Abstract
Background
Bone marrow-derived mesenchymal stem cells (BMSCs) are general progenitor cells of osteoblasts and adipocytes and they are characterized as a fundamental mediator for bone formation. The current research studied the molecular mechanisms underlying circRNA-regulated BMSC osteogenic differentiation.
Methods
Next-generation sequencing (NGS) was employed to study abnormal circRNA and mRNA expression in BMSCs before and after osteogenic differentiation induction. Bioinformatics analysis and luciferase reporting analysis were employed to confirm correlations among miRNA, circRNA, and mRNA. RT-qPCR, ALP staining, and alizarin red staining illustrated the osteogenic differentiation ability of BMSCs.
Results
Data showed that circ-Iqsec1 expression increased during BMSC osteogenic differentiation. circ-Iqsec1 downregulation reduced BMSC osteogenic differentiation ability. The present investigation discovered that Satb2 played a role during BMSC osteogenic differentiation. Satb2 downregulation decreased BMSC osteogenic differentiation ability. Bioinformatics and luciferase data showed that miR-187-3p linked circ-Iqsec1 and Satb2. miR-187-3p downregulation or Satb2 overexpression restored the osteogenic differentiation capability of BMSCs post silencing circ-Iqsec1 in in vivo and in vitro experiments. Satb2 upregulation restored osteogenic differentiation capability of BMSCs post miR-187-3p overexpression.
Conclusion
Taken together, our study found that circ-Iqsec1 induced BMSC osteogenic differentiation through the miR-187-3p/Satb2 signaling pathway.
Similar content being viewed by others
Background
Osteoporosis (OP) is a widespread metabolic bone trait identified by reduced bone mineral density and bone quality, resulting in increased risk regarding bone fracture. It has a hazard ratio of 1.45 and 1.70 [1, 2]. It is broadly recognized that a disproportionate balance between osteoblast-related bone formation and osteoclast-mediated bone absorption in the bone marrow microenvironment leads to OP pathogenesis [3,4,5,6]. Mesenchymal stem cells (MSCs) differentiate directly into osteoblasts and then deposit mineralized extracellular matrix. MSCs have been most broadly investigated and applied in transplantation and therapy, both in basic experiments and in clinical trials [7]. Among these cells, BMSCs belong to the class of adult stem cells with the abilities of self-renewal and multi-directional division potential, which makes them able to transform into adipocytes, chondrocytes, osteoblasts, and nerve cells under different induction conditions [8, 9]. Nevertheless, the regulatory mechanisms are not clear.
Circular RNAs (circRNAs) are newly discovered RNAs that can form closed continuous rings covalently [10]. circRNAs are more stable than linear RNAs because they lack a free end for RNA enzyme-mediated degradation [11]. Research revealed that circRNA can regulate the microenvironment during osteogenic differentiation [6, 12, 13]. Former investigations suggested that circ-0016624 might sponge miR-98 to regulate BMP2 expression during postmenopausal OP [14]. circ-CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells through miR-7/SMAD/GDF5 and p38 MAPK signaling pathways [15]. circ-AFF4 modulates osteogenic differentiation of BMSCs through SMAD1/5 pathway activation via the miR-135a-5p/Irisin/FNDC5 axis [16]. However, circRNA roles in regulating osteoblastic differentiation is unclear. Therefore, more research is needed to investigate the basic molecular mechanisms of the circRNA regulatory network with respect to bone regeneration.
The current study determined that circ-Iqsec1 promoted the induction of osteogenic differentiation and that knockdown of circ-Iqsec1 significantly suppressed osteogenesis (OS) in BMSCs. miR-187-3p expression decreased as differentiation proceeded. In addition, circ-Iqsec1 induced BMSC osteogenic differentiation via the miR-187-3p/Satb2 signaling pathway. These data have proposed novel functions of circ-Iqsec1 during BMSC osteogenic differentiation and highlighted its potential application as a novel therapy target in bone formation-related traits.
