Abstract
Background
Accumulating evidence suggests that BMI1 confers protective effects against Alzheimer’s disease (AD). However, the mechanism remains elusive. Based on recent pathophysiological evidence, we sought for the first time to identify genetic variants in BMI1 as associated with AD biomarkers, including amyloid-β.
Methods
We used genetic, longitudinal cognition, and cerebrospinal fluid (CSF) biomarker data from participants in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort (N = 1565). First, we performed a gene-based association analysis of common single nucleotide polymorphisms (SNPs) (minor allele frequency (MAF) > 5%) located within ± 20 kb of the gene boundary of BMI1, an optimal width for including potential regulatory SNPs in the 5′ and 3′ untranslated regions (UTR) of BMI1, with CSF Aβ1-42 levels. Second, we performed cross-sectional and longitudinal association analyses of SNPs in BMI1 with cognitive performance using linear and mixed-effects models. We replicated association of SNPs in BMI1 with cognitive performance in an independent cohort (N=1084), Religious Orders Study and the Rush Memory and Aging Project (ROS/MAP).
Results
Gene-based genetic association analysis showed that BMI1 was significantly associated with CSF Aβ1-42 levels after adjusting for multiple testing using permutation (permutation-corrected p value=0.005). rs17415557 in BMI1 showed the most significant association with CSF Aβ1-42 levels. Participants with minor alleles of rs17415557 have increased CSF Aβ1-42 levels compared to those with no minor alleles. Further analysis identified and replicated the minor allele of rs17415557 as being significantly associated with slower cognitive decline rates in AD.
Conclusions
Our findings provide fundamental evidence that BMI1 rs17415557 may serve as a protective mechanism related to AD pathogenesis, which supports the results of previous studies linking BMI1 to protection against AD.
Similar content being viewed by others
Introduction
The etiology of non-familial Alzheimer's disease (AD) remains unclear despite extensive research efforts. In terms of genetic risks, researchers have focused on multiple common genetic risk factors with low effect sizes [1]. Recent large-scale genome wide association studies (GWAS) have identified more than 20 AD susceptibility loci [2]. Although common genetic variants have relatively small individual impact, the overall effect of multiple genetic risks can significantly increase the likelihood of developing AD [3].
The BMI1 gene encodes a 37kDa protein BMI1, a component of the polycomb repressive complex 1 (PRC1). BMI1 is involved in cell development, DNA damage response, cellular senescence regulation, stem cell renewal and differentiation, and oncogenesis [4]. In terms of aging, the reduction of BMI1 expression in aging cells was reported in vitro and in vivo studies [5, 6]. This might be related to one of the functions of BMI1, repressing cellular senescence [7]. Furthermore, recent studies have shown that BMI1 expression is reduced in AD brains but not in other types of dementia, such as frontotemporal dementia or dementia with Lewy bodies [8]. In line with that, BMI1 knock-out induced pluripotent stem cell (iPSC)-derived neurons induced pathologic characteristics of AD [8], and a mouse model study showed increased amyloid plaque, total Tau, and p-Tau levels in aged Bmi1-haplodeficient (Bmi1+/-) mice [9].
Although the association between BMI1 and AD in terms of gene expression and protein concentration has been reported [8], the effect of single nucleotide polymorphisms (SNPs) in the BMI1 gene in AD has not been studied. In view of recent pathophysiological evidence, we sought for the first time to investigate whether genetic variation in BMI1 is associated with a core AD biomarker and cognitive decline. Here, we report a genetic association analysis of SNPs in BMI1 with CSF Aβ1-42 levels and longitudinal cognitive performance in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort. We replicated association of SNPs in BMI1 with cognition performance in an independent Religious Orders Study and the Rush Memory and Aging Project (ROS/MAP) cohort.
Materials and methods
Subjects
Data used in the study were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI), a publicly available database (https://adni.loni.usc.edu) [10, 11]. A total of 1565 participants had genetic data. Of those participants, we used 1157 participants with cerebrospinal fluid (CSF) amyloid-β1-42 (Aβ1-42) levels and 1495 participants with longitudinal cognitive performance data (Table 1, Supplementary Figure 1).
Additionally, we used an independent dataset to validate ADNI findings from association analysis between genotype and cognitive function. The dataset is from two large cohorts maintained by investigators at the Rush Alzheimer’s Disease Center: the Religious Orders Study (ROS) and the Rush Memory and Aging Project (MAP) [12]. A total of 1084 participants had both genetic and longitudinal cognitive performance data.
