Abstract
Objectives
DNA Barcoding has proven to be a reliable method for rapid insect identification. The success of this method is based on the amplification of a specific region, the ‘Folmer’ barcode region at the 5´ start of the cytochrome c oxidase 1 gene (cox1), with universal primers. Previous studies showed failures of standard “universal” primers to amplify this region in psyllids. The aim of the study was the design of a new alternative more reliable primer combination for taxa of the superfamily Psylloidea and its comparison with the performance of the standard “universal” Folmer-primers.
Results
A newly designed degenerate forward primer LCOP-F was developed following comparison of the sequence alignment of the priming site of “universal” primer LCO1490 and the standard insect forward primer LepF1. When combined with the “universal” reverse primer, HCO2198, this new primer pairing was able to generate barcode sequence for all 36 species in 20 genera across the five families of psyllids tested in this study, and these primers were found to be more universally reliable across psyllid taxa than other primer pairs tested.
Similar content being viewed by others
Introduction
Over 4000 species of psyllid, or jumping plant-lice, (Hemiptera: Psylloidea) are described worldwide [1] and it has been estimated that twice as many species are still undescribed [2]. Due to their host specific behaviour, some psyllids have been used as biological control agents of exotic plants [3, 4]. Others are among the most devastating pests worldwide due to their ability to transmit plant pathogens of the genus “Candidatus Liberibacter” [5], such as the tomato and potato psyllid (Bactericera cockerelli Šulc) associated with Zebra chip disease, and the Asian and African citrus psyllids (Diaphorina citri Kuwayama and Trioza erytreae Del Gercio, respectively) associated with huanglongbing disease [6,7,8,9]. Rapid and accurate identification of psyllid species is therefore crucial in many applications, including biosecurity and biodiversity assessments. However, morphological identification of psyllids is challenging for three main reasons: the lack of taxonomic keys, the lack of distinguishing morphological features in some immature stages and some adults (especially females in certain groups), and the presence of seasonal dimorphism [10].
Success of the DNA barcoding method, and construction of reference DNA barcode libraries for taxon identification, relies on the efficiency of “universal” primers to amplify the same gene region for many different taxa. The use of standard mitochondrial primers for invertebrate species identification and systematics was proposed by Folmer et al. [11] and Simon et al. [12], among others, with one particular gene, cytochrome c oxidase 1 gene (cox1), promoted as the best candidate region for a standard DNA barcode for animals [13]. In one of the earliest publications on DNA barcoding, Folmer et al. proposed the cox1 gene and a set of potentially “universal” primers (LCO1490/HCO2198) to amplify the 5’ end of cox1 across a diverse group of invertebrate phyla including molluscs, echinoderms, and tardigrades [11]. Subsequently, studies showed that these “universal” primers performed poorly for certain taxonomic groups [14,15,16].
The cox1 DNA barcode has proven to be a highly effective tool for identification in psyllids [10, 17, 18], but in most cases researchers use different sets of primers to amplify slightly different parts of cox1 and often need more than one primer set to obtain amplification across all focal taxa [10, 19]. The option to have a single primer pair that would reliably amplify and sequence across the superfamily Psylloidea as well as maximize the quantity and informativeness of the data has not been available for psyllids. Furthermore, the use of different primer pairs to amplify slightly different regions of cox1 prevents the construction of a comparative worldwide DNA barcode library for psyllids due to variable lengths and different placement of sequence alignments from different studies.
A screen of publicly available sequences of psyllid taxa in BOLD Systems and GenBank reveals that less than 30% of the species are represented by a cox1 sequence with length > 500 bp, which is a length proposed as a requirement for a standard barcode [20]. Furthermore, only 20% of these have been amplified with the “universal” primers proposed by Folmer et al. [11]. The remaining sequences (80%) have been amplified with many different primer pairs such as the modified primer set developed specifically for Lepidoptera [21] that became standard for many insect taxa, LepF1/LepR1 (but accounts for less than 5% of psyllid sequences); or various alternatives designed for specific geographically focused studies [19, 22,23,24,25] (see Additional file 1: Table S1).