Methods
BMSC preparation, culture, and identification
Tibiae and femurs from BALB/c mice were extracted under sterile conditions to expose the bone marrow cavity, which was rinsed with saline. We collected and centrifuged the bone marrow filtrate at 225 × g for 5 min. The supernatant was discarded, and we resuspended the cells in HyClone low glucose (LG)-DMEM at 1×106 cells per 100 μL. We gradually added the cell suspension to mouse lymphocyte separation medium (Sigma-Aldrich) in a 1:1 (v:v) ratio and centrifuged it at 1000 × g for 20 min. A milky turbid mononuclear cell layer was obtained. The cells were resuspended in LG-DMEM without FBS at 1×106 cells per 100 μL before centrifuging at 225 × g for 5 min. The cells that were pelleted were resuspended in LG-DMEM complete medium containing 10% FBS and incubated in 5% CO2-saturated humidity at 37°C. The culture medium was changed every 3 days. The cells were sub-cultured in a 1:3 ratio when the cell confluence achieved 80~90%. BMSCs were passaged 3–4 times and utilized for the next steps. Fluorescein isothiocyanate (FITC-F) or phycoerythrin (PE) was applied for phenotypic analyses. CD44, CD54, CD31, CD29, CD90, integrin-β1, and vWF marker expressions were detected. IgG-matched isotype served as the internal control for all antibodies.
Cell transfection
Satb2 gene overexpression vectors were made by putting Satb2 cDNA into a pcDNA3.1 vector. The miR-187-3p mimic/inhibitor and siRNA against circ-Iqsec1 (si-circ-Iqsec1) were synthesized by Genepharma (Suzhou, China). Lipofectamine 2000 (Invitrogen) was utilized for cell transfection following protocols.
Bioinformatics analysis
We predicted correlations among miRNA, mRNA, and circRNA applying the online website http://starbase.sysu.edu.cn/.
Multilineage bone marrow stem cell (BMSC) differentiation
To characterize BMSC abilities for multilineage differentiation, we cultivated 3rd-passage mouse BMSCs under various differentiation conditions. For adipocyte differentiation, we cultivated BMSCs in adipogenic differentiation medium. After 2 weeks, we determined adipocyte differentiation using Oil Red O staining. For osteoblast differentiation, we cultivated BMSCs in osteogenic differentiation medium and then stained them with alizarin red post 3 weeks to monitor overall survival (OS).
Strand-specific NGS RNA-seq library preparation
Total RNA from BMSCs induced for 0 and 21 days was obtained utilizing TRIzol reagent (Invitrogen, CA, USA). Our group utilized VAHTS with 3 μg RNA from each sample. RNA-seq (H/M/R) library prep kits from Illumina (Vazyme Biotech Co., Ltd, Nanjing, China) were used to remove ribosomal RNA. RNA types, such as mRNAs and ncRNAs, were retained. RNA was treated applying 40 U RNase R (Epicenter) at 37°C for 3 h, followed by TRIzol purification. An RNA-seq library was prepared through KAPA stranded RNA-seq library prep kits (Roche, Basel, Switzerland), which we employed for NGS (Illumina HiSeq 4000 at Aksomics, Inc., Shanghai, China).
