Abstract
Objectives
Pollen is a useful tool for identifying the provenance and complex ecosystems surrounding honey production in Malaysian forests. As native key pollinators in Malaysia, Apis dorsata and Heterotrigona itama forage on various plant/pollen species to collect honey. This study aims to generate a dataset that uncovers the presence of these plant/pollen species and their relative abundance in the honey of A. dorsata and H. itama. The information gathered from this study can be used to determine the geographical and botanical origin and authenticity of the honey produced by these two species.
Results
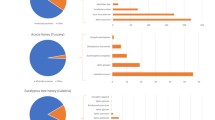
Sequence data were obtained for both A. dorsata and H. itama. The raw sequence data for A. dorsata was 5 Mb, which was assembled into 5 contigs with a size of 6,098,728 bp, an N50 of 15,534, and a GC average of 57.42. Similarly, the raw sequence data for H. itama was 6.3 Mb, which was assembled into 11 contigs with a size of 7,642,048 bp, an N50 of 17,180, and a GC average of 55.38. In the honey sample of A. dorsata, we identified five different plant/pollen species, with only one of the five species exhibiting a relative abundance of less than 1%. For H. itama, we identified seven different plant/pollen species, with only three of the species exhibiting a relative abundance of less than 1%. All of the identified plant species were native to Peninsular Malaysia, especially the East Coast area of Terengganu.
Data description
Our data offers valuable insights into honey’s geographical and botanical origin and authenticity. Metagenomic studies could help identify the plant species that honeybees forage and provide preliminary data for researchers studying the biological development of A. dorsata and H. itama. The identification of various flowers from the eDNA of honey that are known for their medicinal properties could aid in regional honey with accurate product origin labeling, which is crucial for guaranteeing product authenticity to consumers.
Similar content being viewed by others
Objective
Pollen is a useful tool for identifying the provenance and complex ecosystems surrounding honey production in Malaysian forests. As native key pollinators in Malaysia, A. dorsata and H. itama forage on various plant/pollen species to collect honey. This study aims to generate a dataset that uncovers the presence of these plant/pollen species and their relative abundance in the honey of A. dorsata and H. itama. The information gathered from this study can be used to determine the geographical and botanical origin and authenticity of the honey produced by these two species.
Data description
This dataset contains eDNA sequence information from honey samples of A. dorsata and H. itama, collected from the East Coast area of Terengganu, Malaysia in June and July 2022. The samples were located at 4° 57’ 6.48” N and 103° 20’ 25.44” E. Individual DNA sequencing and FASTQ files for both samples are available through the National Centre for Biotechnology Information (NCBI) data repository system. The ITS2 nuclear gene region was amplified using previously described primers [1]. The filtered reads were clustered based on k-mer frequency profile using NanoCLUST [2], followed by consensus generation and error correction with Racon and Medaka v.1.4.1 [3].
For A. dorsata honey eDNA, a total output of 5 Mb was generated, which assembled into 5 OTUs. For H. itama honey eDNA, we obtained 5 contigs with a size of 6,098,728 bp, an N50 of 15,534, and a GC content of 57.42. The operational taxonomic unit (OTU) and FASTA file for this sample are accessible via NCBI (https://dataview.ncbi.nlm.nih.gov/object/SRR21831607) (Table 1). For H. itama, the raw sequence data shows a total size of 6.3 Mb, assembled into 11 contigs with a size of 7,642,028 bp, an N50 of 17,180, and a GC content of 55.38, based on the NCBI genome annotation pipeline. The operational taxonomic unit (OTU) and FASTA file for this sample are accessible via NCBI (https://dataview.ncbi.nlm.nih.gov/object/SRR21831606).
The relative abundance (Ra) of the identified plant and pollen species, along with their taxonomical classification levels (Phylum, Class, Order, Family, Genus, and Species), are presented in Table 2. Each plant species’ individual sequences underwent MEGABLAST analysis to identify highly similar sequences with nearly 100% identity. The complete sequences of selected species were downloaded in FASTA format for subsequent analysis.
