Abstract
Background
Food products of animal origin brought into the EU from third countries, both legally and illegally, can harbor foodborne pathogens such as Salmonella enterica. In this study, we examined five S. enterica isolates recovered either from legally imported chicken meat (n = 3) or from meat products confiscated from air travel passengers arriving in Germany (n = 2). The isolates were serotyped and further characterized by antimicrobial susceptibility testing, PCR-detection and sequencing of genes associated with antimicrobial resistances, and macrorestriction analysis. Transferability of resistance to third-generation cephalosporins was assessed by conjugation experiments and the plasmids tested for their incompatibility groups.
Results
The three isolates from legal imports were identified as S. Heidelberg or as non-flagellated. All three isolates were identified as AmpC producers carrying blaCMY-2 and as non-susceptible to ciprofloxacin. They were additionally resistant to tetracycline and sulfamethoxazole. The blaCMY-2-carrying plasmids were transferable by conjugation and belonged to incompatibility groups IncI1 or IncA/C. The two isolates from illegally imported meat belonged to the serovars Infantis or Weltevreden. The former was phenotypically resistant to five classes of antimicrobial agents while the S. Weltevreden isolate was fully susceptible to all agents tested.
Conclusion
The results of this study demonstrate that meat products imported from third countries, both legally and illegally, can harbor multiresistant Salmonella enterica. Consequently, these imports could constitute a source for the dissemination of antimicrobial resistant isolates, including those resistant to third-generation cephalosporins and fluoroquinolones.
Similar content being viewed by others
Background
Salmonella (S.) enterica is one of the most common bacterial pathogens causing foodborne infections and, thus, constitutes a major healthcare concern worldwide. For 2016, the European Food and Safety Authority (EFSA) reported over 94,000 confirmed cases of human salmonellosis in the European Union (EU) and S. enterica was identified as the bacterial agent responsible for the most foodborne outbreaks [1]. Poultry meat constitutes an important source of Salmonella enterica and infections in humans are often linked to the consumption of improperly cooked poultry meat or a cross-contamination of other foodstuffs [1]. While most non-typhoidal Salmonella spp. infections lead to self-limiting gastrointestinal symptoms, invasive infections can occur and may be life-threatening, particularly in immunocompromised patients [2, 3]. However, the likelihood of severe infections differs between serovars. S. enterica serovar Typhimurium, which is among the most frequently encountered serovars in the EU, has a moderate tendency to cause invasive disease—S. Typhimurium ST313 being a notable exception, frequently associated with invasive disease in Africa [4]. Others, such as S. Heidelberg and narrow host-range serovars like S. Dublin and S. Choleraesuis, are considerably more likely to cause severe infections requiring hospitalization [3, 5]. Previous reports of salmonellosis outbreaks involving multiple countries have raised concerns about the role of international food and animal trade in the spread of Salmonella serovars, especially considering the comparatively high rates of Salmonella detection in some parts of the world [6]. Furthermore, imported meat products have been implicated as a possible source for S. enterica isolates resistant to clinically relevant antimicrobials [7, 8].
In addition to consignments of meat officially imported into the EU, considerable amounts of meat products are brought into the EU illegally, circumventing any controls [9, 10]. To date, very little data are available about serovars present in such products and their characteristics [11, 12].
In this study, S. enterica isolates recovered from meat and meat products imported into the EU, both legally and illegally, were examined. The purpose was to identify the serovars entering the EU via these routes and to further characterize the isolates regarding their antimicrobial resistance phenotypes and genotypes.
Methods
Bacterial isolates
A total of five Salmonella enterica isolates were examined in this study. They were collected in the years 2014 and 2015, in the course of a previous study (Project “ZooGloW”) investigating meat and meat products imported into the European Union both legally and illegally. The procedures for the isolation of Salmonella spp. performed in that study and the sampling of legal poultry meat and pork imports have been described previously [13]. In brief, 516 poultry meat and 136 pork samples, cleared via the border inspection post Hamburg Harbour in 2014 and 2015, were subjected to microbiological analyses. The samples were taken aseptically by the competent authorities, stored in individually sealed bags and were subsequently frozen on-site. They were kept at − 18 °C during storage and transport to the laboratory. Detection of Salmonella spp. was performed according to DIN EN ISO 6579:2002 with an initial enrichment of a 10 g sample. Salmonella spp. isolates were identified in six of these samples from legally imported meat [13].