RNA isolation and real-time PCR
Total RNA was obtained using TRIzol reagent (Invitrogen), followed by cDNA synthesis applying TransScript All-in-One First-Strand cDNA Synthesis SuperMix (Transgen Biotech, Beijing, China). Polymerase chain reaction (PCR) was conducted utilizing a Bio-Rad PCR instrument (Bio-Rad, CA, USA) and 2× Taq PCR master mix (Solarbio, Beijing, China) following all protocols. We calculated fold changes using the 2−ΔΔCt approach. PCR primers were as follows:
circ-Iqsec1: forward 5′-GGCCTAAATCTCTTCAAC-3′ and reverse 5′-GCCAGUCUCGCUGCUGG-3′; miR-187-3p: forward 5′-TCGTGTCTTGTGTTGCAGCC-3′ and reverse 5′-GTGCAGGGTCCGAGGT-3′; RUNX2: forward 5′-ACTACCAGCCACCGAGACCA-3′ and reverse 5′-ACTGCTTGCAGCCTTAAATGACTCT-3′; OCN: forward 5′-AGCCACCGAGACACCATGAGA-3′ and reverse 5′-GGCTGCACCTTTGCTGGACT-3′; Satb2: forward 5′-GCAGTTGGACGGCTCTCTT-3′ and reverse 5′-CACCTTCCCAGCTTGATTATTCC-3′; U6: forward 5′-CGCTTCGGCAGCACATATACTAAAATTGGAAC-3′ and reverse 5′-GCTTCACGAATTTGCGTGTCATCCTTGC-3′; and GAPDH: forward 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse 5′-GGCTGTTGTCATACTTCTCATGG-3′.
Dual-luciferase reporter assay
Our group amplified Satb2 and circ-Iqsec1 3′-UTR and miR-187-3p binding sites through PCR. We inserted sequences into multiple cloning sites in the pMIR-REPORT luciferase miRNA expression reporter vector. We co-transfected HEK293T cells with 0.1-μg luciferase reporter vectors containing wild-type (WT) or mutant-type (MUT) Satb2 or circ-Iqsec1 3′-UTR and miR-187-3p mimic or miR-control utilizing Lipofectamine 2000 (Invitrogen, CA, USA). We computed relative luciferase activity by normalizing firefly luminescence to Renilla luminescence through a dual-luciferase reporter assay system (Promega, WI, USA) following protocols two days post-transfection.
Animals and cell transplantation
We infected BMSCs at the 4th passage utilizing lentivirus (si-circ-Iqsec1, miR-187-3p mimic, or Satb2 overexpression vector) and cultured in osteogenic differentiation medium for 7 days prior to in vivo research. After the BMSCs were trypsinized and directly resuspended in DMEM, we cultured the BMSCs with SynthoGraft (β-tricalcium phosphate; Bicon) for 1 h at 37°C. They were then centrifuged at 150 × g for 5 min and implanted into 2 symmetrical sites in the dorsal subcutaneous space in six 6-week-old BALB/c nude mice. The ethical review committee of Qilu Hospital of Shandong University approved the animal experiments.
Immunohistochemical analyses
We harvested specimens 8 weeks post-transplantation from mice euthanized by CO2 asphyxiation. We decalcified specimens in 10% EDTA (pH 7.4), which we dehydrated and embedded in paraffin. We cultured sections overnight at 4°C with primary antibodies against OCN and RUNX2 and then for 1 h at 37°C with secondary antibodies (Abcam). We stained the sections using 3,3-diaminobenzidine. We counterstained with hematoxylin and examined under an Axiophot light microscope (Zeiss, Oberkochen, Germany).
Statistical analyses
The continuous variables are represented as the mean ± SD. For comparisons, one-way variance of analysis was conducted through GraphPad Prism (GraphPad, CA, USA). P ≤ 0.05 informed statistical significance.
Results
BMSC characterization and differentiation
We made BMSCs from bone marrow obtained from BALB/c mice tibiae and femurs. The cultured BMSCs had a typical morphology with a spindle structure (Fig. 1A). They were positive for known MSC markers CD44, CD90, CD54, CD29, and integrin-β1, yet did not express endothelial cell markers CD31 and vWF (Fig. 1B–I). The study also found that the isolated BMSCs had osteoblast and adipocyte differentiation ability, as demonstrated by Oil Red O (Fig. 1J) and alizarin red (Fig. 1K) staining.
