB, Zhang ZY (2015) Effects of continuous cropping on rhizosphere microecology of flue-cured tobacco and quality of tobacco yield in Yunnan Province. Acta Tabacaria Sinica 21:60–67. https://doi.org/10.16472/j.chinatobacco.2014.151.
Yuan X, Niu D, Gherardi LA, Liu Y, Wang Y, Elser JJ, Fu H (2019) Linkages of stoichiometric imbalances to soil microbial respiration with increasing nitrogen addition: evidence from a long-term grassland experiment. Soil Biol Biochem 138:107580. https://doi.org/10.1016/j.soilbio.2019.107580
Zhang RF (2020) Rhizosphere microorganisms: a promising second genome for plants in greening agriculture. Biotechnol Bull 36:1–2
Zhang K, Yuan L, Shi X, Liang YJ (2010) Effects of different cultivation patterns on yield quality, soil nutrients and enzyme activities of flue-cured tobacco. J Plant Nutr Fertilizers 16:124–128
Zhang Y, Yang Y, Wei QH, Zhao M (2021a) Advances in the characterisation and application of Sphingomonas sp. Chem Bioengineering 38:6–13
Zhang WM, Qiu HZ, Zhang CH, Hai L (2018) Identification of components of potato root secretions and their biological effects in different years of continuous cropping. Chin J Eco-Agricult 26:1811–1818. https://doi.org/10.13930/j.cnki.cjea.180086.
Zhang YJ, Song MY, Zhang YJ, Fang Q, Yang J, Peng DL, et al (2021) Screening of fungi for the prevention and control of cucumber root rot and root knot nematode diseases: the fungi used in this study were Paecilomyces lilacinus and Trichoderma harzianum. Roast Biotechnol Bull 37:40–50. https://doi.org/10.13560/j.cnki.biotech.bull.1985.2020-0872
Zhang WB (2022) Effects of organic and chemical fertilizers allocation on humus binding pattern and microbial community structure in wheat-sweet potato rotation soil. Jiangsu Agricult Sci 50:247–252. https://doi.org/10.15889/j.issn.1002-1302.2022.17.040.
Zheng LW, Zhao YY, Wang YB, Huang YL, Fan FC, Liu SY (2022) Soil properties and microbial diversity of melon planted in different successive cropping years. Microbiology China 49:101–114. https://doi.org/10.13344/j.microbiol.china.210583.
Zhong Z, Li W, Lu X, Gu Y, Wu S, Shen Z, Han X, Yang G, Ren C (2020) Adaptive pathways of soil microorganisms to stoichiometric imbalances regulate microbial respiration following afforestation in the loess plateau. China Soil Biol Biochem 151:108048. https://doi.org/10.1016/j.soilbio.2020.108048
Zhu TC, Ming YF, Li CF, Xiang RY, Jiao SY, Li YQ (2022) Effects of additional organic fertiliser on carbon and nitrogen fractions and microbial communities in saline and alkaline land of the Yellow River Delta. J Soil Water Conserv 36:387–393. https://doi.org/10.13870/j.cnki.stbcxb.2022.06.047.
Acknowledgements
We would like to thank the anonymous reviewers for their valuable comments on this study. The work was supported by the Science and Technology Project of Guizhou Tobacco Company (Grant No. 2021XM21, 2022XM06), and the Science and Technology Project of Yunnan Academy of Tobacco Agricultural Sciences (Grant No. 2021530000241035).
Funding
This study was funded by the Science and Technology Project of Guizhou Tobacco Company (Grant No. 2021XM21, 2022XM06), and the Science and Technology Project of Yunnan Academy of Tobacco Agricultural Sciences (Grant No. 2021530000241035).
Author information
Authors and Affiliations
Contributions
MD: guide the completion of this experiment and revise their papers. BG and YZ: do experiments, data analysis completed the first draft paper. HC and HP: experiment, record data. YH and ZL: participating in the experiment, finished draft paper together with the second author. JB: guide and review manuscript revisions during the submission process. The authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gong, B., He, Y., Luo, Z. et al. Response of rhizosphere soil physicochemical properties and microbial community structure to continuous cultivation of tobacco. Ann Microbiol 74, 4 (2024). https://doi.org/10.1186/s13213-023-01748-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13213-023-01748-1