Following the same procedures, a total of 344 samples of illegally imported meat products, seized from air travel passengers by the competent authorities at Berlin Schönefeld Aiport (SXF) and Frankfurt International Airport (FRA), were examined (unpublished data). These samples included raw and processed products made of pork (n = 203), poultry (n = 127), beef (n = 8), lamb (n = 4), chamois buck (n = 1), rabbit (n = 1) and one dried meat sample of an unknown species. The presence of Salmonella enterica could be confirmed in five samples of raw poultry meat (n = 4) or rabbit meat (n = 1).
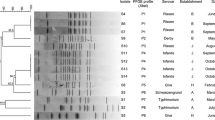
However, three isolates each from legal imports and illegally imported meat products could not be successfully recultured after cryopreservation. Consequently, a total of five of the previously collected Salmonella isolates were available for further analyses in the course of the current study. These included three isolates from legal imports, which were recovered from three separate batches of chicken meat, which traced back to producers operating under different registration numbers each. The remaining two isolates were recovered from illegally imported raw meat, comprising one sample of rabbit cuts (belly, ribs and shoulder) and one sample of marinated poultry meat. An overview of the origin of the samples is given in Fig. 1.
Typing results, resistance phenotypes and genotypes of the five Salmonella isolates. aOnly antimicrobial agents for which CLSI-approved MIC breakpoints are available, are included. BLA β-lactams, CHL chloramphenicol, CIP ciprofloxacin, GEN gentamicin, NAL nalidixic acid, SMX sulfamethoxazole, TET tetracycline, TMP trimethoprim, brackets indicate intermediate resistance
Serotyping and macrorestriction analysis
The isolates were sent to the National Reference Centre for Salmonellae and other Bacterial Enteric Pathogens (Robert Koch Institute, Wernigerode, Germany) for serotyping according to the White-Kauffmann-Le Minor scheme and phage typing of S. Infantis. Phage typing was performed according to the typing system of Miller et al. [14].
The genetic relatedness of the isolates was determined by macrorestriction analysis with XbaI digestion followed by pulsed-field gel electrophoresis (PFGE) according to the PulseNet protocol for Escherichia coli O157:H7, Salmonella, and Shigella [15]. The Bionumerics software version 7.6 (Applied Maths, Sint-Martens-Latem, Belgium) was used for band pattern analysis, applying the Dice coefficient with 0.5% optimization and 1% position tolerance.
Antimicrobial susceptibility testing and detection of resistance genes
Minimum inhibitory concentration values (MICs) were determined according to Clinical and Laboratory Standards Institute (CLSI) documents for broth microdilution and broth macrodilution susceptibility testing [16, 17]. Microtiter plates Micronaut-S for large animals (“Großtiere”), containing 19 antimicrobial agents or combinations, and Micronaut-S β-lactamases (Merlin Diagnostika, Bornheim-Hersel, Germany) were used. Escherichia coli ATCC25922 and ESBL-producing K. pneumoniae ATCC700603 served as quality control strains.
PCR assays targeting genes associated with antimicrobial resistances were carried out with previously described primers and protocols. The PCR assays included β-lactamase encoding genes blaTEM, blaCTX-M, blaCMY-2, blaOXA-1-like, blaOXA-2, and blaSHV [18, 19]; aminoglycoside resistance determinants ant-(2″)-I, aac(3)-II, aac(3)-IV [20]; tetracycline resistance genes tet(A), tet(B), tet(C), tet(D), tet(E), tet(G), tet(H), tet(M), tet(O) [18]; quinolone resistance-associated genes qnrA, qnrB, qnrC, qnrD, qnrS, aac(6′)-Ib-cr, qepA [21, 22]; phenicol resistance genes cmlA, catA1, catA2, catA3, catB2, catB3, cfr, fexA, floR, [18, 23]; as well as sul1, sul2, sul3 and dfrA1, dfrA5/14, dfrA7/17, dfrB1/2 [18], conferring resistance to sulfonamides and trimethoprim, respectively. Additionally, the isolates were tested for the presence of integron classes I, II and III [24, 25]. The quinolone-resistance determining regions of gyrA, gyrB, parC, parE were amplified with previously described primers [26]. Sequencing of amplicons obtained for blaCMY, gyrA, gyrB, parC, parE and the variable regions of integron class I was carried out by Eurofins Genomics (Ebersberg, Germany). Sequences were analyzed using the DNAMAN software version 8 (Lynnon Biosoft, San Ramon, CA, USA) and the BLASTN algorithm [27]. Incompatibility groups of blaCMY-2-carrying plasmids were determined by PCR as described previously [28].