Isolation and identification of differentiation and classical phenotypes of BMSCs. A Phase-contrast images showing BMSC morphology. Scale bars: 20 μm. B–I Surface antigen expression in mouse BMSCs at the 3rd passage. Cells stained positive for mesenchymal stem cell markers CD44, CD29, CD90, CD54, and integrin-β. Endothelial markers CD31 and vWF were used as negative markers. ISO was applied as control. Scale bars: 20 μm. J, K The BMSC differentiation potential was detected through alizarin red (J) and Oil Red O (K) staining. Scale bars: 50 μm
circ-Iqsec1 plays a role during BMSC osteogenic differentiation
To identify the osteogenic differentiation ability, we induced BMSCs with an osteogenic differentiation induction medium for 0, 14, and 24 days. ALP staining (Fig. 2A) and alizarin red staining (Fig. 2B) showed that the osteogenic differentiation ability was time-dependent. High-throughput sequencing showed that osteogenic differentiation resulted in the abnormal expression of circRNA (Fig. 2C). RT-qPCR detection showed nine high-expression circRNAs based on the sequencing data. The data showed that only mmu_circ_0001493 expression increased significantly in the osteogenic differentiation-induced group (Fig. 2D). Furthermore, the RT-qPCR results illustrated that increased mmu_circ_0001493 expression in BMSCs depended on the induction time (Fig. 2E). This suggested that mmu_circ_0001493 functions in the osteogenic differentiation of BMSCs. mmu_circ_0001493 originated by cyclizing two exons from Iqsec1, located at chr6:90639573-90644844. Iqsec1 was 5271 bp, and the spliced mature circRNA was 1539 bp (Fig. 2F). Therefore, mmu_circ_0001493 was also called circ-Iqsec1 by our laboratory.
circ-Iqsec1 plays a role during osteogenic BMSC differentiation. A ALP staining shows the BMSC osteogenic differentiation post osteogenic induction for 0, 2, and 3 weeks. B Alizarin red staining showing BMSC osteogenic differentiation post-osteogenic induction for 0, 2, and 3 weeks. C NGS was used for circRNA expression detection in BMSCs after osteogenic induction for 0 (NC) and 3 (T) weeks. D RT-qPCR detection showing abnormal circRNA expression. Results are denoted by the mean ± SD. **P < 0.01, ***P < 0.001 vs. NC. E RT-qPCR data giving circ-Iqsec1 expression during osteogenic differentiation of BMSCs. Output is denoted as the mean ± SD. **P < 0.01, ***P < 0.001 vs. 0 day. F Genomic loci of Iqsec1 and mmu_circ_0001493
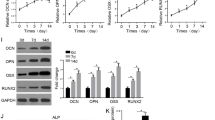
To further identify whether circ-Iqsec1 could participate in BMSC osteogenic differentiation, siRNA against circ-Iqsec1 (si-circ-Iqsec1) was constructed and transfected into BMSCs. The data showed that circ-Iqsec1 expression decreased significantly post-silencing circ-Iqsec1 (Fig. 3A). RT-qPCR data showed that circ-Iqsec1 downregulation inhibited RUNX2 (Fig. 3B) and OCN (Fig. 3C) expression. Immunohistochemical staining for ALP (Fig. 3C) and alizarin red staining for calcium (Fig. 3D) showed that circ-Iqsec1 downregulation decreased BMSC osteogenic differentiation ability.
Downregulation of circ-Iqsec1 significantly decreases BMSC osteogenic differentiation ability. A RT-qPCR data showing circ-Iqsec1 expression in BMSCs after siRNA transfection against circ-Iqsec1 or NC. Outcomes are expressed as the mean ± SD. B, C RT-qPCR data showing RUXN2 and OCN expression after osteogenic induction for 3 weeks. Outcomes are represented as the mean ± SD. D, E Immunohistochemical staining for ALP and alizarin red staining for calcium showing BMSC osteogenic differentiation ability post silencing circ-Iqsec1. ***P < 0.001 vs. NC
Satb2 plays a role during osteogenic differentiation of BMSCs
High-throughput sequencing showed that osteogenic differentiation resulted in the abnormal expression of mRNA (Fig. 4A). RT-qPCR detection and sequencing data showed seven high-expression mRNAs: Satb2, Snx32, Egfl7, Bmp3, Susd5, Clec4a3, and Fmo4. However, only Satb2 expression increased significantly in the osteogenic differentiation-induced group (Fig. 4B). RT-qPCR data further showed that Satb2 expression increased in BMSCs in a time-dependent way following induction (Fig. 4C). This suggests that Satb2 functioned in BMSC osteogenic differentiation.