Genotyping
The ADNI participants were genotyped using several Illumina genotyping platforms. Quality control (QC) procedures for participants and SNPs were performed as described previously [13]. After QC procedures, we selected only non-Hispanic participants of European ancestry and imputed un-genotyped SNPs separately in each platform using Markov Chain Haplotyping with the Haplotype Reference Consortium data as a reference panel [14]. The ROS/MAP whole genome sequencing libraries were prepared using the KAPA Hyper Library Preparation Kit in accordance with the manufacturer’s instructions and sequenced on an Illumina HiSeq X sequencer using pair-end read chemistry and read lengths of 150bp. The paired-end 150bp reads were aligned to the NCBI reference human genome (GRCh37) using the Burrows-Wheeler Aligner (BWA-MEM) [14, 15]. Local alignment was performed around indels to identify putative insertions or deletions in the region using the GATK (version 3.5) indel realignment tool. Base quality score recalibration was performed using the GATK BQSR. Variant calling and QC procedures have been described elsewhere [16]. Briefly, GATK HaplotypeCaller and GenotypeGVCFs modules were used to generate individual genotype calls in genomic VCF and VCF format. Following variant calling, the variant quality recalibration step in the GATK pipeline was used to empirically calibrate high quality variants. Variant-level QC was performed using PLINK, which includes checking genotype concordance using previous GWAS data, excluding variants with excess and/or systematic genotype missingness, examining departure from Hardy-Weinberg Equilibrium, and identifying Mendelian inconsistencies among related individuals.
CSF biomarkers
In ADNI, CSF Aβ1-42 and phosphorylated Tau (p-Tau) levels were measured by validated and highly automated Roche Elecsys electrochemiluminescence immunoassays (Roche Diagnostics, Mannheim, Germany) [17].
Amyloid positivity
Amyloid positivity was determined using CSF Aβ1-42 levels and 18F-Florbetapir positron emission tomography (PET) standardized uptake value ratios (SUVRs). In terms of CSF Aβ1-42, the provisional cut-point of 1073 pg/ml was used and 1.11 was used as the cut-point for 18F-Florbetapir PET SUVR. Participants who tested positive at least once during the follow up were labeled as amyloid positive.
Cognitive performance measures
As a cognitive performance measure for the ADNI participants, we used the AD Assessment Scale-cognitive subscale 13 (ADAS-cog13) [18], which includes 13 items (Word Recall, Naming Objects and Fingers, Commands, Constructional Praxis, Ideational Praxis, Orientation, Word Recognition, Language, Comprehension of Spoken Language, Word Finding Difficulty, Remembering Test Instructions, Delayed Word Recall, Number Cancellation or Maze Task) related to fundamental cognitive functions. The ROS/MAP participants underwent cognitive assessment using a battery of 21 cognitive performance tests. Nineteen of these tests across a range of cognitive abilities including 7 episodic memory tests (Word List Memory, Word List Recall, Word List Recognition, immediate and delayed recall of the East Boston Story and Story A from Logical Memory), 3 semantic memory tests (15-item Boston Naming Test, verbal fluency, 15-item word reading test), 3 working memory tests (Digit Span Forward, Digit Span Backward, Digit Ordering), 2 perceptual orientation tests (Line Orientation, 16-item progressive matrices), and 4 perceptual speed tests (Symbol Digits Modality-oral, Number Comparison, Stroop Color Naming, Stroop Word Naming) were used to construct a global composite measure of cognitive function. Further, information on this composite measure is published elsewhere [19, 20].
Statistical analysis
For gene-based association analysis, we selected common SNPs (MAF >5%) located within ± 20 kb of upstream and downstream regions of the BMI1 gene. We chose the 20 kb window, which provides an optimal width for including potential regulatory SNPs in the 5′ and 3′ untranslated regions (UTR) of BMI1, while controlling for false SNP-to-gene mappings due to larger windows. The gene-based association analysis with additive genetic models was performed using a set-based test in PLINK. Permutation (20,000 permutations) was used to adjust for multiple testing, which calculated an empirical p-value to determine the statistical significance of all SNPs in BMI1 jointly. For CSF Aβ1-42 levels, age, sex, and APOE ε4 carrier status were used as covariates. Furthermore, we performed association analysis of the SNP showing the most significant association with cognitive performance. We used a linear regression model with age, sex, and educational attainment as covariates. Longitudinal association analysis of the SNP with rates of cognitive decline was performed using a linear mixed-effects model under a missing at random hypothesis. The SNP genotype, time, the interaction term (SNP * time), age, sex, and educational attainment were treated as main effects. Random intercepts and slopes for time were used to accommodate individual variation. Because most (≃ 95%) ADNI participants had a follow-up period shorter than 10 years, we used data points up to ten years from baseline to ensure the robustness of our results. For the same reason, we included data points up to 17 years of follow-up for ROS/MAP. We performed sex- and APOE ε4 carrier status- and β-amyloid positivity-stratified analysis. In addition to the stratified analyses, we investigated the interaction between the SNP in BMI1 and grouping variables (sex, APOE ε4 carrier status, β-amyloid positivity). Because CSF Aβ1-42 levels and ADAS-cog13 scores showed a skewed distribution, we normalized the data using log transformation and square root transformation, respectively.