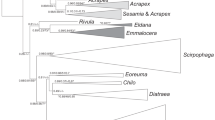
The poor amplification and universality of most of these primers positioned at the start of cox1 is a result of a not particularly conserved sequence in the first 100–200 bp of the gene. For this reason, a number of researchers have selected to use forward primers upstream of the start of the gene (e.g., primer pair C1-J-1718/C1-N-2191 from Simon et al. [12], C1-J-1709/HCO2198 used in Martoni et al. [17], and PsyCOI-F3 from Martoni et al. [19], (Fig. 1), but then yield a shorter total sequence length. We therefore focused on designing a new degenerate forward primer at the start of cox1 that can potentially be paired with a number of proven reverse primers. We tested the performance of this new forward primer on 36 species from 20 genera in five families of Psylloidea. In addition, we compared the efficacy of our new degenerate forward primer against previous standard primers, LCO1490/HCO2198 from Folmer et al. [11] and LepF1/LepR1 from Hebert et al. [21].
Our primary aim in introducing a new degenerate forward primer at the 5′ start of the cox1 gene is to help standardize DNA barcoding in psyllids and promote sequencing of the same primary psyllid barcode, even if in addition to other gene regions.
Methods
Species collection and DNA extraction
A total of 154 specimens from 36 species, in 20 genera across five of the seven families of Psylloidea as classified by Burckhardt et al. [26] were used in this study. Additional sample information is provided in Additional file 2: Table S2. All specimens were morphologically identified to species based on keys in Hodkinson [27], Hodkinson [28], Rapisarda [29] and Bastin et al. [30]. DNA was extracted from whole individuals using a Chelex non-destructive protocol [31] as follows: each specimen was immersed in 10 ml of Proteinase K (10 mg/ml) and 150 µl of Chelex diluted at 10% and incubated in the thermoblock at 55 °C for 12 h. The solution was used directly as the DNA extraction while the specimen was removed and retained as a DNA voucher, preserved in 70% ethanol and deposited in the Instituto Canario de Investigaciones Agrarias (ICIA), Spain.
Primer design, PCR and sequencing
To design the new forward primer, sequences of cox1 and flanking regions from publicly available psyllid data (NCBI) (e.g., from complete mitochondrion) were assembled into an alignment using ClustalW in Mega 7 [32]. Additional sequences of the tRNA-W region upstream of the 5′ end of cox1, were also obtained using the primers tRWF1 [33] and LepR1 [21] (Additional file 2: Table S2). The tRWF1 primer located in the tRNA allowed us to sequence through the LCO primer binding site as the tRNA-W gene is located around 200 bp upstream of cox1 [33]. Altogether, we compared sequences from six of the seven recognized psyllid families (no samples or published sequences of Mastigimatidae were available) to design the new degenerate forward primer. Details of the samples and sources of the sequences used for the design of the forward primer are provided in Additional file 3: Table S3. Alignment of sequences was performed with ClustalW in Mega 7 [32], and then checked and adjusted manually before comparing base calls with published primer sequences. Based on variable sites across the primer length, we designed the new degenerate forward primer, LCOP-F (5′ AGAACWAAYCATAAAAYWATTGG-3′) as a modification of the universal forward primer LCO1490. In contrast, the priming site of the reverse primer, HCO2198, is relatively conserved among psyllid species (see Additional file 4: Fig. S1), and so we decided to pair our new degenerate forward primer with HCO2198, which gives an amplified length of ~ 660 bp.
We compared the formation of reproducible discrete bands after electrophoresis of PCR products of our newly designed primer LCOP-F coupled with HCO2198 with the “universal” primer pair LCO1490/HCO2198 as well as several other published primer combinations used for amplifying cox1 that focus on the 5′ end of the gene typically considered the standard barcode region for insects. Details of primer sequences, annealing temperatures and citations are provided in Table 1; Fig. 1 provides a schematic of the relative primer locations and expected sequence length. The same DNA extracts were used for the different primer pairs for direct comparison (Additional file 2: Table S2). PCR amplifications were then performed in a 25 µl final reaction volume containing 0.4 µM of each primer, 3 mM MgCl2, NH4 buffer (1X), 0.2 mM of each dNTP, 0.4 mg/ml of acetylated bovine serum albumin (BSA), 0.02 unit/µl of Taq-polymerase (Bioline) and 2 µl of DNA extract (concentration not determined). PCRs were carried out in a Swift™ Maxi Thermal Cyclers (ESCO Technologies) applying the following thermal step: initial denaturation for 4 min at 94 °C, followed by 39 cycles of 30 s at 94 °C, 30 s at annealing temperature of 50 °C and 45 s at 72 °C, then a final extension step of 10 min at 72 °C. PCR products were enzymatically cleaned in microtubes with 0.025 unit/µl rAPid alkaline phosphatase (Roche) and 50 unit/ml exonuclease I (BioLabs) for 15 min at 37 °C followed by 15 min at 85 °C. Then the purified products were sequenced in both directions at Macrogen Inc. (Madrid, Spain). Sequences were checked, edited and assembled for both directions with CLUSTALW within the MEGA 7 software [32].
