Abstract
Background
Next-Generation Sequencing (NGS)-based testing in cancer patients has led to increased detection of variants of uncertain significance (VUS). VUS are genetic variants whose impact on protein function is unknown. VUS pose a challenge to clinicians and patients due to uncertainty regarding their cancer predisposition risk. Paucity of data exists on the pattern of VUS in under-represented populations. This study describes the frequency of germline VUS and clinico-pathological features in Sri Lankan hereditary breast cancer patients.
Methods
Data of 72 hereditary breast cancer patients who underwent NGS-based testing between January 2015 and December 2021 were maintained prospectively in a database and analyzed retrospectively. Data were subjected to bioinformatics analysis and variants were classified according to international guidelines.
Results
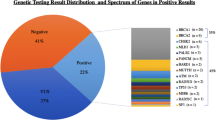
Germline variants were detected in 33/72(45.8%) patients, comprising 16(48.5%) pathogenic/likely pathogenic variants and 17(51.5%) VUS. Distribution of VUS in breast cancer predisposing genes were :APC:1(5.8%), ATM:2(11.7%), BRCA1:1(5.8%), BRCA2:5(29.4%), BRIP1:1(5.8%), CDKN2A:1(5.8%), CHEK2:2(11.7%), FANC1:1(5.8%), MET:1(5.8%), STK11:1(5.8%), NF2:1(5.8%). Mean age at cancer diagnosis in patients with VUS was 51.2 years. Most common tumour histopathology was ductal carcinoma 11(78.6%). 50% of tumours in patients having VUS in BRCA1/2 genes were hormone receptor negative. 73.3% patients had family history of breast cancer.
Conclusions
A significant portion of patients had a germline VUS. Highest frequency was in BRCA2 gene. Majority had family history of breast cancer. This highlights the need to undertake functional genomic studies to determine the biological effects of VUS and identify potentially clinically actionable variants that would be useful for decision-making and patient management.
Similar content being viewed by others
Introduction
Worldwide, the most common cancer in women is breast cancer [1]. The incidence of breast cancer in women in Sri Lanka is gradually rising at a rate of 4% per year [2]. Next-generation sequencing (NGS) is increasingly being used to detect germline variants in cancer predisposing genes in hereditary breast cancer. NGS-based multigene panels and clinical exome sequencing are cost- effective methods of testing for variations in many cancer predisposing genes simultaneously. The likelihood of detection of variants increases with the use of NGS techniques [3].
More than ten genes with breast cancer predisposition have been identified over the past 25 years including the high- penetrant tumor-suppressor genes BRCA1, BRCA2, PTEN, TP53, CDH1, STK11, PALB2 and numerous moderate-penetrant genes like CHEK2, BRIP1 and ATM [4,5,6].
Variants of uncertain significance (VUS) are DNA variations which have an unknown effect on protein function hence, their association with cancer predisposition risk is uncertain [7]. Frequent usage and rapid expansion of NGS-based testing has led to an increment in detection of VUS. Frequency of VUS among breast cancer patients undergoing NGS-based testing is reported to be around 33–54% [8,9,10]. However, there is paucity of data on the pattern of VUS in breast cancer predisposing genes in under-represented populations.
When dealing with VUS in the clinical setting, understanding their actionability and providing appropriate genetic counselling poses a challenge to most clinicians [11,12,13]. In our experience, despite their uncertain significance, such variants create psychological burden to the patients and financial repercussions not only to the patient but to the healthcare system as well due to the sparse and conflicting data on their actionability.
This study aims to describe the frequency and clinico-pathological features of germline VUS identified in a Sri Lankan cohort with hereditary breast cancer and the associated cancer phenotypes in their family members, with a view to building up a genotype-phenotype correlation based on prevailing evidence. We hope this would benefit clinicians in view of deciding management options, arranging family screening and overcoming discrepancies encountered in providing counselling in the context of germline VUS in breast cancer patients.