Results
We analyzed eight common SNPs (MAF > 5%) located within ± 20 kb of the gene boundary of BMI1 from HRC-based imputed ADNI GWAS data. Gene-based association analysis showed that BMI1 was significantly associated with CSF Aβ1-42 levels (permutation-corrected p = 0.005). The significance of associations, genomic locations, and linkage disequilibrium information between the eight SNPs are shown in Supplementary Figure 2. Among the eight SNPs, rs17415557 showed the most significant association with CSF Aβ1-42 levels (β (SE) = 0.116 (0.035), p = 0.001). This SNP was highly correlated with rs72814833 (R2 = 0.99), a 697 base pair upstream (5′) variant of BMI1, which was also significantly associated with CSF Aβ1-42 levels (β (SE) = 0.116 (0.036), p = 0.001). Participants with minor alleles (G) of rs17415557 have higher CSF Aβ1-42 levels, compared to those with no minor alleles (Fig. 1A). Stratified analysis by sex and amyloid positivity showed that this association was pronounced in males (male: β (SE) = 0.151 (0.047), p = 0.001; female: β (SE) = 0.075 (0.052), p = 0.152) and in amyloid-positive participants (amyloid-negative: β (SE) = 0.052 (0.035), p = 0.131; amyloid-positive: β (SE) = 0.099 (0.034), p = 0.004) (Table 2), although no significant interactions were found (p = 0.276 for sex * rs17415557, p = 0.473 for amyloid positivity* rs17415557). The association was significant in both APOE ε4 carrier status groups when stratified. For CSF p-Tau, a tau biomarker for AD, we did not find any significant associations between rs17415557 and CSF p-Tau levels (β (SE) = − 0.020 (0.032), p = 0.521).
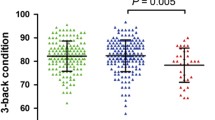
Association of BMI rs17415557 with CSF Aβ1-42 levels and rates of cognitive decline. A Participants with minor alleles (G) of rs17415557 have higher CSF Aβ1-42 levels, compared to those with no minor alleles (β (SE) = 0.05(0.02), p = 0.001). Participants with minor alleles of BMI1 rs17415557 had slower cognitive decline over time compared to those with no minor alleles; for B, ADAS-cog13 (a higher score indicating poor cognitive function) in ADNI, β (SE) = − 0.035 (0.016) and p = 0.024; for C, a global composite measure of cognitive function (a lower score indicating poor cognitive function) in ROS/MAP, β (SE) = 0.024 (0.010) and p = 0.020
For cognition performance at baseline, participants with minor alleles of rs17415557 showed higher ADAS-cog 13 scores in ADNI (β (SE) = 0.194 (0.070), p = 0.006). This association was pronounced in males (male: β (SE) = − 0.210 (0.086), p = 0.014; female: β (SE) = − 0.166 (0.117), p = 0.158) and in amyloid-positive participants (amyloid-negative: β (SE) = − 0.081 (0.099), p = 0.413; amyloid-positive: β (SE) = − 0.255 (0.095), p = 0.008). However, none of these cross-sectional association results of cognitive performance was replicated in ROS/MAP.