The eDNA sequence analysis of honey from A. dorsata revealed frequent identification of plant species such as Corynandra viscosa (42.02%) and Syzygium cumini (40.11%). C. viscosa, locally known as Maman pasir, is an erect herb that can reach a height of 1.2 m. It features attractive yellow-colored flowers with a petiole length of 4.5 cm [4]. On the other hand, the genus Syzygium comprises over 1,200 species of trees or shrubs with sessile flowers ranging from 7 to 12 cm in height [5]. Every pollen species detected in the honey sample belonged to flowering plants, except for Mallotus paniculatus (known locally as Balik Angin), which accounted for less than 1% compared to other flowering plants/pollen species. Additional identified species included Scaevola taccada (10.17%), known locally as Merambong, and Syzygium claviflorum (7.66%), known locally as Bangkoh. It is worth noting that the identified pollen species in the eDNA sequence are native flowering plants found in the Peninsular Malaysia region where the sample was collected. These species have been previously reported in various studies, such as C. viscosa [6], S. cumini [7], and S. taccada [8].
For H. itama the eDNA sequence of honey analysis revealed a significant presence of various plant species. The most abundant species were M. paniculatus (Balik angin) (42%) and Cleome rudisperma (41%), locally called Maman ungu. M. paniculatus is a medicinal plant native to the East Coast of Malaysia [9]. C. rudisperma, on the other hand, is a flowering plant reported to be native to Malaysia [10]. Additional plant species identified in the eDNA analysis included Richardia brasiliensis (0.53%) [11], Ludwigia hyssopifolia (0.42%) (known locally as Lakum air), Eleucine indica (0.56%) (known locally as Rumput sambau) [12], Mimosa pudica (2.46%) (known locally as Semalu) [13], and Acacia mangium (14.49%) (known locally as Manga hutan) [14] (Table 2). Apart from our findings, another study reported a higher abundance of pollen from the phylum Spermatophyta [15]. Specifically, four species, namely Garcinia oblongifolia, Muntingia calabura, Mallotus pellatus, and Pinus squamata, were found to occur abundantly and were consumed by H. itama in all populations.
Limitations
Sample size: A small sample size may not be representative of the larger population and may limit the generalizability of the findings.
Regional specificity: The study focuses on honey samples from the Peninsular Malaysia region, which may limit the generalizability of the findings to other regions or countries.
Identification methods: The study uses eDNA sequencing and pollen analysis to identify plant species in the honey samples.
Honey production: honey was collected from multiple hives in one area. This could affect the diversity and abundance of plant species present in the honey samples.
Honey age: The age of honey can affect the diversity and abundance of plant species present in the sample.
References
Timpano EK, Scheible MK, Meiklejohn KA. Optimization of the second internal transcribed spacer (ITS2) for characterizing land plants from soil. PLoS ONE. 2020;15(4):0231436.
Rodríguez-Pérez H, Ciuffreda L, Flores C. NanoCLUST: a species-level analysis of 16S rRNA nanopore sequencing data. Bioinformatics. 2021;37(11):1600–1.
Vaser R, et al. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017;27(5):737–46.
Rao VH, et al. The status assessment of Corynandra viscosa subsp. nagarjunakondensis (Magnoliopsida: Cleomaceae), endemic to Nagarjunakonda, Andhra Pradesh. India J Threatened Taxa. 2018;10(9):12210–7.
Tagane S, et al. Five new species of Syzygium (Myrtaceae) from Indochina and Thailand. Phytotaxa. 2018;375(4):247–60.
Tamboli AS, et al. Phylogenetic analysis, genetic diversity and relationships between the recently segregated species of Corynandra and Cleoserrata from the genus Cleome using DNA barcoding and molecular markers. CR Biol. 2016;339(3–4):123–32.
Azima AS, Noriham A, Manshoor N. Phenolics, antioxidants and color properties of aqueous pigmented plant extracts: Ardisia colorata var. elliptica, Clitoria ternatea, Garcinia mangostana and Syzygium cumini. J Funct Foods. 2017;38:232–41.
Rojas-Sandoval J, Acevedo-Rodríguez P. Scaevola taccada (beach naupaka)’, CABI Compendium. CABI Int. 2022. https://doi.org/10.1079/cabicompendium.48817.ad.
Bahaman N, et al. Medicinal properties screening of Mallotus paniculatus extract. IIUM Med J Malaysia. 2020;19:1.
Motmainna M, et al. Bioherbicidal properties of Parthenium hysterophorus, Cleome rutidosperma and Borreria alata extracts on selected crop and weed species. Agronomy. 2021;11(4):643.