Methods
We included 72 consecutive breast cancer affected patients from families with hereditary cancer who underwent germline genetic testing through NGS analysis between January 2015 and December 2021.Their data were maintained prospectively in an anonymized database and analyzed retrospectively. NGS data were subjected to bioinformatics analysis and variants were classified according to American College of Medical Genetics and Genomics and Association for Molecular Pathology standards and guidelines. Clinicopathological data of patients harboring VUS including demographic data, tumour histopathology and receptor status as well as the cancer phenotypes in their first-, second- and third-degree relatives were also analyzed using standard statistical methods. Ethical approval for the study was obtained from the Ethics Review Committee, Faculty of Medicine, University of Colombo [EC-13-182]. Informed written consent was obtained from all patients who underwent germline genetic testing for the participation in the study and for publication of data.
Results
Germline genetic variants were identified in 33/72 (45.8%) patients. Pathogenic/likely pathogenic variants were detected in 16/33 (48.5%) patients. VUS were identified in 17/33 (51.5%). All the patients harboring VUS were females.
One (5.8%) VUS was novel which was detected in the CDKN2A gene and the remaining 16 (94.2%) were reported variants, all were missense variants. The distribution of gene-specific VUS detected in the breast cancer cohort is shown in Table 1. The highest frequency was noted in the high-penetrant BRCA2 gene 5 (29.4%), followed by moderate-penetrant ATM 2 (11.7%) and CHEK2 2 (11.7%) genes. Other high-penetrant cancer genes BRCA1, APC, STK11 and moderate-penetrant genes BRIP1, CDKN2A, FANCI and MET had a similar frequency 1 (5.8%). A VUS was detected in the NF2 gene, in a young breast cancer patient. NF2 gene is not reported to be a well-established breast cancer predisposing gene.
The age at cancer diagnosis, tumour histopathological types and receptor status in the breast cancer patients in relation to the gene-specific VUS are shown in Table 2. The highest frequency of patients was observed in the 40–59 years age group (47.1%), followed by the above 60 years age group (29.4%). Youngest patient was aged 28 years and the oldest was 82 years old. Mean age of the cohort was 51.2 years. The most common histopathological type detected was ductal carcinoma. Out of the 14 available histopathology reports, 11 (78.6%) showed ductal carcinoma type. Two (18.2%) among them were detected at the carcinoma in-situ stage while the remaining (81.8%) were invasive type at diagnosis. In the 6 patients with a VUS in the BRCA genes whose tumour receptor status reports were available, 50% (3/6) were estrogen (ER) and progesterone (PR) receptor negative. Triple negative tumour was observed only in one patient harboring a BRCA2 VUS. ER positivity was noted in all other patients with a VUS in the ATM, CDKN2A, CHEK2, MET, STK11 and NF2 genes.
The distribution of cancers in close relatives (up to third-degree) of patients with VUS is shown in Table 3. Family history of breast cancer was observed either in first-, second- or third-degree relatives in all patients except in the patients carrying a VUS in the APC, BRIP1 and NF2 genes. Family history of gastrointestinal malignancies was observed in patients in whom a VUS was detected in the APC, ATM and BRIP1 genes. Family history of leukemia was observed in patients carrying a VUS in the ATM, BRCA1 and BRCA2 genes.
On follow up, the first patient with ATM variant (rs531617441) developed stage 1 endometrial cancer after four years. Her mother also developed breast cancer at the age of 80 years. The patient with NF2 variant (rs749326764) developed liver and vertebral metastasis after one year despite surgical and hormonal therapy and is currently receiving chemotherapy with poor response. The patient with CHEK2 variant (rs375507194) passed away due to progression of her breast cancer after 2 years. All the other patients are on regular follow up and doing well.