In order to investigate the effect of rs17415557 on rates of cognitive decline, we performed a longitudinal analysis of cognitive performance in two independent cohorts. The longitudinal analysis identified and replicated the significant association of rs17415557 with rates of cognitive decline (Fig. 1B, C). Participants with minor alleles of BMI1 rs17415557 had slower cognitive decline over time compared to those with no minor alleles, for ADAS-cog13 (a higher score indicating poor cognitive function) in ADNI, β (SE) = − 0.035 (0.016) and p = 0.024; for a global composite measure of cognitive function (a lower score indicating poor cognitive function) in ROS/MAP, β (SE) = 0.024 (0.010) and p = 0.020. The sex-stratified analysis showed that the impact of rs17415557 on rates of cognitive decline was stronger in females in both cohorts (for ADNI, male: β (SE) = − 0.021 (0.019), p = 0.277; female: β (SE) = − 0.051(0.025), p = 0.044; for ROS/MAP, male: β (SE) = 0.007 (0.017), p = 0.670; female: β (SE) = 0.032 (0.013), p = 0.012) (Table 2). However, the interaction between sex and the rate of cognitive decline (sex * rs17415557 * time) was not significant (p= 0.539).
We identified rs72814833, which is closely correlated with rs17415557 (R2 = 0.99) and a 697 base pair upstream (5′) variant of BMI1, as significantly associated with CSF Aβ1-42 levels, cognitive performance at baseline, and rates of cognitive decline, which was replicated in ROSMAP, as expected due to the strong correlation between rs72814833 and rs17415557 (Supplementary table 1).
Discussion
Here, we investigated the influence of genetic variants in BMI1 on CSF Aβ1-42 levels and rates of cognitive decline. Our gene-based association analysis showed that BMI1 was significantly associated with CSF Aβ1-42 levels. BMI1 rs17415557 with the most significant association with CSF Aβ1-42 levels was also significantly associated with rates of cognitive decline, which was replicated in an independent cohort. Notably, the T to G substitution of rs17415557 was associated with higher CSF Aβ1-42 levels and slower cognitive decline over time. In a recent large-scale GWAS from the International Genomics of Alzheimer’s Project [1], the major allele (T) of rs17415557 was nominally associated with AD (β (SE) = 0.071 (0.035), p = 0.045). These results imply that the minor allele (G) of rs17415557 may have a protective effect against AD.
Our first major finding was that BMI1 rs17415557 is associated with CSF Aβ levels. Interestingly, these findings were prominent within amyloid-positive subjects. This result is in line with a previous study where BMI1 gene expression levels were decreased only in AD and not in other dementias [8], since amyloid positivity is a hallmark of AD. One possible explanation for the relationship between BMI1 and AD suggested in a previous study is that BMI1 deficiency leads to increased p53 and GSK3β levels, which can impair proteasome function [8]. However, the role and molecular mechanism that BMI1 rs17415557 polymorphism may specifically play in the pathogenesis of AD warrants further investigation. CSF phosphorylated tau (pTau) levels were not significantly associated with BMI1 rs17415557, although the direction of association was consistent with that of CSF Aβ levels. Previous studies showed significant associations between BMI1 and pTau levels based on gene or transcript levels. Further investigations are needed to validate our finding for the association of CSF pTau levels with BMI1 rs17415557.
Another major finding was that the minor allele (G) of rs17415557 had a significant protective effect on cognitive decline over time. AD is a chronic progressive disorder showing substantial individual variation in the time-course of cognitive decline [21], making it crucial to predict the clinical trajectory [22]. rs17415557 appears to contribute to this clinical course variability. Notably, the protective effect of the G allele on cognitive decline is most prominent among females. It is not unusual for a SNP to have a differential effect between sexes. The APOE ε4 allele is also differentially associated with cognitive decline in males and females, particularly having a more significant impact on females [23].
BMI1 rs72814833, which is a 697 base pair upstream (5′) variant of BMI1 and highly correlated with rs17415557, is also significantly associated with CSF Aβ1-42 levels and rates of cognitive decline. BMI1 rs72814833 is known to bind with Egr1 protein as determined by the HaploReg v4.1 online tool (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) (Supplementary Table 2). Egr1 is a member of a zinc-finger transcription factor family, and a previous mouse model study showed that Egr1 −/− hematopoietic stem cells exhibited significantly elevated levels of BMI1 expression [24]. Although speculative, polymorphisms in the protein binding site might have influenced the action of Egr1, which could have contributed to our results. Our analysis for 3D chromatin interactions near BMI1 rs17415557 showed that several distant genes, including ARMC3, DNAJC1, and SKIDA1, also interacted with regions near rs17415557 at 3D chromatin, and their expression might also be regulated by rs17415557.
Limitations
A few limitations of this study should be noted. First, replication studies with independent larger samples are needed to confirm the association of BMI1 with CSF Aβ1-42 levels. Second, the mechanism by which the identified SNPs in BMI1 affect the BMI1 gene and hence Aβ level is unknown. Besides the protein-binding properties of the identified SNPs found on a public database, further functional studies are needed to determine the specific biochemical mechanism. Third, for cognitive performance, ADAS-cog13 was used in ADNI, whereas a different global composite measure of cognitive function was used in ROS/MAP. Nevertheless, it is noteworthy that this is the first study to show that rs17415557 and rs72814833, genetic variants located in the upstream region of the BMI1 gene, may play a protective role against AD.