Conjugation experiments
Isolates with resistance to third-generation cephalosporins were subjected to conjugation experiments with plasmid-free, rifampicin- and streptomycin-resistant Escherichia coli HK225 as recipient strain [29]. Donor and recipient strain were grown overnight in 10 ml LB medium containing ampicillin (50 µg/ml) or rifampicin (100 µg/ml), respectively. Following centrifugation and resuspension of the pellet in LB without antibiotics, the cultures were mixed in a 1:5 ratio (donor:recipient) based on their optical density. The mixture was centrifuged, the pellet transferred onto LB agar and incubated overnight at 35 °C. The following day, the conjugation mixture was recovered from the agar plate and suspended in 1 ml LB broth. Serial dilutions were plated on LB agar, LB agar supplemented with rifampicin (100 µg/ml), LB agar supplemented with ampicillin (50 µg/ml), and LB agar supplemented with rifampicin and ampicillin (100 µg/ml and 50 µg/ml, respectively). Transconjugants were taken from the double selective plates and confirmed as E. coli by PCR targeting the gadA gene [30]. Plasmids were extracted using a modification of the alkaline lysis method as described previously [31]. Plasmid sizes were estimated by agarose gel electrophoresis and visual comparison to the 94 kilobase pairs (kb) plasmid of S. Typhimurium stain LT2 (Fig. 2) [32]. One representative transconjugant per isolate, carrying a single plasmid, was chosen for further analyses.
Results
Serotypes and antimicrobial resistances
Among the isolates recovered from legally imported samples, two were identified as S. Heidelberg whereas the third was a non-flagellated, d-tartrate-positive isolate of O group B (O:4). However, all three isolates showed a high similarity of PFGE band patterns of up to > 90% (Fig. 1). The two isolates from illegally imported meat showed less than 50% similarity with the three isolates from legal imports and each other. The isolate from illegally imported meat from Vietnam was identified as S. Weltevreden. The second isolate, recovered from marinated poultry meat illegally introduced from Egypt, belonged to the serovar Infantis. This isolate showed a lysis pattern that did not conform to any recognized S. Infantis phage type and was designated “reacts but does not conform” (RDNC).
All isolates recovered from legal imports and one isolate from illegally imported meat were resistant to three or more classes of antimicrobial agents and were classified as multiresistant [33]. The respective resistance phenotypes and genotypes are shown in Table 1. The three isolates from legal imports were AmpC-producers and carried blaCMY-2. They were further resistant to nalidixic acid, sulfamethoxazole, and tetracycline and either resistant (n = 2) or intermediate resistant (n = 1) to ciprofloxacin (MICs of 1 µg/ml or 0.5 µg/ml ciprofloxacin, respectively), according to CLSI breakpoints. Decreased quinolone susceptibility was attributed to mutations resulting in a Ser83Phe substitution in the deduced amino acid sequence of the GyrA protein and a Thr57Ser substitution in ParC (Fig. 1). No isolate carried the genes qnrA, qnrB, qnrC, qnrD, qnrS, aac(6′)-Ib-cr, or qepA, associated with plasmid-mediated quinolone resistance.
The S. Infantis isolate from illegally imported meat also showed multiple phenotypic resistances. It was resistant to nalidixic acid, ciprofloxacin, tetracycline, gentamicin, sulfamethoxazole, trimethoprim, chloramphenicol and additionally showed elevated MIC values for florfenicol (MIC = 32 µg/ml). Furthermore, this isolate harbored a class I integron containing an aadA1 gene cassette showing 100% nucleotide sequence identity to previously published sequences (e.g. GenBank KF921523). The isolate did not produce positive results with any of the primer pairs targeting trimethoprim, gentamicin and phenicol resistance genes included in this study, however. In contrast to this, the S. Weltevreden isolate was susceptible to all antimicrobial agents tested.