Discussion
In this study, the frequency of germline VUS was 51.5%, which is in keeping with data from previous studies [8,9,10]. The highest number of VUS were detected in the BRCA2 gene. Similar findings were observed in published studies [14,15,16]. Similar to the findings in this cohort, a relatively high frequency of VUS in the ATM gene was reported in several previous studies [14,15,16]. In contrast to our findings, other studies report a lower frequency of VUS in the CHEK2 gene [14,15,16]. This may be attributed to the small sample size in this study. An interesting finding is that though we detected a VUS in the NF2 gene, no previous studies had reported any VUS in this gene in association with breast cancer [14,15,16].
The observation of the highest frequency of patients with the age of cancer diagnosis in the 40–59-year age group rather than in the age group over 60 years, contrasts with the observation in a recent Sri Lankan study done based on the national breast cancer database. This observation points towards the hereditary cancer predisposition of individuals in this cohort [2]. Ductal carcinoma was the commonest histopathological type observed and is compatible with the findings from the previous Sri Lankan study [2].
It is well established that variants in the APC gene are implicated in the regulation of the intracellular level of beta-catenin through the Wingless/Wnt signal transduction pathway and have been implicated in carcinogenesis through loss of tumour suppressor activity [17]. APC gene variants have therapeutic implications as well by imparting chemotherapy resistance highlighting the importance of detecting APC variants in cancer patients [18, 19]. The contribution of APC variants in colorectal cancer is also well established. The overall APC gene variation rate in breast cancer patients ranges between 0.4 and 18% [20, 21]. In this cohort, the patient harboring the APC gene VUS at c.1564 A > G had a significant family history of esophageal cancer, melanoma and thyroid cancer in first degree relatives. APC variants are considered to contribute to esophageal cancer [22,23,24,25,26] and melanoma [27, 28] and are known to have prognostic and therapeutic implications as well. Previous studies have shown thyroid cancer incidence to be increased in patients with APC variants [29, 30].
Heterozygosity for ATM gene variants increases the risk of development of breast cancer by 2-3-fold compared to the general population [31, 32]. The other cancers seen in the pedigrees of the 2 patients with an ATM gene VUS at c.7502 A > G were leukemia [33,34,35,36] in a third-degree relative and colorectal cancer in second- and third-degree relatives [37, 38]. These cancer types have previously been reported in association with ATM gene variants and are considered to have therapeutic implications as well.
BRCA gene variants are well recognized in association with breast and ovarian cancers. However, one of the patients with a BRCA2 gene VUS at c.8417 C > T in our cohort had a family history of leukemia and endometrial carcinoma. Several studies have reported leukemia developing after chemotherapy in patients with BRCA pathogenic variants but evidence on direct correlation of BRCA gene variations with hematological malignancy is not well established [39,40,41,42]. Several studies have reported the development of endometrial carcinoma in carriers of BRCA gene variants; however, this is largely confounded by Tamoxifen usage in hormone receptor positive breast cancer patients [43,44,45].
Association of BRIP1 gene variants with breast cancer is reported [6], however some upcoming controversial data is questioning its significance in the context of breast cancer development [46]. Its predisposition to ovarian cancer is well established in several studies [46, 47]. However, our patient with a VUS in BRIP1 gene at c.3103 C > T did not have a personal or family history of ovarian cancer. Colorectal cancer development in carriers of BRIP1 gene variants has been reported and a similar occurrence was observed in our patient’s pedigree [48, 49].
The pedigree of the patient with the novel VUS in the CDKN2A gene at c.377 A > G clearly depicts a hereditary breast cancer syndrome pattern. Variants in this tumour suppressor gene have been reported previously to be associated with breast cancer development [50, 51].
Germline variations in the CHEK2 tumour suppressor gene are known to be associated with carcinogenesis [52]. High risk of breast cancer development is observed in individuals carrying CHEK2 gene variants especially in the context of family history of breast cancer, which is clearly demonstrated in the pedigree of our patient with a VUS in the CHEK2 gene [53].
Pathogenic variants in the FANCI, MET and STK11 have been reported in association with breast cancer [54,55,56]. Though the evidence for tumour development with NF2 gene variants is sparse except for the nerve sheath tumours, there is now growing evidence that it may be implicated in the development of several other cancers including breast cancer [57, 58].