Conclusion
In conclusion, the findings of our study from two independent well-characterized cohorts provide fundamental evidence that BMI1 and SNPs (rs17415557 and rs72814833) in BMI1 are associated with CSF Aβ1-42, a hallmark biomarker of AD, and cognitive decline rates. These findings support results of previous studies linking BMI1 to protection against AD [4, 5, 8]. Further studies, including animal models, are needed to investigate the molecular mechanisms underlying our findings.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the ADNI repository, (https://ida.loni.usc.edu/login.jsp?project=ADNI#) and AMP-AD Knowledge Portal (https://adknowledgeportal.synapse.org/).
Change history
20 January 2022
A Correction to this paper has been published: https://doi.org/10.1186/s13195-022-00960-6
Abbreviations
- AD:
-
Alzheimer’s disease
- CSF:
-
Cerebrospinal fluid
- ADNI:
-
Alzheimer’s disease neuroimaging initiative
- SNP:
-
Single nucleotide polymorphism
- ROS/MAP:
-
Religious Orders Study and the Rush Memory and Aging Project
- PRC1:
-
Polycomb repressive complex 1
- iPSC:
-
Induced pluripotent stem cell
- Aβ:
-
Amyloid-β
- NCBI:
-
National Center for Biotechnology Information
- QC:
-
Quality control
- PET:
-
Positron emission tomography
- SUVR:
-
Standardized uptake value ratio
- ADAS-cog13:
-
Alzheimer’s Disease Assessment Scale-cognitive subscale 13
- MAF:
-
Minor allele frequency
- SE:
-
Standard error
References
Kunkle BW, Grenier-Boley B, Sims R, et al. Genetic meta-analysis of diagnosed Alzheimer's disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet. 2019;51:414–30.
Andrews SJ, Fulton-Howard B, Goate A. Interpretation of risk loci from genome-wide association studies of Alzheimer's disease. Lancet Neurol. 2020;19:326–35.
Desikan RS, Fan CC, Wang Y, et al. Genetic assessment of age-associated Alzheimer disease risk: development and validation of a polygenic hazard score. PLoS Med. 2017;14:e1002258.
Bhattacharya R, Mustafi SB, Street M, Dey A, Dwivedi SK. Bmi-1: At the crossroads of physiological and pathological biology. Genes Dis. 2015;2:225–39.
Abdouh M, Chatoo W, El Hajjar J, David J, Ferreira J, Bernier G. Bmi1 is down-regulated in the aging brain and displays antioxidant and protective activities in neurons. PLoS One. 2012;7:e31870.
Itahana K, Zou Y, Itahana Y, et al. Control of the replicative life span of human fibroblasts by p16 and the polycomb protein Bmi-1. Mol Cell Biol. 2003;23:389–401.
Roe C, Behrens M, Xiong C, Miller J, Morris J. Alzheimer disease and cancer. Neurology. 2005;64:895–8.
Flamier A, El Hajjar J, Adjaye J, Fernandes KJ, Abdouh M, Bernier G. Modeling late-onset sporadic Alzheimer’s disease through BMI1 deficiency. Cell Rep. 2018;23:2653–66.
El Hajjar J, Chatoo W, Hanna R, et al. Heterochromatic genome instability and neurodegeneration sharing similarities with Alzheimer's disease in old Bmi1+/- mice. Sci Rep. 2019;9:594.
Hendrix JA, Finger B, Weiner MW, et al. The worldwide Alzheimer's disease neuroimaging initiative: an update. Alzheimers Dement. 2015;11:850–9.
Saykin AJ, Shen L, Yao X, et al. Genetic studies of quantitative MCI and AD phenotypes in ADNI: progress, opportunities, and plans. Alzheimers Dement. 2015;11:792–814.
Bennett DA, Buchman AS, Boyle PA, Barnes LL, Wilson RS, Schneider JA. Religious orders study and rush memory and aging project. J Alzheimers Dis. 2018;64:S161–89.
Park YH, Hodges A, Risacher SL, et al. Dysregulated Fc gamma receptor-mediated phagocytosis pathway in Alzheimer's disease: network-based gene expression analysis. Neurobiol Aging. 2020;88:24–32.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8.
