Abstract
Objectives
Male infertility accounts for approximately 30% of cases of reproductive failure. The characterization of genetic variants using cytogenomic techniques is essential for the adequate clinical management of these patients. We aimed to conduct a cytogenetic investigation of numerical and structural rearrangements and a genomic study of Y chromosome microdeletions/microduplications in infertile men derived from a single centre with over 14 years of experience.
Results
We evaluated 151 infertile men in a transversal study using peripheral blood karyotypes and 15 patients with normal karyotypes through genomic investigation by multiplex ligation-dependent probe amplification (MLPA) or polymerase chain reaction of sequence-tagged sites (PCR-STS) techniques. Out of the 151 patients evaluated by karyotype, 13 presented chromosomal abnormalities: two had numerical alterations, and 11 had structural chromosomal rearrangements. PCR-STS detected a BPY2 gene region and RBMY2DP pseudogene region microdeletion in one patient. MLPA analysis allowed the identification of one patient with CDY2B_1 and CDY2B_2 probe duplications (CDY2B and NLGN4Y genes) and one patient with BPY2_1, BPY2_2, and BPY2_4 probe duplications (PRY and RBMY1J genes).
Similar content being viewed by others
Introduction
Infertility is the inability of a sexually active couple to generate and maintain a pregnancy that results in a live foetus after trying for one year [1]. Male infertility (MI) accounts for approximately 30% of cases of reproductive failure [2], thus justifying its investigation.
The diagnosis of MI involves a series of laboratory and imaging tests. Among the genetic tests available, the G-banding karyotype is considered a valid technique for identifying numerical and structural alterations greater than 5 Mb [3, 4]. In addition, modern genetic tolls allow the identification of minor DNA alterations. On the long arm of the Y chromosome, for instance, the azoospermia factor (AZF) regions have multiple genes associated with fertility [5], that are susceptible to microdeletion and/or microduplication due to their ampliconic sequences organized as palindromes prone to nonallelic homologous recombination [6].
In addition to pregestational infertility, which is related to failure to conceive, including azoospermic and oligozoospermic men, there is gestational infertility when the couple is able to conceive but the embryo/foetus stops growing [7]. Thus, in addition to studying the genotype-phenotype relationship of patients with spermatic failure, it is also important to evaluate cases of repeated abortion presenting normal spermograms. There is strong evidence showing that normozoospermia is not synonymous with male fertility [8].
In this study, we present data from 2006 to 2019 from males with history of infertility, who attended our medical genetics Unit. Initial screening was performed using classical cytogenetics, and in selected cases, multiplex ligation-dependent probe amplification (MLPA) or polymerase chain reaction of sequence-tagged sites (PCR-STS) was performed to investigate single gene microduplications/microdeletions on the Y chromosome to assist in the genetic counseling of patients for infertility.
Main text
Subjects and methods
Patients
We present data from 151 patients attending the Medical Genetics Service of the Hospital do Servidor Público do Estado de São Paulo in Instituto de Assistência Médica do Servidor Público do Estado (HSPE-IAMSPE) from January 1, 2006, to November 30, 2019. These patients were aged between 27 and 49 years. These patients/couples were referred by the gynaecology, urology, and assisted reproduction units of the hospital with the complaint of infertility of unknown aetiology after performing clinical imaging, and laboratory tests.
This study retrospectively analysed the medical records of infertile patients between 2006 and 2017. Additional data were obtained from laboratory tests performed between 2018 and 2019.
In total, 15 patients were selected for Y chromosome genomic investigation by MLPA and/or PCR-STS methods. Among these patients, we included pregestational infertile men (with azoospermia or oligozoospermia) and gestational infertile men (with a history of recurrent miscarriages).
Eligibility criteria for the genomic investigation encompassed couples that did not have a successful full-term pregnancy by natural methods for a minimum of one year. The men had a normal karyotype, sex hormone patterns within the reference values, and at least one spermogram. Their wives/partners had laboratory tests and imaging exams showing normal features of the reproductive system, a normal karyotype, and adequate hormonal patterns, without any apparent or diagnosed cause, that would justify infertility. Some of these women reported the birth of children with a previous partner. Therefore, only those male subjects with a history of infertility were included, where possible confounding factors for infertility of the female partners were ruled out.