This study does not provide data on segregation analysis or functional studies on the biological effects of the VUS. Availability of such data would provide some additional evidence regarding the potential actionability of the variants. Hence, the findings of this study may provide an intriguing line of future research to determine the clinical actionability of the VUS reported herein.
Limitations
-
Small sample size and unavailability of functional data and segregation analysis to assess the biological effects of the VUS.
-
Other modifiable risk factors for cancer development were not considered when analyzing the data pertaining to each individual.
-
Unavailability of data in few patients pertaining to tumour histopathology, hormone receptor status and family history of cancer.
Conclusions
The analysis of the cancer phenotypes associated with each VUS including the phenotypic expression in close relatives and comparison with pre-existing data reported before suggests that some of the germline VUS detected in this cohort might be contributing to the cancer development, though currently existing standard classification criteria categorize them as VUS. In the context of VUS, these findings highlight the importance of considering the cancer phenotype of each patient in an individualized manner and incorporating data on cancer expression in other family members in analyzing and interpreting the potential actionability of germline variants in cancer patients which would aid in overcoming to some extent, the discrepancies and conflicts encountered in the process of clinical decision-making.
Data availability
The datasets generated and analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
References
International Agency for Research on Cancer. (2008). GLOBOCAN 2008: cancer incidence and mortality worldwide in 2008. http://globocan.iarc.fr/
Fernando A, Jayarajah U, Prabashani S, Fernando EA, Seneviratne SA. Incidence trends and patterns of breast cancer in Sri Lanka: an analysis of the national cancer database. BMC Cancer. 2018;18(1):1–6. https://doi.org/10.1186/s12885-018-4408-4.
Walsh T, Lee MK, Casadei S, Thornton AM, Stray SM, Pennil C, …, King MC. (2010). Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proceedings of the National Academy of Sciences, 107(28), 12629–12633.https://doi.org/10.1073/pnas.1007983107.
Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, King MC. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250(4988):1684–9. https://doi.org/10.1126/science.2270482.
Wooster R, Neuhausen SL, Mangion J, Quirk Y, Ford D, Collins N, …, Stratton MR. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science. 1994;265(5181):2088–90. https://doi.org/10.1126/science.8091231.
Easton DF, Pharoah PD, Antoniou AC, Tischkowitz M, Tavtigian SV, Nathanson KL, …, Foulkes WD. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med. 2015;372(23):2243–57. https://doi.org/10.1056/NEJMsr1501341.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, …, Rehm HL. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Sci. 2015;17(5):405–23. https://doi.org/10.1038/gim.2015.30.
O’leary E, Iacoboni D, Holle J, Michalski ST, Esplin ED, Yang S, Ouyang K. Expanded gene panel use for women with breast cancer: identification and intervention beyond breast cancer risk. Ann Surg Oncol. 2017;24(10):3060–6. https://doi.org/10.1245/s10434-017-5963-7.
Beitsch PD, Whitworth PW, Hughes K, Patel R, Rosen B, Compagnoni G, …, Nussbaum RL. Underdiagnosis of hereditary breast cancer: are genetic testing guidelines a tool or an obstacle? J Clin Oncol. 2019;37(6):453. https://doi.org/10.1200%2FJCO.18.01631.
Kurian AW, Ward KC, Hamilton AS, Deapen DM, Abrahamse P, Bondarenko I, …, Katz SJ. Uptake, results, and outcomes of germline multiple-gene sequencing after diagnosis of breast cancer. JAMA Oncol. 2018;4(8):1066–72. https://doi.org/10.1001/jamaoncol.2018.0644.
Eccles BK, Copson E, Maishman T, Abraham JE, Eccles DM. Understanding of BRCA VUS genetic results by breast cancer specialists. BMC Cancer. 2015;15(1):1–9. https://doi.org/10.1186/s12885-015-1934-1.