Localization of cephalosporin resistance genes on transferable plasmids
Mating experiments demonstrated inter-genus transferability of the plasmid bearing blaCMY-2 for all three AmpC-producing isolates. The blaCMY-2 genes of isolates 253 and 417 were located on an IncA/C plasmid with an estimated size of over 100 kb, which further contained sul2 and tet(A), rendering E. coli HK225 transconjugants phenotypically resistant to tetracycline and sulfonamides in addition to β-lactams. The efficiency of conjugation of these plasmids was rather low, however, with 1.23 × 10−7 and 3.8 × 10−8 transconjugants per donor for isolates 253 and 417, respectively. The plasmid of the third isolate belonged to incompatibility group IncI1 and was transferred at a markedly higher frequency (5 × 10−3). This plasmid was slightly smaller with an estimated size of over 95 kb, based on visual comparison to the 94 kb plasmid of S. Typhimurium LT2. No co-transfer of other resistance determinants was observed for this plasmid.
Discussion
In recent years, the number of air travel passengers has increased continuously and is expected to rise to an estimated 7.8 billion travelers in 2036 [34]. Consequently, the total amount of meat and meat products illegally transported in passenger luggage can be expected to rise accordingly. To date, little data are available about the characteristics of salmonellae recovered from such food items. In the present study, the two isolates from illegally imported meat belonged to the serovars Infantis or Weltevreden. Both isolates were recovered from raw meat products, in one case seasoned with a marinade of unknown composition, transported in air passenger luggage. In contrast to this, a study conducted in Spain reported the following seven different serovars among eleven isolates from food of animal origin confiscated at a Spanish airport: monophasic serovar 4, 12: d: − , Rauform, Anatum, Oranienburg, Enteritidis, Newport and Typhimurium [11]. Notably, these were mostly of South American origin and included isolates detected in cheese samples. Counting only meat and meat products, 6 of the 122 samples examined in that study were positive for S. enterica [11]. In another study, a total of four isolates were detected on illegally introduced sausages of Russian origin out of 367 samples of illegally imported meat and meat products (1.1%) [12]. Two isolates each belonged to the serovars S. Infantis and S. Enteritidis. In their study, as well as in the course of the sampling preceding the current study, samples were kept frozen before analyses [12, 13]. Previous research demonstrates that freezing does not fully eliminate S. enterica, but provides differing results on survival rates during frozen storage ranging from no significant changes in bacterial counts to minimal survival [35, 36]. Consequently, structural injury to the bacterial cells during freezing and thawing of the samples prior to analyses may have contributed in part to the low detection of Salmonella spp.. One study reported a relatively constant population of Salmonella spp. isolates in artificially inoculated breaded chicken strips and nuggets during 16 weeks of storage at − 20 °C. However, enumeration on selective agar resulted in a statistically significant reduced recovery of salmonellae compared to non-selective media, indicating structural injury to the cells [35]. In contrast to this, the authors of a study conducted in Mexico observed a considerable decline of the Salmonella population present on naturally contaminated pork during storage at − 15 °C [36]. In a recent study, a lower detection rate of salmonellae in frozen chicken carcasses was determined compared to chilled and non-chilled samples, though the difference was not statistically significant [37]. Due to different storage conditions and meat types used in individual studies, the comparability of the results is limited. Additionally, many further factors were suggested to influence the survival of Salmonella spp. during frozen storage, such as pH and fat content of the meat, which were not evaluated for the samples examined prior to the current study [35].
S. Weltevreden is a common serovar in Southeast Asia and the isolate in the present study was recovered from illegally imported rabbit meat from Vietnam [38, 39]. The isolate did not show resistances to any of the antimicrobial agents included in the test panel. This is in accordance with previous studies that found a low prevalence of antimicrobial resistances for this serovar [40, 41]. Of the 11 S. enterica isolates from illegally imported food products of animal origin examined in the study by Rodríguez-Lázaro and collegues, most isolates did not show phenotypic resistances to any of the antimicrobial agents tested, either. The remaining isolates were each resistant to a single class of antimicrobials [11].
In contrast to this, the S. Infantis in the present study was resistant to substances belonging to five different classes of antimicrobials. In recent years, resistances to multiple antimicrobial agents have been observed comparatively frequently in S. Infantis. Together with S. Kentucky, this serovar currently accounts for a significant proportion of multiresistant non-typhoidal S. enterica isolates in the EU [42].