Exclusion criteria for genomic analysis consisted of patients presenting (1) chromosomal alterations identified by karyotype examination, (2) obstruction of the urogenital pathways, (3) previous disease that could justify infertility, such as varicocele, mumps, or any other condition indicated by the urologist, (4) no sperm test, and (5) normozoospermia, without the occurrence of recurrent spontaneous abortions. In addition, we excluded patients selected for cytogenetic screening but could not be contacted for genomic analysis (in the retrospective selection of patients: between 2006 and 2017).
Study design is presented in Fig. 1.
We carefully investigated patients before cytogenomic evaluation on their clinical condition, age, outcome of reproductive methods, and spontaneous and recurrent abortions. All procedures were carried out in accordance with the Declaration of Helsinki.
Methods
We performed cytogenetic analysis through the G-bands technique by acetic saline using Wright. At least 20 metaphases were analysed for each patient using light microscopy.
We performed genomic analysis by the MLPA technique using the SALSA MLPA probe-mix P360 version B1 and PCR-STS in an affiliated laboratory to evaluate, AZFa, AZFb, and AZFc regions (sY84, sY86, sY127, sY134, sY254, and sY255).
Results and discussion
Karyotype analysis
Out of 151 male patients with history of infertility, 13 were found with chromosomal anomalies; two with numerical and 11 with structural anomalies or polymorphism.
Klinefelter syndrome (KS) is the most frequently identified genetic cause of MI [9]. We detected this syndrome in two patients with numerical chromosomal alterations: one was pure Klinefelter with a 47,XXY karyotype, and the other was a mosaic mos 47,XXY[46]/46,XY[04] karyotype.
Amongst the structural anomalies observed, the chromosome 9 inversion was found in seven patients with karyotype 46,XY,inv(9)(p12;q13)[20]. The other structural alterations were 46,XX[100], 46,XY,t(3;4)(q13;q34)[20], 46,XY,t(X;3)(p22;p11)[20], and 46,XY,inv(Y)(p11?;q11?)[20], each of which was identified in one different patient.
Structural chromosomal alterations, such as reciprocal translocations, Robertsonian translocations, and chromosome 9 inversions, play a significant role in MI, similar to polymorphic structural alterations that affect fertility [10,11,12,13,14].
Genomic analysis
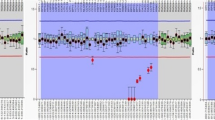
Regarding the genomic analysis, of the 11 cases with sperm failure (pregestational infertility), six patients had azoospermia, four had oligozoospermia, and one had oligoasthenozoospermia. Among these 11 cases, one azoospermic patient presented a duplication of the CDY2B gene probes (CDY2B_1 and CDY2B _2) analysed by the MLPA technique (Fig. 2A). Another azoospermic patient presented a partial microdeletion of the AZFc region detected by PCR-STS at loci sY254 and sY255 (in the BPY2 gene and RBMY2DP pseudogene region). Of the four cases with normozoospermia and miscarriages (gestational infertility), one patient had Y chromosome duplications in BPY2 gene probes. This patient presented two spermograms with normal counts (58.0 and 61.2 million/mL), motility (66 and 38%), and viability (70 and 79%). He had a history of approximately six years of reproductive failure and four spontaneous abortions, all of which occurred in the gestational period of 4 to 8 weeks. The patient presented duplications in three of the five probes of the BPY2 gene (Fig. 2B).
MLPA abnormal results. (A) Genomic analysis of the pregestational infertile patient with CDY2B gene probes region duplication. Histogram of the results of each probe containing the CDY2B_1 and CDY2B_2 probes above the reference values. MLPA is a comparative test, and the analysis was normalized to the number of suitable copies (x2) considering that all probes presented as duplicates have more than two copies of the gene. (B) Genomic analysis of the gestational infertile patient with BPY2 probes region duplication. Histogram of the results of each probe containing probes BPY2_1, BPY2_2 and BPY_4 above the reference values. MLPA analysis was normalized to the number of suitable copies (x3) considering that all probes presented as duplicates have more than three copies – Coffalyser software
The clinical characteristics and genomic analysis of patients with alterations on the Y chromosome are shown in Table 1.
It is interesting to point out that the Y chromosome microduplications observed would not be detected using PCR-STS technique, considered to be the gold standard method for genomic analysis [15,16,17].
Although Y chromosome gene microdeletions are well established as one of the most common causes of male infertility [18], the consequences of its microduplications have not yet been fully established [19].
Noordam et al. (2011) found decreased sperm count and motility in men with primary AZFc microduplications (no microdeletions) [16]. Johansson et al. (2015) suggested that microduplications of regions on the Y chromosome could interfere with fertility by altering gene dosage [20]. Singh et al. (2019) reported that an overdose of genes on the Y chromosome and on specific autosome regions may impair spermatogenesis [21].