Results and discussion
PCR amplification products were obtained with the new degenerate forward primer, LCOP-F, coupled with reverse primer HCO2198 from Folmer et al. [11] for 146 of the 154 specimens, resulting in a 95% success rate. This primer combination amplified the first 658 bp of cox1 for all 36 species tested, which represents five of the seven recognized families [26]. Sequences have been deposited in GenBank database (Accession numbers: OR027185-OR027257, OR029451-OR029453). None of these sequences showed frame-shifts or stop-codon. The result of a primer efficiency comparison is shown in Table 2, and Additional file 2: Table S2 and Additional file 5: Table S5, respectively. As expected, the lowest amplification success was obtained with the insect barcode primer pairs: LepF1 with LepR1 and LCO1490 with HCO2198, with only 13% and 35% of the species successfully amplified respectively. According to Hajibabaei et al. [34], new primer design should be considered if less than 95% success rate is obtained with existing primers for a broad range of species in the target group. Alignment of our sequences with 31 species obtained from GenBank revealed nucleotide mismatches with two of the commonly used barcoding forward primers, LCO1490 and LepF1 (see Fig. 2), that likely explains the failure of these universal primers to generate PCR amplicons for most psyllid species tested. This is consistent with recent studies that have revealed high sequence variability in the primer site of the Folmer forward primer LCO1490 [33, 35]. The primer pair, tRWF1/LepR1 successfully amplified 78% of species tested (47 of the 60 specimens), while the pair C1-J-1718/H7005P-R (the latter a degenerate reverse primer from Percy & Cronk [36]) successfully amplified all but one species tested (119 of the 143 specimens tested, 83%). However, although showing a good success rate, this latter primer pair avoids the variable 5′ end of cox1 and therefore does not recover the start of cox1 which is deemed important for a standard barcode region (Fig. 1). Our new degenerate forward primer, LCOP-F paired with HCO2198 gives an amplified length of ~ 660 bp, but it should be possible to extend this sequence length by pairing LCOP-F with other proven reverse primers (e.g., H7005P-R). Combining the two degenerate primers LCOP-F/H7005P-R provides a sequence length of ~ 1000 bp that could provide additional information for phylogenetical studies, but the efficacy of this primer combination has not been extensively tested in this study.
Although our sampling includes only one member of family Carsidaridae, and the two smallest families are unrepresented (Calophyidae and Mastigimatidae), our sampled dataset spans the maximum phylogenetic distance within the Psylloidea, with the unsampled families phylogenetically nested among those families sampled. For many other Hemiptera groups (e.g., Auchenorrhyncha and Heteroptera) the LCO1490 forward primer sequence provides a 100% match, but from a survey of GenBank sequences some taxa in other Sternorrhyncha groups (e.g., Aphidomorpha, Aleyrodoidea, Coccoidea) and some of the wider hemipteroid groups (e.g., Psocoptera and Thysanoptera) there are similar mismatches at the primer binding site to those found in Psylloidea and therefore the new LCOP-F primer may also prove effective in these groups.
Conclusion
The new degenerate primer coupled with one of the original “universal” primers successfully amplified the cox1 DNA barcode region for all species tested representing five of the seven recognised families in Psylloidea. In addition, these primers were able to successfully sequence the DNA barcode for all species tested. This study also confirmed that the forward priming site at the start of cox1 is not particularly conserved in psyllids, which likely explains the failure of standard barcode primers to amplify this 5’ region of cox1. Finally, the new degenerate forward primer will improve the implementation of DNA barcoding in psyllids by providing a standard option, and will also facilitate the establishment of a DNA barcode library for rapid and accurate identification of psyllid species and more effective comparison of sequence data from different studies.
Limitations
Although the new degenerate forward primer has been tested on a phylogenetically broad representation of psyllids, further work is needed to confirm its efficacy in a larger sample of psyllid species, as well as in some of the tropical groups not or only poorly represented in this study (e.g., in families Calophyidae, Carsidaridae and Mastigimatidae), and also its potential utility for other Hemiptera and a wider range of insects.
Availability of data and materials
Sequences generated for this study are deposited in GenBank: Accession numbers OR027185-OR027257 and OR029451-29453. All other data generated or analysed during this study are included in this published article and its additional files.
Abbreviations
- BOLD:
-
Barcode of Life Data System
- PCR:
-
Polymerase chain reaction
References
Ouvrard D. The World Psylloidea database (from psyl’list) [Data set resource]. Nat History Museum. 2015;13:434-54.