The isolates from legally imported chicken meat in the current study included one non-flagellated isolate. However, this isolate appeared to be closely related to the two S. Heidelberg isolates recovered from legal imports, based on the high similarity in PFGE band patterns and the shared O-group. Flagella play an important role as virulence factors and enhance adhesion to and invasion of host cells. However, this mostly affects the early stages of infection and non-flagellated strains do not appear notably less virulent if an infection reaches the systemic phase [43, 44].
S. Heidelberg is not commonly associated with human cases of salmonellosis in the EU and did not rank among the 20 most frequent serovars in an EFSA report covering the years 2014–2016 [1]. Data on national cases of human salmonellosis, released by the Robert Koch Institute, also indicate a low prevalence of S. Heidelberg infections in Germany [45]. According to these data, the four serovars accounting for the vast majority of cases in 2016 were S. Enteritidis and S. Typhimurium, followed by S. Infantis and S. Derby. Particularly in North America, however, S. Heidelberg is often the cause of human salmonellosis and was reported to be the sixth most frequent serovar in the US in 2015 [46, 47].
Notably, all isolates from legal imports in this study were recovered from fresh poultry meat. Regarding Salmonella, commission regulation (EC) 2073/2005, amended by commission regulation (EU) 1086/2011, only specifies the absence of S. Enteritidis and S. Typhimurium (including its monophasic variant) as food safety criteria for fresh poultry meat. Other serovars are not covered, even though they might be more common in other countries such as Brazil – the largest exporter of chicken meat supplying the EU [8, 48,49,50].
All three isolates from legally imported poultry meat in this study were AmpC producers carrying blaCMY-2 and they were additionally resistant to ciprofloxacin. Like third-generation cephalosporins, fluoroquinolones are important choices for the treatment of severe cases of salmonellosis [51]. Consequently, co-resistance to both is of particular concern. The overall prevalence of ESBL- and AmpC-β-lactamases among Salmonella enterica isolates is still low in the EU, however, the AmpC phenotype in particular has been comparatively often seen in S. Heidelberg isolates [52, 53]. S. Heidelberg carrying blaCMY-2 have previously been detected on imported poultry meat of Brazilian origin and such imports were suggested to be involved in the increase of third-generation cephalosporin-resistant S. Heidelberg in the Netherlands [7, 8]. The blaCMY-2 genes of the isolates in the present study were located on large conjugative plasmids belonging either to IncA/C or IncI1. Plasmids of these incompatibility groups have previously been associated with AmpC-producing S. Heidelberg from various sources and likely play an important role in the spread of resistance to third-generation cephalosporins in S. Heidelberg and other Salmonella serovars [7, 8, 47, 54].
Conclusion
In conclusion, this study reports the presence of multiresistant Salmonella enterica isolates in meat products imported into Germany either legally or illegally. In particular, the co-resistance to third generation cephalosporins and fluoroquinolones in all three isolates from legally imported meat is worrying. Despite the limited number of isolates examined, the results of this study demonstrate how meat and meat products entering the EU could constitute a route for the dissemination of such isolates and provides further evidence that serovars other than Enteritidis and Typhimurium on fresh poultry meat should not be disregarded.
References
European Food Safety Authority (EFSA) and the European Centre for Disease Prevention and Control (ECDC). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food‐borne outbreaks in 2016. EFSA J. 2017;15(12):e05077.
Chen HM, Wang Y, Su LH, Chiu CH. Nontyphoid Salmonella infection: microbiology, clinical features, and antimicrobial therapy. Pediatr Neonatol. 2013;54(3):147–52.
Crump JA, Sjölund-Karlsson M, Gordon MA, Parry CM. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin Microbiol Rev. 2015;28(4):901–37.
Ramachandran G, Panda A, Higginson EE, Ateh E, Lipsky MM, Sen S, et al. Virulence of invasive Salmonella Typhimurium ST313 in animal models of infection. PLoS Negl Trop Dis. 2017;11(8):e0005697.
Foley SL, Nayak R, Hanning IB, Johnson TJ, Han J, Ricke SC. Population dynamics of Salmonella enterica serotypes in commercial egg and poultry production. Appl Environ Microbiol. 2011;77(13):4273–9.