The CDY2B gene probes region variation is classified as a variant of uncertain significance (VUS) for spermatogenesis (score 0) by the Franklin tool database (http://franklin.genoox.com) due to a lack of information associated with the region of the variant using ACMG 2020 criteria [22]. This region embraces CDY2B and NLGN4Y genes. Ghorbel et al. (2014) found significantly decreased spermatogenesis in infertile men with CDY1B gene deletions compared with fertile control individuals [23]. Machev et al. (2004) suggested a strong association between CDY1 gene deletions and infertility [24]. Thus, alterations in the CDY gene family may affect fertility. Although we cannot claim that the duplication of the CDY2B gene detected in the present study is related to azoospermia, we believe that this alteration may have an effect on spermatogenesis. An increase in histone hyperacetylation in sperm DNA, a known function of the CDY2B gene [25], may promote increased chromatin decondensation, disrupting normal DNA condensation during meiosis.
Previous studies have found significant differences between infertile and fertile patients, indicating that Y chromosome microduplications may interfere with normal spermatogenesis [19, 26,27,28]. However, those reports on azoospermic and oligozoospermic men described partial AZFc microduplications of regions including several genes in clusters. Moreover, normozoospermic infertile men, such as those with a history of repeated spontaneous abortions, are not frequently reported in the literature. It is now known that male infertility is related to several factors, in addition to abnormal seminal parameters, since normozoospermic men may still be infertile [10].
The BPY2 probes region variation is regarded as a VUS for infertility by the Franklin tool due to a lack of information associated with the region of the variant using ACMG criteria [22]. One of our patients presenting duplication of the BPY2 probes region had a history of miscarriage. There are two genes in this region: RBMY1J and PRY. Although we cannot say that duplications of these genes directly affect male fertility, we cannot exclude the possibility that this situation occurs because it has been suggested that PRY gene has a function in apoptosis [29]. Thus, if the apoptosis occurs in excess, it may affect the course of gestation and result in embryo defects incompatible with growth and/or spontaneous abortion. The patient with this alteration showed duplication of three out of the five BPY2 gene probes used in the MLPA kit, revealing duplication of PRY and RBMY1J genes. Microduplications in regions of the PRY gene identified in the present study may be related to embryo development arrest due to excess of apoptosis.
Another patient presented a microdeletion in the BPY2 gene region, which may be associated with his azoospermia. This is because the absence of the BPY2 gene may affect male fertility by decreasing catalysis processes during spermatogenesis. This gene encodes a protein that interacts with ubiquitin ligase E3A (UBE3A) in the testes [24]. UBE3A promotes ubiquitination, a normal process that catalyses molecules that are no longer needed and plays a crucial role in metabolizing substituted histones in late spermatids. Additionally, the lack of degradation of histones released in the process results in the absence of adequate sperm formation or maturation, as observed in a testicular biopsy of this patient (data not shown).
In addition to the Y chromosome, several autosomal genes are crucial for human spermatogenesis and embryo/fetal development [30, 31]. Genes on the Y chromosome may modulate or regulate the function of autosomal genes, as their functions are not yet perfectly understood. Microduplications on the CDY2B gene may have a regulatory role on different genes, leading to spermatogenesis failure. On the other hand, microduplications on the PRY gene may affect genes essential for normal embryo or fetal development resulting in repeated abortions. More studies on gene expression and sperm DNA fragmentation are necessary to understand the individual role of genes on Y chromosome in male fertility. Epigenetic alterations and noncoding RNAs may also interfere with male fertility [32]. DNA methylation profile and noncoding RNA expression studies would help to clarify specific male infertility conditions. Moreover, the recent report of the entire sequence of the Y chromosome highlights the relevance of more studies of genomic variants related to male infertility, using the new reference data set T2T-CHM13 + Y [33].
In conclusion, our results describe the presence of new Y chromosome genomic abnormalities in infertile male patients.
Limitations
The limitations of the study are that it was limited to the use of two genomic techniques and they were employed in a reduced number of infertile male patients.
Data availability
The genomic datasets generated during the current study are publicly available at ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/submitters/509274): accession number SUB13879430.