Ruth K. Additions to the weed flora of Peninsular Malaysia. Malay Nat J. 2009;61(2):133–42.
Zakri ZHM, et al. Eleusine indica for Food and Medicine. J Agrobiotechnology. 2021;12(2):68–87.
Parker C. Mimosa pudica (sensitive plant). CABI Compendium. 2017. https://doi.org/10.1079/cabicompendium.34202.
Jamilah MS, Nur-Faiezah AG, Siti Kehirah A, Siti Mariam MN, Razali MS. Woody plants on dune landscape of Trengganu, Peninsular Malaysia. J Trop for Sci. 2014;26(2):267–74.
Fahimee J, et al. Metabarcoding in diet assessment of Heterotrigona itama based on trnL marker towards domestication program. Insects. 2021;12(3):205.
Hameed F et al. Jamun. Antioxidants in Fruits: Properties and Health Benefits. 2020; 615–637.
Jeffrey C. On the nomenclature of the strand Scaevola species (Goodeniaceae). Kew Bull. 1980;34(3):537–45.
Craven LA, Biffin E. An infrageneric classification of Syzygium (Myrtaceae). Blumea-Biodiversity Evol Biogeogr Plants. 2010;55(1):94–9.
Sierra SEC, Van Welzen PC. A taxonomic revision of Mallotus section Mallotus (Euphorbiaceae) in Malesia. Blumea. 2005;50:249–74.
Rojas-Sandoval J. P.A.-R., Cleome rutidosperma (fringed spiderflower). CABI digital library. 2014.
Marianne J, Datiles PA-R. Acacia mangium (brown salwood). CABI digital library. 2017.
Moran G, Muona O, Bell J. Acacia mangium: a tropical forest tree of the coastal lowlands with low genetic diversity. Evolution. 1989;43(1):231–5.
Adoho ACC, et al. Review of the literature of Eleusine indica: phytochemical, toxicity, pharmacological and zootechnical studies. J Pharmacognosy Phytochemistry. 2021;10(3):29–33.
Pinto DS, et al. Secondary metabolites isolated from Richardia brasiliensis Gomes (Rubiaceae). Revista Brasileira de Farmacognosia. 2008;18:367–72.
POWO, Plants of the World Online. Facilitated by the Royal Botanic Gardens. Kew. Published on the Internet. 2021.
WFO, Richardia brasiliensis Gomez. 2023.
Parker C. Ludwigia hyssopifolia (water primrose). CABI Compendium. 2012.
Acknowledgements
The authors would like to express their sincere gratitude to the Big BEE Honey Sdn. Bhd. of Kg. Jambu Bongkok, Merchang, Marang, Terengganu for providing the raw materials directly from the beehives. The first author self-financed their PhD work. Finally, the authors acknowledge the University Malaysia Sabah (UMS) for the APC grant provided for this High Impact Publication.
Funding
This study was supported by the Universiti Malaysia Sabah for payment of APC.
Author information
Authors and Affiliations
Contributions
Nurul Huda: Resources, Writing – review & editing, Funding acquisition; Saeed Ullah: Data curation, Formal analysis, Investigation, Software, Writing – original draft; Roswanira Abdul Wahab: Supervision, Validation, Resources, Writing – review & editing; Mohd Nizam Lani: Data curation, Supply raw materials; Nur Hardy Abu Daud: Conceptualization, Software, Writing – review & editing; Amir Husni Mohd Shariff: Data curation, Formal analysis, Investigation, Methodology; Norjihada Izzah Ismail: Data curation, Formal analysis, Investigation, Methodology; Azzmer Azzar Abdul Hamid: Supervision, Conceptualization, Software, Writing; Mohd Azrul Naim Mohamad: Resources, Writing – review & editing; Fahrul Huyop: Supervision, Funding acquisition, Project administration, Validation, Writing – review & editing.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The author declared that they have no known competing financial interest that could have appeared to influence the work presented in this paper.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Huda, N., ullah, S., Wahab, R.A. et al. The first ITS2 sequence data set of eDNA from honey of Malaysian giant honeybees (Apis dorsata) and stingless bees (Heterotrigona itama) reveals plant species diversity. BMC Res Notes 16, 211 (2023). https://doi.org/10.1186/s13104-023-06495-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13104-023-06495-9