Antunes P, Mourão J, Campos J, Peixe L. Salmonellosis: the role of poultry meat. Clin Microbiol Infect. 2016;22(2):110–21.
Liakopoulos A, Geurts Y, Dierikx CM, Brouwer MS, Kant A, Wit B, et al. Extended-spectrum cephalosporin-resistant Salmonella enterica serovar Heidelberg strains, the Netherlands. Emerg Infect Dis. 2016;22(7):1257–61.
Campos J, Mourão J, Silveira L, Saraiva M, Correia CB, Maçãs AP, et al. Imported poultry meat as a source of extended-spectrum cephalosporin-resistant CMY-2-producing Salmonella Heidelberg and Salmonella Minnesota in the European Union, 2014–2015. Int J Antimicrob Agents. 2017;51(1):151–4.
Jansen W, Merkle M, Daun A, Flor M, Grabowski NT, Klein G. The quantity and quality of illegally imported products of animal origin in personal consignments into the European Union seized at two German airports between 2010 and 2014. PLoS ONE. 2016;11(2):e0150023.
Müller A, Seinige D, Jansen W, Klein G, Ehricht R, Monecke S, et al. Variety of antimicrobial resistances and virulence factors in Staphylococcus aureus isolates from meat products legally and illegally introduced to Germany. PLoS ONE. 2016;11(12):e0167864.
Rodríguez-Lázaro D, Ariza-Miguel J, Diez-Valcarce M, Stessl B, Beutlich J, Fernandez-Natal I, et al. Identification and molecular characterization of pathogenic bacteria in foods confiscated from non-EU flights passengers at one Spanish airport. Int J Food Microbiol. 2014;209:20–5.
Beutlich J, Hammerl JA, Appel B, Nöckler K, Helmuth R, Jöst K, et al. Characterization of illegal food items and identification of foodborne pathogens brought into the European Union via two major German airports. Int J Food Microbiol. 2015;209:13–9.
Jansen W, Woudstra S, Müller A, Grabowski N, Schoo G, Gerulat B, et al. The safety and quality of pork and poultry meat imports for the common European market received at border inspection post Hamburg Harbour between 2014 and 2015. PLoS ONE. 2018;13(2):e0192550.
Miller T, Braun PG, Fehlhaber K, Prager R, Pfeifer Y, Rabsch W. Typing of Salmonella enterica serovar Infantis isolates from 51 outbreaks in Germany between 1974 and 2009 by a novel phage-typing scheme. Epidemiol Infect. 2014;142(1):75–83.
Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3(1):59–67.
Clinical and Laboratory Standards Institute (CLSI). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—ninth edition CLSI document M07-A9. CLSI: Wayne; 2012.
Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. Twenty-fourth informational supplement. CLSI document M100-S24. CLSI: Wayne; 2014.
Prüller S, Rensch U, Meemken D, Kaspar H, Kopp PA, Klein G, et al. Antimicrobial susceptibility of Bordetella bronchiseptica isolates from swine and companion animals and detection of resistance genes. PLoS ONE. 2015;10(8):e0135703.
Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65(3):490–5.
Sandvang D, Aarestrup FM. Characterization of aminoglycoside resistance genes and class 1 integrons in porcine and bovine gentamicin-resistant Escherichia coli. Microb Drug Resist. 2000;6(1):19–27.
Kehrenberg C, Friederichs S, de Jong A, Michael GB, Schwarz S. Identification of the plasmid-borne quinolone resistance gene qnrS in Salmonella enterica serovar Infantis. J Antimicrob Chemother. 2006;58(1):18–22.
Vredenburg J, Varela AR, Hasan B, Bertilsson S, Olsen B, Narciso-da-Rocha C, et al. Quinolone-resistant Escherichia coli isolated from birds of prey in Portugal are genetically distinct from those isolated from water environments and gulls in Portugal, Spain and Sweden. Environ Microbiol. 2014;16(4):995–1004.
Arcangioli MA, Leroy-Sétrin S, Martel JL, Chaslus-Dancla E. A new chloramphenicol and florfenicol resistance gene flanked by two integron structures in Salmonella typhimurium DT104. FEMS Microbiol Lett. 1999;174(2):327–32.