Scherr CL, Lindor NM, Malo TL, Couch FJ, Vadaparampil ST. Genetic counselors’ practices and confidence regarding variant of uncertain significance results and reclassification from BRCA testing. Clin Genet. 2015;88(6):523–9. https://doi.org/10.1111/cge.12563.
Eccles DM, Mitchell G, Monteiro ANA, Schmutzler R, Couch FJ, Spurdle AB, …, Goldgar D. BRCA1 and BRCA2 genetic testing—pitfalls and recommendations for managing variants of uncertain clinical significance. Ann Oncol. 2015;26(10):2057–65. https://doi.org/10.1093/annonc/mdv278.
Henrie A, Hemphill SE, Ruiz-Schultz N, Cushman B, DiStefano MT, Azzariti D, …, Eilbeck K. ClinVar miner: demonstrating utility of a web‐based tool for viewing and filtering ClinVar data. Hum Mutat. 2018;39(8):1051–60. https://doi.org/10.1002/humu.23555.
Guindalini RSC, Viana DV, Kitajima JPFW, Rocha VM, López RVM, Zheng Y, …, Folgueira MAAK. Detection of germline variants in brazilian breast cancer patients using multigene panel testing. Sci Rep. 2022;12(1):1–12. https://doi.org/10.1038/s41598-022-07383-1.
Wong ES, Shekar S, Met-Domestici M, Chan C, Sze M, Yap YS, …, Lee AS. Inherited breast cancer predisposition in Asians: multigene panel testing outcomes from Singapore. NPJ genomic medicine. 2016;1(1):1–9. https://doi.org/10.1038/npjgenmed.2015.3.
Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer. 2001;1(1):55–67. https://doi.org/10.1038/35094067.
Stefanski CD, Keffler K, McClintock S, Milac L, Prosperi JR. APC loss affects DNA damage repair causing doxorubicin resistance in breast cancer cells. Neoplasia. 2019;21(12):1143–50. https://doi.org/10.1016/j.neo.2019.09.002.
King TD, Suto MJ, Li Y. The wnt/β-catenin signaling pathway: a potential therapeutic target in the treatment of triple negative breast cancer. J Cell Biochem. 2012;113(1):13–8. https://doi.org/10.1002/jcb.23350.
Sørlie T, Bukholm I, Børresen-Dale AL. Truncating somatic mutation in exon 15 of the APC gene is a rare event in human breast carcinomas. Mutations in brief no. 179. Online. Hum Mutat. 1998;12(3):215–5. https://europepmc.org/article/med/10660330.
Furuuchi K, Tada M, Yamada H, Kataoka A, Furuuchi N, Hamada JI, …, Moriuchi T. Somatic mutations of the APC gene in primary breast cancers. Am J Pathol. 2000;156(6):1997–2005. https://doi.org/10.1016/s0002-9440(10)65072-9.
Kawakami K, Brabender J, Lord RV, Groshen S, Greenwald BD, Krasna MJ, …, Meltzer SJ. Hypermethylated APC DNA in plasma and prognosis of patients with esophageal adenocarcinoma. J Natl Cancer Inst. 2000;92(22):1805–11. https://doi.org/10.1093/jnci/92.22.1805.
Usadel, H., Brabender, J., Danenberg, K. D., Jerónimo, C., Harden, S., Engles, J.,… Sidransky, D. (2002). Quantitative adenomatous polyposis coli promoter methylation analysis in tumor tissue, serum, and plasma DNA of patients with lung cancer. Cancer research, 62(2),371–375. https://aacrjournals.org/cancerres/article/62/2/371/509230/Quantitative-Adenomatous-Polyposis-Coli-Promoter.
Powell SM, Papadopoulos N, Kinzler KW, Smolinski KN, Meltzer SJ. APC gene mutations in the mutation cluster region are rare in esophageal cancers. Gastroenterology. 1994;107(6):1759–63. https://doi.org/10.1016/0016-5085(94)90818-4.