Abbreviations
- AZF:
-
azoospermia factor
- HSPE - IAMSPE:
-
Hospital do Servidor Público do Estado de São Paulo in Instituto de Assistência Médica do Servidor Público do Estado
- KS:
-
Klinefelter syndrome
- MI:
-
male infertility
- MLPA:
-
multiplex ligation-dependent probe amplification
- PCR-STS:
-
polymerase chain reaction of sequence-tagged sites
- VUS:
-
variant of uncertain significance
References
Zegers-Hochschild F, Adamson GD, de Mouzon J, Ishihara O, Mansour R, Nygren K et al. International Committee for Monitoring Assisted Reproductive Technology (ICMART) and the World Health Organization (WHO) revised glossary of ART terminology, 2009. Fertil Steril. 2009;92(5):1520-4.
Agarwal A, Mulgund A, Hamada A, Chyatte MR. A unique view on male infertility around the globe. Reprod Biol Endocrinol. 2015;13:37.
Seabright M. A rapid banding technique for human chromosomes. Lancet. 1971;2(7731):971–2.
Sanchez O, Escobar JI, Yunis JJ. A simple G-banding technique. Lancet. 1973;2(7823):269.
Lin YM, Lin YH, Teng YN, Hsu CC, Shinn-Nan Lin J, Kuo PL. Gene-based screening for Y chromosome deletions in Taiwanese men presenting with spermatogenic failure. Fertil Steril. 2002;77(5):897–903.
Krausz C, Chianese C, Giachini C, Guarducci E, Laface I, Forti G. The Y chromosome-linked copy number variations and male fertility. J Endocrinol Invest. 2011;34(5):376–82.
Li D, Zhang H, Wang R, Zhu H, Li L, Liu R. Chromosomal abnormalities in men with pregestational and gestational infertility in northeast China. J Assist Reprod Genet. 2012;29(8):829–36.
Krausz C. Male infertility: pathogenesis and clinical diagnosis. Best Pract Res Clin Endocrinol Metab. 2011;25(2):271–85.
Rutherford TR, Matson P. Klinefelter syndrome: phenotype, testicular function and infertility treatment. J Reprod Biotechnol Fertil. 2019;8:66–79.
Practice Committee of the American Society for Reproductive M. Diagnostic evaluation of the infertile male: a committee opinion. Fertil Steril. 2015;103(3):e18–25.
Xie X, Li F, Tan W, Tang J. Analysis of the clinical features of pericentric inversion of chromosome 9. J Int Med Res. 2020;48(9):300060520957820.
Elhady GM, Kholeif S, Nazmy N. Chromosomal aberrations in 224 couples with recurrent pregnancy loss. J Hum Reprod Sci. 2020;13(4):340–8.
Serapinas D, Valantinaviciene E, Machtejeviene E, Bartkeviciute A, Bartkeviciene D. Evaluation of chromosomal structural anomalies in Fertility disorders. Med (Kaunas). 2021;57(1).
Rawal L, Kumar S, Mishra SR, Lal V, Bhattacharya SK. Clinical manifestations of chromosomal anomalies and polymorphic variations in patients suffering from Reproductive failure. J Hum Reprod Sci. 2020;13(3):209–15.
Krausz C, Hoefsloot L, Simoni M, Tuttelmann F. European Academy of A, European Molecular Genetics Quality N. EAA/EMQN best practice guidelines for molecular diagnosis of Y-chromosomal microdeletions: state-of-the-art 2013. Andrology. 2014;2(1):5–19.
Noordam MJ, Westerveld GH, Hovingh SE, van Daalen SK, Korver CM, van der Veen F, et al. Gene copy number reduction in the azoospermia factor c (AZFc) region and its effect on total motile sperm count. Hum Mol Genet. 2011;20(12):2457–63.
Foresta C, Moro E, Ferlin A. Y chromosome microdeletions and alterations of spermatogenesis. Endocr Rev. 2001;22(2):226–39.
Zhang YS, Dai RL, Wang RX, Zhang HG, Chen S, Liu RZ. Analysis of Y chromosome microdeletion in 1738 infertile men from northeastern China. Urology. 2013;82(3):584–8.
Lu C, Jiang J, Zhang R, Wang Y, Xu M, Qin Y, et al. Gene copy number alterations in the azoospermia-associated AZFc region and their effect on spermatogenic impairment. Mol Hum Reprod. 2014;20(9):836–43.
Johansson MM, Van Geystelen A, Larmuseau MH, Djurovic S, Andreassen OA, Agartz I, et al. Microarray analysis of Copy Number variants on the human Y chromosome reveals novel and frequent duplications overrepresented in specific haplogroups. PLoS ONE. 2015;10(8):e0137223.
