Abstract
Background
For many putative Salmonella enterica subsp. enterica virulence genes, functional characterization across serovars has been limited. Cytolethal distending toxin B (CdtB) is an incompletely characterized virulence factor that is found not only in Salmonella enterica subsp. enterica serovar Typhi (Salmonella Typhi) and dozens of Gram negative bacterial pathogens, but also in non-typhoidal Salmonella (NTS) serovars.
Methods
A comparative genomics approach was performed to characterize sequence conservation of the typhoid toxin (TT), encoded in the CdtB-islet, between Salmonella Typhi and NTS serovars. The cytotoxic activity of representative Salmonella enterica subsp. enterica serovars Javiana, Montevideo and Schwarzengrund strains and their respective isogenic cdtB mutants was determined in human intestinal epithelial Henle-407 cells by assessment of cell cycle progression of infected cells using fluorescence-activated cell sorting (FACS). Two-way analysis of variance (ANOVA) was used to determine whether cdtB deletion had a significant (p < 0.05) effect on the percentage of Henle-407 cells at each stage of the cell cycle.
Results
Here we show that a CdtB-islet encoding the cytolethal distending toxin B (CdtB), pertussis-like toxin A (PltA), and pertussis-like toxin B (PltB) is present in a dozen NTS serovars and that these proteins have a high level of sequence conservation and each form monophyletic clades with corresponding Salmonella Typhi genes. Human epithelial Henle-407 cells infected with three representative CdtB-encoding NTS serovars displayed G2/M phase cell cycle arrest that was absent in cells infected with corresponding isogenic cdtB null mutants (p < 0.0001 for the factor ∆cdtB deletion).
Conclusion
Our results show that CdtB encoded by NTS serovars has a genomic organization, amino acid sequence conservation and biological activity similar to the TT, and thus, may contribute to disease pathogenesis.
Similar content being viewed by others
Background
The cytolethal distending toxin B (CdtB) is a recently recognized virulence factor of Salmonella enterica subsp. enterica serovar Typhi [1], as well as a wide range of other Gram negative bacterial pathogens in the Gamma and Epsilon classes of Proteobacteria [2]. Host cells intoxicated with CdtB display a DNA damage response (DDR) characterized by irreversible cell cycle arrest, with persistent DDR leading to cell death by apoptosis [2]. Multi-locus sequence analysis data reported by our group have previously shown an unambiguous subdivision of S. enterica subsp. enterica into at least two populations that we designated clades A and B [3]; this subdivision has been confirmed by other studies [4, 5]. Clades A and B not only represents well supported subdivisions, but also show distinct genomic features that strongly suggests niche specialization of these subpopulations; for example clade B strains contain several clade specific genes or operons, including a ß-glucuronidase operon, a S-fimbrial operon, and clade B specific cell surface related genes [3]. The study by den Bakker et al. [3] also identified an islet encoding the cytolethal distending toxin B (CdtB-islet) in the genomes of 56 non-typhoidal Salmonella isolates, including (i) 37/38 clade B isolates, 14/115 non-typhoidal clade A isolates, and five isolates that did not clearly group into clade A or B [3]. Despite these observations and the important role of non-typhoidal Salmonella as foodborne and zoonotic pathogens, the role of CdtB in non-typhoidal serovars has remained understudied; studies that explore the functionality of CdtB in non-typhoidal serovars are thus essential.
In Salmonella Typhi, the CdtB-islet includes five genes, namely pltA, pltB, ttsA, sty1887, in addition to cdtB. pltA and pltB encode homologs of pertussis toxin components, which are responsible for ADP-ribosylation of a host protein [6] and export of CdtB from the Salmonella containing vacuole as well as from infected host cells. CdtB, PltA, and PltB are the three subunits that form the so called “typhoid toxin” in Salmonella Typhi. ttsA encodes a phage-origin muramidase necessary for the secretion of the PltA/CdtB/PltB toxin [7]. sty1887 encodes a putative homolog of a phage tail protein; deletion of this gene in Salmonella Typhi did not affect secretion of CdtB [7]. While other Gram-negative bacteria also contain cdtB, these other species typically do not contain homologs of pltA and pltB. In these other Gram-negative species toxin import into host cell compartments and into the cytosol appears to typically be facilitated by CdtA and CdtC; in these bacteria CdtA, CdtB and CdtC are protein subunits and assemble into a single holotoxin [2]. Importantly, CdtA and CdtC show no homologies with PltA and PltB.