Clément G, Braunschweig R, Pasquier N, Bosman FT, Benhattar J. Methylation of APC, TIMP3, and TERT: a new predictive marker to distinguish Barrett’s oesophagus patients at risk for malignant transformation. J Pathology: J Pathological Soc Great Br Irel. 2006;208(1):100–7. https://doi.org/10.1002/path.1884.
Zare M, Jazii FR, Alivand MR, Nasseri NK, Malekzadeh R, Yazdanbod M. Qualitative analysis of adenomatous Polyposis Coli promoter: hypermethylation, engagement and effects on survival of patients with esophageal cancer in a high risk region of the world, a potential molecular marker. BMC Cancer. 2009;9(1):1–12. https://doi.org/10.1186/1471-2407-9-24.
Karachaliou, G. S., Alkallas, R., Carroll, S. B., Caressi, C., Zakria, D., Patel,N. M., … Moschos, S. J. (2022). The clinical significance of adenomatous polyposis coli (APC) and catenin Beta 1 (CTNNB1) genetic aberrations in patients with melanoma.BMC cancer, 22(1), 1–14. https://doi.org/10.1186/s12885-021-08908-z.
Worm J, Christensen C, Grønbæk K, Tulchinsky E, Guldberg P. Genetic and epigenetic alterations of the APC gene in malignant melanoma. Oncogene. 2004;23(30):5215–26. https://doi.org/10.1038/sj.onc.1207647.
Uchino, S., Ishikawa, H., Miyauchi, A., Hirokawa, M., Noguchi, S., Ushiama, M., …Sakai, T. (2016). Age-and gender-specific risk of thyroid cancer in patients with familial adenomatous polyposis. The Journal of Clinical Endocrinology & Metabolism, 101(12), 4611–4617. https://doi.org/10.1210/jc.2016-2043.
Jarrar, A. M., Milas, M., Mitchell, J., Laguardia, L., Berber, E., Siperstein, A.,… Church, J. M. (2011). Screening for thyroid cancer in patients with familial adenomatous polyposis. Annals of surgery, 253(3), 515–521. https://doi.org/10.1097/SLA.0b013e3181fcba8a.
Renwick, A., Thompson, D., Seal, S., Kelly, P., Chagtai, T., Ahmed, M., … Rahman,N. (2006). ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nature genetics, 38(8), 873–875. https://doi.org/10.1038/ng1837.
Easton, D. F., Pharoah, P. D., Antoniou, A. C., Tischkowitz, M., Tavtigian, S. V.,Nathanson, K. L., … Foulkes, W. D. (2015). Gene-panel sequencing and the prediction of breast-cancer risk. New England Journal of Medicine, 372(23), 2243–2257. https://doi.org/10.1056/NEJMsr1501341.
Bullrich, F., Rasio, D., Kitada, S., Starostik, P., Kipps, T., Keating, M., … Croce,C. M. (1999). ATM mutations in B-cell chronic lymphocytic leukemia. Cancer research, 59(1), 24–27. https://aacrjournals.org/cancerres/article/59/1/24/505073/ATM-Mutations-in-B-Cell-Chronic-Lymphocytic.
Schaffner C, Stilgenbauer S, Rappold GA, Döhner H, Lichter P. Somatic ATM mutations indicate a pathogenic role of ATM in B-cell chronic lymphocytic leukemia. Blood The Journal of the American Society of Hematology. 1999;94(2):748–53. https://doi.org/10.1182/blood.V94.2.748.
Guarini, A., Marinelli, M., Tavolaro, S., Bellacchio, E., Magliozzi, M., Chiaretti,S., … Foà, R. (2012). ATM gene alterations in chronic lymphocytic leukemia patients induce a distinct gene expression profile and predict disease progression. Haematologica, 97(1), 47. https://doi.org/10.3324%2Fhaematol.2011.049270.
Rose-Zerilli, M. J., Forster, J., Parker, H., Parker, A., Rodríguez, A. E., Chaplin,T., … Strefford, J. C. (2014). ATM mutation rather than BIRC3 deletion and/or mutation predicts reduced survival in 11q-deleted chronic lymphocytic leukemia: data from the UK LRF CLL4 trial. haematologica, 99(4), 736. https://doi.org/10.3324%2Fhaematol.2013.098574.