Kadlec K, Kehrenberg C, Schwarz S. Molecular basis of resistance to trimethoprim, chloramphenicol and sulphonamides in Bordetella bronchiseptica. J Antimicrob Chemother. 2005;56(3):485–90.
Senda K, Arakawa Y, Ichiyama S, Nakashima K, Ito H, Ohsuka S, et al. PCR detection of metallo-beta-lactamase gene (bla IMP) in Gram-negative rods resistant to broad-spectrum β-lactams. J Clin Microbiol. 1996;34(12):2909–13.
O’Regan E, Quinn T, Pagès JM, McCusker M, Piddock L, Fanning S. Multiple regulatory pathways associated with high-level ciprofloxacin and multidrug resistance in Salmonella enterica serovar Enteritidis: involvement of ramA and other global regulators. Antimicrob Agents Chemother. 2009;53(3):1080–7.
Nucleotide BLAST, provided by the National Center for Biotechnology Information, U.S. National Library of Medicine. https://blast.ncbi.nlm.nih.gov/Blast.cgi. Accessed 13 Mar 2018.
Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63(3):219–28.
Kayser FH, Morenzoni G, Homberger F. Activity of cefoperazone against ampicillin-resistant bacteria in agar and broth dilution tests. Antimicrob Agents Chemother. 1982;22(1):15–22.
Doumith M, Day MJ, Hope R, Wain J, Woodford N. Improved multiplex PCR strategy for rapid assignment of the four major Escherichia coli phylogenetic groups. J Clin Microbiol. 2012;50(9):3108–10.
Kehrenberg C, Schwarz S. Occurrence and linkage of genes coding for resistance to sulfonamides, streptomycin and chloramphenicol in bacteria of the genera Pasteurella and Mannheimia. FEMS Microbiol Lett. 2001;205(2):283–90.
McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413(6858):852–6.
Schwarz S, Silley P, Simjee S, Woodford N, van Duijkeren E, Johnson AP, et al. Assessing the antimicrobial susceptibility of bacteria obtained from animals. Vet Microbiol. 2010;141(1–2):1–4.
International Air Transport Associatoin (IATA) Press Release No. 55: 2036 Forecast Reveals Air Passengers Will Nearly Double to 7.8 Billion. 2017. http://www.iata.org/pressroom/pr/Pages/2017-10-24-01.aspx. Accessed 30 Jul 2018.
Dominguez SA, Schaffner DW. Survival of Salmonella in processed chicken products during frozen storage. J Food Prot. 2009;72(10):2088–92.
Escartı́n EF, Lozano JS, Garcı́a OR. Quantitative survival of native Salmonella serovars during storage of frozen raw pork. Int J Food Microbiol. 2000;54(1):19–25. https://doi.org/10.1016/S0168-1605(99)00149-X.
Khan AS, Georges K, Rahaman S, Abdela W, Adesiyun AA. Prevalence and serotypes of Salmonella spp. on chickens sold at retail outlets in Trinidad. PLoS ONE. 2018;13(8):e0202108.
Domingues AR, Vieira AR, Hendriksen RS, Pulsrikarn C, Aarestrup FM. Spatio-temporal analysis of Salmonella surveillance data in Thailand. Epidemiol Infect. 2014;142(8):1614–24.
Lettini AA, Vo Than T, Marafin E, Longo A, Antonello K, Zavagnin P, et al. Distribution of Salmonella serovars and antimicrobial susceptibility from poultry and swine farms in central Vietnam. Zoonoses Public Health. 2016;63(7):569–76.
Li B, Yang X, Tan H, Ke B, He D, Wang H, et al. Whole genome sequencing analysis of Salmonella enterica serovar Weltevreden isolated from human stool and contaminated food samples collected from the southern coastal area of China. Int J Food Microbiol. 2018;266:317–23.
Makendi C, Page AJ, Wren BW, Le Thi Phuong T, Clare S, Hale C, et al. A phylogenetic and phenotypic analysis of Salmonella enterica serovar Weltevreden, an emerging agent of diarrheal disease in tropical regions. PLoS Negl Trop Dis. 2016;10(2):e0004446.
Hindermann D, Gopinath G, Chase H, Negrete F, Althaus D, Zurfluh K, et al. Salmonella enterica serovar Infantis from food and human infections, Switzerland, 2010–2015: poultry-related multidrug resistant clones and an emerging ESBL producing clonal lineage. Front Microbiol. 2017;8:1322.