On the basis of the well-documented role of CdtB in host-pathogen interactions in Salmonella Typhi and other bacterial pathogens, we hypothesized that CdtB may also play a critical role in host cell interactions of non-typhoidal Salmonella serovars newly identified as encoding CdtB. We used a comparative genomics approach to characterize sequence conservation of pltA, pltB, and cdtB among S. Typhi and non-typhoidal Salmonella serovars. To confirm the biological activity of CdtB in non-typhoidal Salmonella serovars, we created cdtB null mutants in three representative non-typhoidal Salmonella strains and assessed the cell cycle of infected Henle-407 human epithelial cells.
Results
Phylogenetic analysis revealed that CdtB encoded in the genomes of NTS serovars has a high level of sequence conservation when compared to S. Typhi’s CdtB
A maximum likelihood (ML) based phylogeny of CdtB amino acid sequences showed that homologs of this gene are widely distributed among Gram-negative bacteria (Fig. 1), including 11 serovars classified into S. enterica subsp. enterica clade B (13 isolates, Fig. 1), S. enterica subsp. enterica serovar Inverness (FSL R8-3668), S. enterica subsp. arizonae (RSK2980), and S. bongori (NCTC 12419). All S. enterica genomes that contained CdtB also encoded PltA and PltB, these genes are characteristic of the CdtB-islet, which encodes the components of the S. Typhi typhoid holotoxin complex [8]. In 18/20 cdtB positive S. enterica strains, we also identified paralogs of pltA and pltB in a genomic region that was not within the CdtB-islet. These paralogs have previously been annotated as the artA/artB operon in S. Typhimurium DT104 [9] and encode an AB5 toxin [10]. S. Typhi CdtB, PltA, and PltB show high levels of homology with the corresponding proteins in non-typhoidal S. enterica subsp. enterica isolates (with the exception of S. Inverness) with 99.3–100% amino acid (aa) sequence identity for CdtB (Fig. 1), 98.3–100% aa sequence identity for PltA (Additional file 1), and 94.9–100% aa sequence identity for PltB (Additional file 2). The high level of sequence conservation for CdtB is further confirmed by the high (>98%) aa sequence identity among the 12 S. enterica subsp. enterica serovars (including Typhi and Paratyphi A) (Fig. 1). Moreover, functionally critical residues (i.e., PltA Cys 214, CdtB Cys 269) within the typhoid holotoxin proteins, as determined by Song et al. [8], were conserved in all 14 non-typhoidal Salmonella genomes examined (Additional file 3).
Henle-407 human epithelial cells infected with typhoid toxin-producing NTS strains displayed cell arrest in G2/M phase of the cell cycle
Given the high level of sequence conservation for all three typhoid toxin subunits (i.e., CdtB, PltA, PltB) including conservation of functionally critical aa residues, we hypothesized that a functional typhoid holotoxin is produced by non-typhoidal salmonellae. Therefore, we constructed isogenic mutants with deletions of cdtB in three non-typhoidal Salmonella enterica strains, classified in clade B, for further phenotypic characterization. One representative strain of Salmonella serovars Javiana, Montevideo, and Schwarzengrund was selected because these serovars have been responsible for several large outbreaks in the last 10 years [11–14] in the United States. In addition, S. Javiana and S. Montevideo are among the six most frequently reported serovars isolated from humans in the US [15]. When Henle-407 cells were infected with parent strains and corresponding isogenic mutants, a clear cell cycle arrest was observed at 72 h post-inoculation, using a fluorescence-activated cell sorter (FACS). Specifically, Henle-407 cells infected with Salmonella parent strains were arrested in the G2/M phase of the cell cycle (Fig. 2); 60.3% of Henle cells infected with the parent strains were in the G2/M phase (average across all three serovars), which is significantly higher as compared to the Henle cells infected with the cdtB mutant strains (24.3% average across all three serovars; p < 0.0001; see Fig. 2). By contrast, 18.6% of Henle cells infected with the parent strains (average across all three serovars) were in G1 phase, which is significantly (p < 0.0001; Fig. 2) lower as compared to Henle cells infected with the corresponding Salmonella cdtB null mutants (61.2% average across all three serovars) and compared to uninfected cells (60.0%). Serovar did not have a significant effect, and there were no significant interactions between cdtB status (mutant vs. wild type) and serovar. These findings show that non-typhoidal Salmonella serovars produce a functional typhoid toxin that causes G2/M cell cycle arrest in human epithelial cells. While some may argue that complementation of the cdtB mutants would be needed to confirm these findings, the fact that we found consistent phenotypes across three distinct serovars provides strong evidence that the effects shown are due to the mutations introduced. Although S. Typhi was not included as a control in our experiments, the patterns of cell cycle arrest were similar to those previously reported; for example, Spanò et al. [6] showed that 84% of Typhi infected Henle cells were in G2/M phase as compared to 16% of cells infected with Typhi ΔcdtB and 14% of untreated cells.