Singh V, Bala R, Chakraborty A, Rajender S, Trivedi S, Singh K. Duplications in 19p13.3 are associated with male infertility. J Assist Reprod Genet. 2019;36(10):2171–9.
Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med. 2020;22(2):245–57.
Ghorbel M, Baklouti-Gargouri S, Keskes R, Chakroun N, Sellami A, Fakhfakh F, et al. Deletion of CDY1b copy of Y chromosome CDY1 gene is a risk factor of male infertility in Tunisian men. Gene. 2014;548(2):251–5.
Machev N, Saut N, Longepied G, Terriou P, Navarro A, Levy N, et al. Sequence family variant loss from the AZFc interval of the human Y chromosome, but not gene copy loss, is strongly associated with male infertility. J Med Genet. 2004;41(11):814–25.
Lahn BT, Tang ZL, Zhou J, Barndt RJ, Parvinen M, Allis CD, et al. Previously uncharacterized histone acetyltransferases implicated in mammalian spermatogenesis. Proc Natl Acad Sci U S A. 2002;99(13):8707–12.
Lin YW, Hsu LC, Kuo PL, Huang WJ, Chiang HS, Yeh SD, et al. Partial duplication at AZFc on the Y chromosome is a risk factor for impaired spermatogenesis in Han Chinese in Taiwan. Hum Mutat. 2007;28(5):486–94.
Ye JJ, Ma L, Yang LJ, Wang JH, Wang YL, Guo H, et al. Partial AZFc duplications not deletions are associated with male infertility in the Yi population of Yunnan Province, China. J Zhejiang Univ Sci B. 2013;14(9):807–15.
Yang B, Ma YY, Liu YQ, Li L, Yang D, Tu WL, et al. Common AZFc structure may possess the optimal spermatogenesis efficiency relative to the rearranged structures mediated by non-allele homologous recombination. Sci Rep. 2015;5:10551.
Stouffs K, Lissens W, Verheyen G, Van Landuyt L, Goossens A, Tournaye H, et al. Expression pattern of the Y-linked PRY gene suggests a function in apoptosis but not in spermatogenesis. Mol Hum Reprod. 2004;10(1):15–21.
Okutman O, Rhouma MB, Benkhalifa M, Muller J, Viville S. Genetic evaluation of patients with non-syndromic male infertility. J Assist Reprod Genet. 2018;35(11):1939–51.
Vallet-Buisan M, Mecca R, Jones C, Coward K, Yeste M. Contribution of semen to early embryo development: fertilization and beyond. Hum Reprod Update. 2023;29(4):395–433.
Gunes S, Arslan MA, Hekim GNT, Asci R. The role of epigenetics in idiopathic male infertility. J Assist Reprod Genet. 2016;33(5):553–69.
Rhie A, Nurk S, Cechova M, Hoyt SJ, Taylor DJ, Altemose N et al. The complete sequence of a human Y chromosome. Nature. 2023.
Acknowledgements
We are grateful to all patients, the IAMSPE staff and collaborators that took part in the study.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
ATD, LDK and JW conceived and designed the study. MRGA recruited the subjects for MLPA tests and with AB gathered patient data and undertook karyotypes. AB analysed and interpreted the karyotypes. FARM, GFSC, AMN and MRGA performed MLPA experiments and helped to analyse and interpret MLPA results. ATD with EAZ and BW analysed and interpreted MLPA results. MRGA and ABM performed critical discussion and text draft and ATD, ABM and LDK contributed to drafting the final version of the manuscript. All authors contributed to this research, revising it critically, approved the final manuscript to the submission and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The protocol for the research for genomic analysis project was approved by the Instituto de Assistência Médica ao Servidor Público Estadual (IAMSPE) Ethics Committee, São Paulo, Brasil under the number CAAE 79306117.7.0000.5463, and written informed consent was obtained from all participants. The retrospective study of medical records was approved under the number CAAE 36467620.1.0000.5463 by the IAMSPE Ethics Committee. All the procedures were followed in accordance with the ethical standards of the responsible committee on human experimentation and with the Helsinki Declaration of 1964 and its later amendments.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Adriano, M.R.G., Bortolai, A., Madia, F.A.R. et al. Cytogenetics investigation in 151 Brazilian infertile male patients and genomic analysis in selected cases: experience of 14 years in a public genetic service. BMC Res Notes 17, 67 (2024). https://doi.org/10.1186/s13104-024-06710-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13104-024-06710-1