Maillet P, Chappuis PO, Vaudan G, Dobbie Z, Müller H, Hutter P, Sappino AP. A polymorphism in the ATM gene modulates the penetrance of hereditary non-polyposis colorectal cancer. Int J Cancer. 2000;88(6):928–31. https://doi.org/10.1002/1097-0215. :6%3C928::AID-IJC14%3E3.0.CO;2-P.
Vitiello, P. P., Martini, G., Mele, L., Giunta, E. F., De Falco, V., Ciardiello, D.,… Martinelli, E. (2021). Vulnerability to low-dose combination of irinotecan and niraparib in ATM-mutated colorectal cancer. Journal of Experimental & Clinical Cancer Research, 40(1), 1–15. https://doi.org/10.1186/s13046-020-01811-8.
Friedenson B. The BRCA1/2 pathway prevents hematologic cancers in addition to breast and ovarian cancers. BMC Cancer. 2007;7(1):1–11. https://doi.org/10.1186/1471-2407-7-152.
Cole M, Strair R. Acute myelogenous leukemia and myelodysplasia secondary to breast cancer treatment: case studies and literature review. Am J Med Sci. 2010;339(1):36–40. https://doi.org/10.1097/MAJ.0b013e3181bedb74.
Fruscalzo A, Damante G, Calcagno A, Di Loreto C, Marchesoni D. Four primary malignancies successively occurred in a BRCA2 mutation carrier: a case report. Cancer Invest. 2006;24(6):611–4. https://doi.org/10.1080/07357900600894872.
Hall MJ, Li L, Wiernik PH, Olopade OI. BRCA2 mutation and the risk of hematologic malignancy. Leukemia & lymphoma. 2006;47(4):765–7. https://europepmc.org/article/med/16886281.
Beiner, M. E., Finch, A., Rosen, B., Lubinski, J., Moller, P., Ghadirian, P., … Hereditary Ovarian Cancer Clinical Study Group. (2007). The risk of endometrial cancer in women with BRCA1 and BRCA2 mutations. A prospective study. Gynecologic oncology, 104(1), 7–10. https://doi.org/10.1016/j.ygyno.2006.08.004.
Segev, Y., Iqbal, J., Lubinski, J., Gronwald, J., Lynch, H. T., Moller, P., … Hereditary Breast Cancer Study Group. (2013). The incidence of endometrial cancer in women with BRCA1 and BRCA2 mutations: an international prospective cohort study. Gynecologic oncology, 130(1), 127–131. https://doi.org/10.1016/j.ygyno.2013.03.027.
Shu, C. A., Pike, M. C., Jotwani, A. R., Friebel, T. M., Soslow, R. A., Levine, D.A., … Kauff, N. D. (2016). Uterine cancer after risk-reducing salpingo-oophorectomy without hysterectomy in women with BRCA mutations. JAMA oncology, 2(11), 1434–1440. https://doi.org/10.1001/jamaoncol.2016.1820.
Weber-Lassalle, N., Hauke, J., Ramser, J., Richters, L., Gross, E., Blümcke, B., …Hahnen, E. (2018). BRIP1 loss-of-function mutations confer high risk for familial ovarian cancer, but not familial breast cancer. Breast Cancer Research, 20(1), 1–6. https://doi.org/10.1186/s13058-018-0935-9.
Suszynska M, Ratajska M, Kozlowski P. BRIP1, RAD51C, and RAD51D mutations are associated with high susceptibility to ovarian cancer: mutation prevalence and precise risk estimates based on a pooled analysis of ~ 30,000 cases. J ovarian Res. 2020;13(1):1–11. https://doi.org/10.1186/s13048-020-00654-3.