Cell cycle analysis of human epithelial Henle-407 cells infected with non-typhoidal Salmonella wild type (WT) strains and their corresponding isogenic cdtB mutants. Cells infected with Salmonella WT strains and their isogenic mutants were analyzed by FACS at 72 h of post-infection. Data shown represent the averages of two independent biological replicates with two technical replicates (two infected wells per experiment) each. Y axis indicates percentage of cells in G1, S, and G2/M phase (as indicated on the X axis); the bars indicate the range of values. Cells infected with Salmonella WT parent strains (i.e., S. Montevideo, S. Schwarzengrund, and S. Javiana) showed significantly higher percentage cells in G2/M phase compared to the corresponding isogenic ∆cdtB mutants (p < 0.0001 for the factor ∆cdtB deletion; ANOVA). Conversely, cells infected with the isogenic ∆cdtB mutants showed significantly higher percent cells in G1 phase (p < 0.0001; ANOVA) compared to the corresponding parent WT strains.
Salmonella serovars that showed a higher proportion of cases with invasive disease were significantly (p < 0.05) more likely to be cdtB positive
To determine whether cdtB positive serovars are more likely to be associated with invasive disease, we utilized previously reported epidemiological data on association of disease severity with different Salmonella serovars; this study used data for 46,639 human cases reported in the US between 1996 and 2006 [16]. Serovars were classified as cdtB positive if all isolates of a given serovar characterized by den Bakker et al. [3] were reported to be positive for cdtB. Among the serovars in this study, 13 showed a significantly higher proportion of invasive disease cases (as compared to Salmonella Typhimurium), while 35 showed a lower or equal proportion of invasive disease cases. When serovars were classified as cdtB positive or negative, based on data reported by den Bakker et al. [3], 8 of the 13 serovars that showed a higher proportion of invasive disease cases could be classified as cdtB positive serovars (i.e., Oranienburg, Poona, Schwarzengrund, Panama, Sandiego, Brandenburg, Muenster, Urbana). Among the 35 serovars that showed an equal or lower proportion of invasive disease cases, only eight were classified as cdtB positive (i.e., Javiana, Miami, Montevideo, Rubislaw, Gaminara, Kiambu, Johannesburg and Give). A Fisher’s exact test showed that serovars that showed a higher proportion of cases with invasive disease were significantly (p < 0.05) more likely to be cdtB positive as compared to serovars that showed a lower proportion of cases with invasive disease.