Haiko J, Westerlund-Wikström B. The role of the bacterial flagellum in adhesion and virulence. Biology (Basel). 2013;2(4):1242–67.
Lockman HA, Curtiss R. Salmonella typhimurium mutants lacking flagella or motility remain virulent in BALB/c mice. Infect Immun. 1990;58(1):137–43.
RKI. Infektionsepidemiologisches Jahrbuch meldepflichtiger Krankheiten für, Infectious disease epidemiology annual report for 2016. Berlin: Robert Koch-Institut; 2016. p. 2017.
Centers for Disease Control and Prevention (CDC). National enteric disease surveillance: Salmonella annual report, 2015. Atlanta: US Department of Health and Human Services, CDC, 2017. https://www.cdc.gov/nationalsurveillance/salmonella-surveillance.html.
Edirmanasinghe R, Finley R, Parmley EJ, Avery BP, Carson C, Bekal S, et al. A whole-genome sequencing approach to study cefoxitin-resistant Salmonella enterica serovar Heidelberg isolates from various sources. Antimicrob Agents Chemother. 2017;61(4):e01919.
Voss-Rech D, Vaz CS, Alves L, Coldebella A, Leão JA, Rodrigues DP, et al. A temporal study of Salmonella enterica serotypes from broiler farms in Brazil. Poult Sci. 2015;94(3):433–41.
European Commission. EU Market Situation for Poultry. Committee for the Common Organisation of the Agricultural Markets Brussels, European Commision, Directorate General for Agriculture and Rural Development. 2018. https://circabc.europa.eu/sd/a/cdd4ea97-73c6-4dce-9b01-ec4fdf4027f9/19.04.2017-Poultry.pptfinal.pdf. Accessed 19 Jul 2018.
Commission European. Commission Regulation (EU) N° 1086/2011 of 27 October 2011 amending Annex II to Regulation (EC) N° 2160/2003 of the European Parliament and of the Council and Annex I to Commission Regulation (EC) N° 2073/2005 as regards Salmonella in fresh poultry meat. Off J Eur Union L. 2011;281:7–11.
European Centre for Disease Prevention and Control (ECDC), European Food Safety Authority (EFSA), European Medicines Agency (EMA), Scientific Committee on Emerging and Newly Identified Health Risks (SCENIHR). Joint opinion on antimicrobial resistance (AMR) focused on zoonotic infections. EFSA J. 2009;7(11):1372.
Rodríguez I, Barownick W, Helmuth R, Mendoza MC, Rodicio MR, Schroeter A, et al. Extended-spectrum β-lactamases and AmpC β-lactamases in ceftiofur-resistant Salmonella enterica isolates from food and livestock obtained in Germany during 2003-07. J Antimicrob Chemother. 2009;64(2):301–9.
European Food Safety Authority (EFSA) and the European Centre for Disease Prevention and Control (ECDC). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2015. EFSA J. 2017;15(2):e04694.
Mataseje LF, Baudry PJ, Zhanel GG, Morck DW, Read RR, Louie M, et al. Comparison of CMY-2 plasmids isolated from human, animal, and environmental Escherichia coli and Salmonella spp. from Canada. Diagn Microbiol Infect Dis. 2010;67(4):387–91.
Authors’ contributions
AM prepared the manuscript, performed the experiments and analyzed the data. CK coordinated the project, analyzed the data and revised the manuscript. WJ and NG analyzed results and reviewed the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to thank Iris Oltrogge, Inna Pahl and Rouwen Stucke for expert technical assistance. Furthermore the authors thank M. Wahnfried for serotyping, S. Kulbe and D. Busse for phage typing and Dr. W. Rabsch (all Robert Koch Institute in Wernigerode).
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was in part supported by the German Federal Ministry of Education and Research within the research program for civil security, Project ZooGloW, FKZ 13N12697. This had no influence on the design of the study and collection, analysis, and interpretation of data or the writing of the manuscript.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Müller, A., Jansen, W., Grabowski, N.T. et al. Characterization of Salmonella enterica serovars recovered from meat products legally and illegally imported into the EU reveals the presence of multiresistant and AmpC-producing isolates. Gut Pathog 10, 40 (2018). https://doi.org/10.1186/s13099-018-0268-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13099-018-0268-3