Ali M, Delozier CD, Chaudhary U. BRIP-1 germline mutation and its role in colon cancer: presentation of two case reports and review of literature. BMC Med Genet. 2019;20(1):1–5. https://doi.org/10.1186/s12881-019-0812-0.
Martín-Morales, L., Garre, P., Lorca, V., Cazorla, M., Llovet, P., Bando, I., … Caldés,T. (2021). BRIP1, a gene potentially implicated in Familial Colorectal Cancer Type X. Cancer Prevention Research, 14(2), 185–194. https://doi.org/10.1158/1940-6207.CAPR-20-0316.
Spitzwieser M, Entfellner E, Werner B, Pulverer W, Pfeiler G, Hacker S, Cichna-Markl M. Hypermethylation of CDKN2A exon 2 in tumor, tumor-adjacent and tumor-distant tissues from breast cancer patients. BMC Cancer. 2017;17(1):1–16. https://doi.org/10.1186/s12885-017-3244-2.
Aftab A, Shahzad S, Hussain HMJ, Khan R, Irum S, Tabassum S. CDKN2A/P16INK4A variants association with breast cancer and their in-silico analysis. Breast Cancer. 2019;26(1):11–28. https://doi.org/10.1007/s12282-018-0894-0.
Cybulski, C., Gorski, B., Huzarski, T., Masojć, B., Mierzejewski, M., Dębniak, T.,… Lubiński, J. (2004). CHEK2 is a multiorgan cancer susceptibility gene. The American Journal of Human Genetics, 75(6), 1131–1135. https://doi.org/10.1086/426403.
Cybulski C, Wokolorczyk D, Jakubowska A, et al. Risk of breast cancer in women with a CHEK2 mutation with and without a family history of breast cancer. J Clin Oncol. 2011;29(28):3747–52.
Gordiev, M., Brovkina, O., Shigapova, L. H., Shagimardanova, E., Enikeev, R. F., Nikitin,A., … Sakaeva, D. (2019). Heterozygous mutation in fanconi anemia genes associated with hereditary breast cancer. Annals of Oncology, 30, iii10. https://doi.org/10.1093/annonc/mdz095.028.
Gastaldi S, Comoglio PM, Trusolino L. The Met oncogene and basal-like breast cancer: another culprit to watch out for? Breast Cancer Res. 2010;12(4):1–10. https://doi.org/10.1186/bcr2617.
Clements A, Robison K, Granai C, Steinhoff MM, Scalia-Wilbur J, Moore RG. A case of Peutz-Jeghers syndrome with breast cancer, bilateral sex cord tumor with annular tubules, and adenoma malignum caused by STK11 gene mutation. Int J Gynecologic Cancer. 2009;19(9). https://doi.org/10.1111/IGC.0b013e3181ae3f71.
Petrilli AM, Fernández-Valle C. Role of Merlin/NF2 inactivation in tumor biology. Oncogene. 2016;35(5):537–48. https://doi.org/10.1038/onc.2015.125.
Arakawa H, Hayashl N, Nagase H, Ogawa M, Nakamura Y. Alternative splicing of the NF2 gene and its mutation analysis of breast and colorectal cancers. Hum Mol Genet. 1994;3(4):565–8. https://doi.org/10.1093/hmg/3.4.565.
Acknowledgements
The authors acknowledge all the patients who participated in the study.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by KG, NDS, GA, NN and VHWD. The first draft of the manuscript was written by KG, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All procedures performed in the study involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Ethical approval for the study was obtained from the Ethics Review Committee, Faculty of Medicine, University of Colombo, Sri Lanka [EC-13-182]. Informed written consent was obtained from all patients who underwent germline genetic testing for the participation in the study.
Consent for publication
Not Applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Gunawardena, K., Sirisena, N.D., Anandagoda, G. et al. Germline variants of uncertain significance, their frequency, and clinico-pathological features in a cohort of Sri Lankan patients with hereditary breast cancer. BMC Res Notes 16, 95 (2023). https://doi.org/10.1186/s13104-023-06365-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13104-023-06365-4