Discussion
A comparative genomic study by den Bakker et al. [3] previously revealed the presence of a CdtB-islet within the genomes of 56 non-typhoidal Salmonella isolates, of which approximately 70% belonged to a restricted subpopulation of S. enterica subsp. enterica (clade B). Subsequently, Desai et al. [17] reported the presence of cdtB (referred to as typhoid toxin in that study) in the genomes of 2 S. enterica subsp. diarizoniae and 2 S. enterica subsp. arizoniae strains. Consistent with previous analyses [18], the present comparative genomic analyses confirmed that homologs of Salmonella Typhi cdtB are widely distributed among Gram-negative bacterial pathogens. Contribution of CdtB to disease pathogenesis has been reported in dozens of bacterial pathogens, including reduced cytotoxicity of cdtB null mutants (e.g., H. ducreyi, H. hepaticus, C. jejuni) and CdtB mediated G2/M cell cycle arrest (e.g., E. coli, A. actinomycetemcomitans, H. ducreyi, H. hepaticus) [2, 19]. Importantly, our sequence analysis shows that the genes encoded by the CdtB-islet of S. Typhi and non-typhoidal serovars are highly conserved and share a common phylogenetic ancestor, providing new evidence of a functional CdtB in a large subset of pathogenic non-typhoidal Salmonella.
Phenotypic characterization of cdtB null mutants in three different serovar backgrounds confirmed that CdtB is functional across different non-typhoidal Salmonella and required for induction of G2/M phase cell cycle arrest of intoxicated human intestinal epithelial Henle-407 cells. While our findings are consistent with previous reports that S. Typhi CdtB is directly responsible for G2/M phase cell cycle arrest in infected eukaryotic host cells [1, 6–8], our data provide important new evidence that indicate a contribution of CdtB in disease pathogenesis across a large number of human non-typhoidal Salmonella strains. Importantly CdtB-islet positive strains may also cause cell cycle arrest in the infected target tissues, which may increase the risk of long-term sequelae. This is consistent with Lara-Tejero & Galán [20] who suggested that exposure to CdtB and the associated DDR may predispose individuals infected with C. jejuni to intestinal cancer. Consistent with our results, a recent study also confirmed the presence of a functional CdtB in a strain of Salmonella Javiana [21]; this study also showed that the cell cycle arrest pattern observed in cells infected with the Salmonella Typhi control strain was similar to that observed for the Salmonella Javiana test strain. Another recent study also showed that a Salmonella Javiana cdtB mutant strain did not adhere more to mice macrophages when compared to the wild type (WT) [22]. Previous studies with Salmonella Typhi [1] have also demonstrated that CdtB is only synthesized when Salmonella is within the intracellular compartment. As IgeR, which has been found to repress Salmonella Typhi cdtB transcription in extracellular environments by binding to the cdtB promoter, is conserved across typhoidal and non-typhoidal Salmonella serovars, IgeR may also downregulates cdtB transcription in extracellular environments in non-typhoidal serovars. Future studies are needed though to evaluate regulation of CdtB expression in non-typhoidal serovars in order further understand the specific role of CdtB in non-typhoidal Salmonella serovars.
Conclusion
Our findings highlight the possibility that non-typhoidal Salmonella may represent distinct CdtB-islet positive and negative subgroups, which differ in pathogenetic mechanisms and host interactions. Further experimental work, including characterization of cdtB mutants in animal models, is clearly needed though to more completely define the contributions of CdtB and the other proteins encoded in the CdtB-islet, as well as the artAB operon, to pathogenesis of a growing list of non-typhoidal Salmonella serovars recognized as encoding these virulence factors; our mutants are freely available to other researchers for these types of experiments. Given the association of several CdtB-producing bacterial pathogens with cancer [23, 24] and the ability of CdtB to cause a DDR in a wide range of eukaryotic cells, it would also be important to explore whether CdtB-producing bacteria can promote cancer development in their respective hosts. As an analysis of previously reported data on distribution of invasive and non-invasive disease among patients infected with different Salmonella serovars provided preliminary support that cdtB positive isolates may be more likely to cause invasive disease, future epidemiological studies are also needed to determine whether cdtB-positive Salmonella isolates are associated with different or more severe disease outcomes in human hosts, in addition to animal studies on the role of CdtB in invasive infection by non-typhoidal Salmonella.
Methods
PltA, PltB, and CdtB phylogeny and amino acid sequence analysis
PltA, PltB and CdtB amino acid sequences of S. Typhi CT18 were obtained from GenBank. Salmonella genomes (both finished and draft, obtained from the nr/nt and WGS databases at NCBI) were queried for homologs of S. Typhi CT18 PltA, PltB and CdtB using both nucleotide and protein blast searches (blastp and blastn, respectively). Nucleotide sequences obtained from these searches were translated into amino acid sequences and aligned using MAFFT [25]. Maximum likelihood phylogenies were created with PhyML (version 20130708), using the WAG model of amino acid substitution and a gamma distribution of variable sites. To assess the robustness of the inferred phylogeny 250 bootstrap replicates were performed for each analysis.