Burckhardt D, Queiroz DL. Neotropical jumping plant-lice (Hemiptera, Psylloidea) associated with plants of the tribe Detarieae (Leguminosae, Detarioideae). Zootaxa. 2020;33:4733.
Wineriter SA, Halbert SE. Boreioglycaspis melaleucae (Hemiptera: Psyllidae), an Introduced Biocontrol Agent of Melaleuca quinquenervia (Myrtaceae) in Florida. Entomol Circ. 2002;410:1–4.
Syrett P, Fowler SV, Harman HM, Hayes LM, Memmott J, Sheat JJ. Establishment of Arytainilla Spartiophila Förster (Hemiptera: Psyllidae), a new biological control agent for broom, Cytisus scoparius, in New Zealand. New Zeal Entomol. 2007;30:53–62.
Munyaneza JE. Psyllids as vectors of emerging bacterial Diseases of annual crops. Southwest Entomol. 2010;35:471–7.
Bové JM, Huanglongbing. A destructive, newly-emerging, century-old Disease of citrus. J Plant Pathol. 2006.
Gottwald T. Citrus Huanglongbing: the Pathogen and its impact. Plant Health Progress; 2007.
Hansen AK, Trumble JT, Stouthamer R, Paine TD. A new huanglongbing species “Candidatus Liberibacter psyllaurous”, found to infect Tomato and Potato, is vectored by the Psyllid Bactericera cockerelli (Sulc). Appl Environ Microbiol. 2008;74:5862–5.
Munyaneza JE, Crosslin JM, Upton JE. Association of Bactericera cockerelli (Homoptera: Psyllidae) with zebra chip, a new potato Disease in southwestern United States and Mexico. J Econ Entomol. 2007;100:656–63.
Cho G, Malenovský I, Burckhardt D, Inoue H, Lee S. DNA barcoding of pear psyllids (Hemiptera: Psylloidea: Psyllidae), a tale of continued misidentifications. Bull Entomol Res. 2020;110:521–34.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–9.
Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 1994;87:651–701.
Hebert PDN, Cywinska A, Ball SL, DeWaard JR. Biological identifications through DNA barcodes. Proc R Soc B Biol Sci. 2003;270:313–21.
Park D-S, Suh S-J, Hebert PDN, Oh H-W, Hong K-J. DNA barcodes for two scale insect families, mealybugs (Hemiptera: Pseudococcidae) and armored scales (Hemiptera: Diaspididae). Bull Entomol Res. 2011;101:429–34.
Che J, Chen H, Yang J, Jin J, Jiang K, Yuan Z, et al. Universal COI primers for DNA barcoding amphibians. Mol Ecol Resour. 2012;12:247–58.
Van Steenkiste N, Locke SA, Castelin M, Marcogliese DJ, Abbott CL. New primers for DNA barcoding of digeneans and cestodes (Platyhelminthes). Mol Ecol Resour. 2015;15:945–52.
Martoni F, Bulman S, Pitman A, Taylor G, Armstrong K. DNA barcoding highlights cryptic diversity in the New Zealand Psylloidea (Hemiptera: Sternorrhyncha). Diversity. 2018;10:1–18.
Suh S-J, Choi D-S. Using DNA barcoding for identification of some psyllids (Hemiptera: Psylloidea) intercepted at South Korea ports-of-entry. Insecta Mundi. 2020;0816:1–10.
Martoni F, Taylor GS, Blacket MJ. Illuminating insights into the biodiversity of the Australian psyllids (Hemiptera: Psylloidea) collected using light trapping. Insects. 2020;11:1–13.
Ratnasingham S, Hebert PDN. BOLD: the barcode of life data system: barcoding. Mol Ecol Notes. 2007;7:355–64.
Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci U S A. 2004;101:14812–7.
Pérez-Rodríguez J, Krüger K, Pérez-Hedo M, Ruíz-Rivero O, Urbaneja A, Tena A. Classical biological control of the African citrus psyllid Trioza Erytreae, a major threat to the European citrus industry. Sci Rep. 2019;9:9440.
Oettl S, Schlink K. Molecular identification of two vector species, cacopsylla melanoneura and cacopsylla picta (Hemiptera: Psyllidae), of apple proliferation Disease and further common psyllids of Northern Italy. J Econ Entomol. 2015;108:2174–83.
Kang AR, Baek JY, Lee SH, Cho YS, Kim WS, Han YS, et al. Geographic homogeneity and high gene flow of the pear psylla, Cacopsylla pyricola (Hemiptera: Psyllidae), detected by mitochondrial COI gene and nuclear ribosomal internal transcribed spacer 2. Anim Cells Syst (Seoul). 2012;16:145–53.