De Jager PL, Ma Y, McCabe C, et al. A multi-omic atlas of the human frontal cortex for aging and Alzheimer's disease research. Sci Data. 2018;5:180142.
Bittner T, Zetterberg H, Teunissen CE, et al. Technical performance of a novel, fully automated electrochemiluminescence immunoassay for the quantitation of β-amyloid (1–42) in human cerebrospinal fluid. Alzheimers Dement. 2016;12:517–26.
Rosen WG, Mohs RC, Davis KL. A new rating scale for Alzheimer's disease. Am J Psychiatry. 1984;141;1356–1364.
Wilson RS, Barnes LL, Krueger KR, Hoganson G, Bienias JL, Bennett DA. Early and late life cognitive activity and cognitive systems in old age. J Int Neuropsychol Soc. 2005;11:400.
Wilson RS, Boyle PA, Yu L, et al. Temporal course and pathologic basis of unawareness of memory loss in dementia. Neurology. 2015;85:984–91.
Busse A, Hensel A, Gühne U, Angermeyer M, Riedel-Heller S. Mild cognitive impairment: long-term course of four clinical subtypes. Neurology. 2006;67:2176–85.
Jang H, Park J, Woo S, et al. Prediction of fast decline in amyloid positive mild cognitive impairment patients using multimodal biomarkers. NeuroImage Clin. 2019;24:101941.
Buckley RF, Mormino EC, Amariglio RE, et al. Sex, amyloid, and APOE ε4 and risk of cognitive decline in preclinical Alzheimer's disease: Findings from three well-characterized cohorts. Alzheimers Dement. 2018;14:1193–203.
Min IM, Pietramaggiori G, Kim FS, Passegué E, Stevenson KE, Wagers AJ. The transcription factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell Stem Cell. 2008;2:380–91.
Acknowledgements
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01AG024904) and DODADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Funding
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). The Religious Orders and the Rush Memory and Aging studies were supported by the National Institute on Aging grants P30AG10161, R01AG15819, R01AG17917, U01AG46152, and U01AG61356. Additional support for data analysis was provided by NLM R01 LM012535, NIA R03 AG063250, NIA R01 AG19771, NIA P30 AG072976, NIA U01 AG068057, NIA U01 AG072177, NLM R01 LM013463, DOD W81XWH-14-2-0151, NIGMS P50GM115318, and KHIDI HI19C1088.
Author information
Authors and Affiliations
Consortia
Contributions
JPK contributed to the conception and design of the study, analysis of data, and drafting of the manuscript. BK contributed to the design of the study and drafting of the manuscript. BJP contributed to the drafting of the manuscript. SWS contributed to the drafting of the manuscript. DAB contributed to the drafting of the manuscript. AJS contributed to the conception and design of the study, acquisition of data, and drafting of the manuscript. KN contributed to the conception and design of the study, analysis of data, and drafting of the manuscript. The authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The institutional review boards at all participating centers approved this study, and informed consent was obtained from the patients and caregivers.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised: the authors noticed that an 8 was inserted between CS and F of the word CSF in the article title.
Supplementary Information
Additional file 1: Supplementary Figure 1.
Flowchart for inclusion and exclusion of participants. ADNI = Alzheimer’s Disease Neuroimaging Initiative, GWAS = Genome-wide association study, CSF = Cerebrospinal Fluid, ADAS-cog = Alzheimer’s Disease Assessment Scale-cognitive subscale.
Additional file 2: Supplementary Figure 2.
Visualization of genomic locations, associations with Aβ, and linkage disequilibrium of eight SNPs. (A) Association map of the eight SNPs within 20kb of BMI1 gene (B) Linkage disequilibrium statistics (D’) between SNPs are shown.
Additional file 3: Supplementary Table 1.
Association of BMI1 rs72814833 with CSF Aβ42 and global cognitive function. Results of cross-sectional and longitudinal association analysis regarding BMI1 rs72814833.
Additional file 4: Supplementary Table 2.
Regulatory effects of the two SNPs of the BMI1 gene (HaploReg, v4.1, update 05.11.2015). Annotation results of BMI1 rs17415557 and rs72814833 from HaploReg v4.1web tool.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kim, J.P., Kim, BH., Bice, P.J. et al. BMI1 is associated with CSF amyloid-β and rates of cognitive decline in Alzheimer’s disease. Alz Res Therapy 13, 164 (2021). https://doi.org/10.1186/s13195-021-00906-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13195-021-00906-4