Bacterial strains
Three non-typhoidal Salmonella strains representing serovars Javiana (isolate FSL S5-0395), Schwarzengrund (FSL R6-0879), and Montevideo (FSL R8-4841) were obtained from the New York State Department of Health; all strains were from humans with clinical symptoms of salmonellosis. The presence of pltA, pltB, and cdtB in these strains was confirmed by TaqMan® assays (Life Technologies) as previously described [3]. Isogenic ∆cdtB mutants were constructed by using the Lambda Red system as previously described [26] and cdtB deletions were confirmed by PCR and sequencing of the deletion allele (see Additional file 4).
Henle-407 cell infection
Human epithelial Henle-407 cells (ATCC CCL-6) were grown at 37°C in 5.0% CO2 atmosphere and Dulbecco’s Modified Eagle Medium (DMEM; Corning) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals). For infection studies, bacterial cultures were prepared as previously described [1], with some modifications; all Salmonella growth steps were performed in Lysogeny Broth (LB, commonly referred to as Luria–Bertani broth) with 0.3 M NaCl at 37°C, without shaking. Briefly, overnight Salmonella cultures were diluted 1:100 in fresh LB-NaCl broth and incubated at 37°C until they reached OD600 of 0.4. Then, these cultures were diluted 1:100 into Nephelo culture flasks with 50 mL LB-NaCl broth, and incubated at 37°C until the cultures reached an OD600 of 0.4, followed by incubation for an additional 3 h to yield a final density of approx. 1 × 109 CFU/mL. Infection studies were performed with Henle-407 cells seeded in 6-well plates and incubated for 24 h before inoculation; media was replaced with fresh DMEM-FBS 30 min before inoculation with Salmonella at an MOI of 50. After incubation at 37°C and 5.0% CO2 for 1 h, the cells were washed with phosphate buffered saline (PBS) followed by incubation in fresh DMEM-FBS containing gentamicin (100 μg/mL) for 1 h. After 1 h, the Henle-407 cells were washed 3 times with PBS and fresh DMEM-FBS with gentamicin (10 μg/mL) was added, followed by incubation for another 72 h. At the end of the incubation period, the uninfected control and infected Henle-407 cells were processed for cell cycle analysis with a fluorescence-activated cell sorter (FACS), as described below. All assays were performed in duplicate in two separate biological replicates with each Salmonella strain.
Cell cycle analysis
The cell cycle of uninfected control and infected Henle-407 cells was determined by FACS analysis as described previously [20], with some modifications. Briefly, Henle-407 cells were washed, trypsinized, and centrifuged at 1,500 rpm for 5 min at room temperature. The supernatant was removed, the cells were fixed by adding cold 70% ethanol (while vortexing at slow speed) and kept at –20°C for at least 1 h before adding PBS containing 0.1% (v/v) Tween 20 and 1% (w/v) BSA (PBST). Then the cells were incubated for 10 min at room temperature, washed three times in PBST before re-suspending in propidium iodide (PI; Sigma-Aldrich) staining solution [40 μg of PI/mL, 100 μg of RNase A/mL (Sigma-Aldrich)], followed by incubation at room temperature in the dark for an additional 10 min. Subsequent DNA content analysis of approximately 3 × 104 cells was performed with a LSRII Flow Cytometer (BD-Biosciences). The percentages of Henle-407 cells in G1, S, and G2/M phase of the cell cycle were calculated after quantifying the mean percentages of the cells detected in manually adjusted gates for 2N, 3N, and 4N DNA contents.
Statistical analysis
Data were imported into a commercially available statistical software program (SAS, version 9.2; SAS Institute Inc., Cary, NC, USA) for analysis. Two-way analysis of variance (ANOVA) was used to determine whether cdtB deletion had a significant (p < 0.05) effect on the percentage of Henle-407 cells at the G1, S, and G2/M stages of the cell cycle; each measurement represented the mean of two technical replications. CdtB status (mutant vs. wild type) and serovar were included as factors in the analysis, and their interaction was also evaluated. Tukey’s test was used to investigate any differences in the means.