Guidolin AS, Fresia P, Cônsoli FL. The genetic structure of an invasive pest, the Asian citrus psyllid Diaphorina citri (Hemiptera: Liviidae). PLoS One. 2014;9:1–17.
Burckhardt D, Ouvrard D, Percy DM. An updated classification of the jumping plant-lice (Hemiptera: Psylloidea) integrating molecular and morphological evidence. Eur J Taxon. 2021;2021:137–82.
Hodkinson ID. Status and taxonomy of the Trioza (Bactericera) Nigricornis Förster complex (Hemiptera: Triozidae). Bull Entomol Res. 1981;71:671–9.
Hodkinson D. New species of psyllid from the Canary Islands and Madeira (Homoptera: Psylloidea). Eos (Washington DC). 1990;66:29–35.
Rapisarda C. Contributo alla conoscenza degli psilloidei viventi su Rhamnus alaternus L. in Italia (Homoptera: Psylloidea). Phytophaga. 1989;3:21–60.
Bastin S, Burckhardt D, Reyes-Betancort JA, Hernández-Suárez E, Ouvrard D. A review of the jumping plant-lice (Hemiptera: Psylloidea) of the Canary Islands, with descriptions of two new genera and sixteen new species. Zootaxa. 2023;5313:1–98.
Casquet J, Thebaud C, Gillespie RG. Chelex without boiling, a rapid and easy technique to obtain stable amplifiable DNA from small amounts of ethanol-stored spiders. Mol Ecol Resour. 2012;12:136–41.
Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Park DS, Suh SJ, Oh HW, Hebert PDN. Recovery of the mitochondrial COI barcode region in diverse Hexapoda through tRNA-based primers. BMC Genomics. 2010;11:1–7.
Hajibabaei M, DeWaard JR, Ivanova NV, Ratnasingham S, Dooh RT, Kirk SL, et al. Critical factors for assembling a high volume of DNA barcodes. Philos Trans R Soc B Biol Sci. 2005;360:1959–67.
Sharma P, Kobayashi T. Are universal DNA primers really universal? J Appl Genet. 2014;55:485–96.
Percy DM, Cronk QCB. Psyllid honeydew as a Bombus food source in the boreal landscape. Ecol Entomol. 2022;47:713–8.
Acknowledgements
Saskia Bastin is recipient of a 2019–2023 PhD grant from the Agencia Canaria de Investigación, Innovación y Sociedad de la Información (ACIISI), Consejería de Economía, Industria, Comercio y Conocimiento of the Gobierno de Canarias and the European Social Fund. She is enrolled in the PhD program in Biodiversity and Conservation at the University of La Laguna (ULL). We thank one anonymous reviewer for helpful comments on an earlier version of this manuscript.
Funding
This research was carried out with financial support from the research project CUARENTAGRI (MAC2/1.1a/231).
Author information
Authors and Affiliations
Contributions
SB, DMP and FS conceived the study. SB performed the analysis, and all molecular laboratory work, and drafted the manuscript. DP and FS provided critical feedback and supervised the study. DP contributed to the writing of the manuscript. FS provided resources. The corresponding author is FS. All authors read, revised and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Publicly available sequences of psyllid taxa present in BOLD Systems with barcode length, primers used, length of the DNA barcode sequences > 500bp (Assessed on 19.01.2023).
Additional file 2: Table S2.
Information of the samples used in this study with the results for each primer pair. Results: blank = non-tested; yes/no = formation or not of reproducible discrete bands after electrophoresis of PCR amplifications
Additional file 3: Table S3.
Details of the samples and sequences used for the design of the forward primer.
Additional file 4: Fig. S1.
Snap shot of the multiple alignment of the reverse primer binding site of the psyllid cox 1 sequences obtained from GenBank and during this study including also the sequence of the "universal" primer HCO2198 and the standard insect reverse primer LepR1.
Additional file 5: Table S4.
Results of PCR amplifications of the 36 psyllid species analysed (20 genera and 5 families) with the different pairs of primers tested. X = no amplification; Yes/No = formation or not of reproducible discrete bands after electrophoresis of PCR amplifications
Additional file 6. References.
Bibliographic references of supplementary information.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Bastin, S., Percy, D.M. & Siverio, F. Establishing reliable DNA barcoding primers for jumping plant lice (Psylloidea, Hemiptera). BMC Res Notes 16, 322 (2023). https://doi.org/10.1186/s13104-023-06585-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13104-023-06585-8