References
Haghjoo E, Galán JE (2004) Salmonella Typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proc Natl Acad Sci USA 101:4614–4619
Jinadasa RN, Bloom SE, Weiss RS, Duhamel GE (2011) Cytolethal distending toxin: a conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages. Microbiology 157:1851–1875
Den Bakker HC, Moreno Switt AI, Govoni G, Cummings CA, Ranieri ML, Degoricija L et al (2011) Genome sequencing reveals diversification of virulence factor content and possible host adaptation in distinct subpopulations of Salmonella enterica. BMC Genomics 12:425
Didelot X, Bowden R, Street T, Golubchik T, Spencer C, McVean G et al (2011) Recombination and population structure in Salmonella enterica. PLoS Genet 7:e1002191
Timme RE, Pettengill J, Allard MW, Strain E, Barrangou R, Wehnes C et al (2013) Phylogenetic diversity of the enteric pathogen Salmonella enterica subsp. enterica inferred from genome-wide reference-free SNP characters. Genome Biol Evol 5:2109–2123
Spanò S, Ugalde JE, Galán JE (2008) Delivery of a Salmonella Typhi exotoxin from a host intracellular compartment. Cell Host Microbe 3:30–38
Hodak H, Galán JE (2013) A Salmonella Typhi homologue of bacteriophage muramidases controls typhoid toxin secretion. EMBO Rep 14:95–102
Song J, Gao X, Galán JE (2013) Structure and function of the Salmonella Typhi chimaeric A2B5 typhoid toxin. Nature 499:350–354
Uchida I, Ishihara R, Tanaka K, Hata E, Makino S, Kanno T et al (2009) Salmonella enterica serotype Typhimurium DT104 ArtA-dependent modification of pertussis toxin-sensitive G proteins in the presence of [32P]NAD. Microbiology 155:3710–3718
Wang H, Paton JC, Herdman BP, Rogers TJ, Beddoe T, Paton AW (2013) The B subunit of an AB5 toxin produced by Salmonella enterica serovar Typhi up-regulates chemokines, cytokines, and adhesion molecules in human macrophage, colonic epithelial, and brain microvascular endothelial cell lines. Infect Immun 81:673–683
Bidol SA, Daly ER, MPH, Rickert RE, S. Newport Investigation Team 2005, S. Braenderup Investigation Team 2005 et al (2007) Multistate outbreaks of Salmonella infections associated with raw tomatoes eaten in restaurants–United States, 2005–2006. Morb Mortal Wkly Rep 56:909–911
Ferraro A, Deasy M, Dato V, Moll, M, Sandt C, Tait J et al (2008) Multistate outbreak of human Salmonella infections caused by contaminated dry dog food–United States, 2006–2007. Morb Mortal Wkly Rep 57:521–524
Lienau EK, Strain E, Wang C, Zheng J, Ottesen AR, Keys CE et al (2011) Identification of a salmonellosis outbreak by means of molecular sequencing. N Engl J Med 364:981–982
Centers for Disease Control and Prevention (2013) An atlas of Salmonella in the United States, 1968–2011. 1–248. http://www.cdc.gov/salmonella/pdf/salmonella-atlas-508c.pdf
Centers for Disease Control and Prevention (2014) National Center For Emerging and Zoonotic Infectious Diseases: National Enteric Disease Surveillance. Salmonella annual report, 2012, pp 1–21
Jones TF, Ingram LA, Cieslak PR, Vugia DJ, Tobin-D’ Angelo M, Hurd S et al (2008) Salmonellosis outcomes differ substantially by serotype. J Infect Dis 198:109–114
Desai PT, Porwollik S, Long F, Cheng P, Wollam A, Clifton SW et al (2013) Evolutionary genomics of Salmonella enterica subspecies. MBio 4:00579
Gargi A, Reno M, Blanke SR (2012) Bacterial toxin modulation of the eukaryotic cell cycle: are all cytolethal distending toxins created equally? Front Cell Infect Microbiol 2:1–11
Liyanage NPM, Dassanayake RP, Kuszynski CA, Duhamel GE (2013) Contribution of Helicobacter hepaticus cytolethal distending toxin subunits to human epithelial cell cycle arrest and apoptotic death in vitro. Helicobacter 18:433–443
Lara-Tejero M, Galán JE (2000) A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science 290:354–357
Mezal EH, Bae D, Khan AA (2014) Detection and functionality of the cdtB, pltA and pltB from Salmonella enterica serovar Javiana. Pathog Dis 72:95–103
Williams K, Gokulan K, Shelman D, Akiyama T, Khan A, Khare S (2015) Cytotoxic mechanism of cytolethal distending toxin in nontyphoidal Salmonella serovar (Salmonella Javiana) during macrophage infection. DNA Cell Biol 34:113–124
Ericsson AC, Myles M, Davis W, Ma L, Lewis M, Maggio-Price L et al (2010) Noninvasive detection of inflammation-associated colon cancer in a mouse model. Neoplasia 12:1054–1065
Fox JG (2007) Helicobacter bilis: bacterial provocateur orchestrates host immune responses to commensal flora in a model of inflammatory bowel disease. Gut 56:897–898
Katoh K, Toh H (2008) Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9:286–298
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645
Authors’ contributions
LDR-R, MW, and GED conceived the study. BMB constructed the mutants. LDR-R conducted the all experiments in this manuscript. HCdB performed the phylogenetic analysis. LDR-R wrote the paper with input from HCdB, GED and MW. All authors read and approved the final manuscript.
Acknowledgements
We would like to acknowledge Dr. Rasika N. Jinadasa and Lavanya Gowri Sayam for technical assistance with FACS analysis. This work was supported in part by a United States Department of Agriculture, National Institute of Food and Agriculture Special Research Grant (2010-34459-20756) to M.W. and by funds provided by the National Institutes of Health, National Institute of Allergy and Infectious Diseases R21 Grant (1AI085332) to G.E.D.
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1:
Amino acid based maximum likelihood phylogeny of PltA. Non-typhoidal Salmonella enterica subsp. enterica serovars are colored green, S. Paratyphi A accessions are colored orange, and S. Typhi accessions are colored red. Values on or next to the branches are bootstrap values based on 250 bootstrap replicates.
Additional file 2:
Amino acid based maximum likelihood phylogeny of PltB. Non-typhoidal Salmonella enterica subsp. enterica serovars are colored green, S. Paratyphi A accessions are colored orange, and S. Typhi accessions are colored red. Values on or next to the branches are bootstrap values based on 250 bootstrap replicates.
Additional file 3:
Salmonella genomes and sequences used for cdtB, pltA, and pltB analyses. Table in a Microsoft excel file listing the accession numbers of the genomes used for phylogenetic analysis.
Additional file 4:
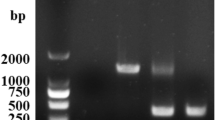
Sequence of cdtB and surrounding region, indicating that deletion is internal to cdtB; cdtB start and stop codons are boxed. Primer sequences in bold. Primer sequence homologous to pKD4 template primer P1 and P2 are italicized. ‘Scar’ sequence from pKD4 in orange. The sequence shown is for the serovar Javiana mutant; the serovar Schwarzengrund and Montevideo mutants showed equivalent sequences. (B) Organization of the region that contains pltB, pltA, and cdtB, which indicates that the cdtB deletion shown in (a) will not disrupt pltA or pltB. (C) Gel that shows the PCR product for the cdtB deletion alleles in serovars Schwarzengrund (lane 1), Montevideo (lane 4), and Javiana (lane 7); these PCR products were used to generate the sequencing data shown in part (A).
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Rodriguez-Rivera, L.D., Bowen, B.M., den Bakker, H.C. et al. Characterization of the cytolethal distending toxin (typhoid toxin) in non-typhoidal Salmonella serovars. Gut Pathog 7, 19 (2015). https://doi.org/10.1186/s13099-015-0065-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13099-015-0065-1