Satb2 plays a role in osteogenic differentiation of BMSCs. A NGS was applied for mRNA expression detection in BMSCs after osteogenic induction for 0 (NC) and 3 (T) weeks. B RT-qPCR results showing abnormal mRNA expression. Data are denoted as the mean ± SD. **P < 0.01, ***P < 0.001 vs. NC. C RT-qPCR detection giving Satb2 expression during BMSC osteogenic differentiation. Data are denoted as the mean ± SD. **P < 0.01, ***P < 0.001 vs. 0 day. D RT-qPCR detection giving Satb2 expression in BMSCs after siRNA transfection against Satb2 (si-Satb2) or negative control (NC). Data are expressed as the mean ± SD. E, F RT-qPCR data giving RUXN2 and OCN expression after osteogenic induction for 3 weeks. Outputs are expressed as the mean ± SD. G Immunohistochemical staining for ALP and alizarin red staining showing BMSC osteogenic differentiation ability post silencing Satb2. ***P < 0.001 vs. NC
To further identify whether Satb2 participates in BMSC osteogenic differentiation, siRNA against Satb2 (si-Satb2) was constructed and transfected into BMSCs. The result showed that Satb2 expression decreased significantly after silencing of Satb2 (Fig. 4D). RT-qPCR detection showed that downregulation of Satb2 inhibited RUNX2 (Fig. 4E) and OCN (Fig. 4F) expression. Immunohistochemical staining for ALP and alizarin red staining for calcium showed that Satb2 downregulation reduced BMSC osteogenic differentiation ability (Fig. 4G). This suggested that Satb2 functioned during BMSC osteogenic differentiation.
miR-187-3p links Satb2 and circ-Iqsec1
Studies have found that miRNA mediates circRNA and mRNA regulation [17, 18]. Target miRNAs connecting Satb2 and circ-Iqsec1 were sought. Bioinformatics analyses found that circ-Iqsec1 interacts with many miRNAs such as miR-1224-5p, miR-666-3p, miR-3068-5p, miR-3075-5p, miR-764-3p, miR-132-5p, miR-187-3p, miR-485-5p, miR-3081-3p, miR-3076-3p, miR-3110-5p, miR-674-5p, and miR-3473d. Further bioinformatics analysis found that the 3′-UTR of Satb2 could interact with miR-15b-5p, miR-23b-3p, miR-101a-3p, miR-124-3p, miR-128-3p, miR-132-3p, miR-140-3p, miR-144-3p, miR-153-3p, and miR-187-3p. Venn diagram analysis showed that only nine miRNAs could interact with both 3′-UTR-Satb2 and circ-Iqsec1 (Fig. 5A). RT-qPCR detection showed that miR-187-3p expression decreased significantly in BMSCs in the osteogenic differentiation-induced group (Fig. 5B).
The miR-187-3p links Satb2 and circ-Iqsec1. A Venn diagram showing that miRNA interacts with both Satb2 and circ-Iqsec1. B RT-qPCR data giving nine miRNA expressions in BMSCs post osteogenic induction for 0 (NC) and 3 (T) weeks. C Bioinformatics analyses predicting miR-187-3p binding sites in circ-Iqsec1. MUT version regarding circ-Iqsec1 is provided. D Relative luciferase activity 2 d post HEK293T cell transfection with miR-187-3p mimic/NC or circ-Iqsec1 WT/MUT. E miR-187-3p binding site predictions regarding Satb2 3'-UTR. Mutant 3′-UTR-Satb2 version is given. F Relative luciferase activity 2 days post-HEK293T cell transfection through miR-187-3p mimic/NC or 3′-UTR-Satb2 WT/Mut. G–I RT-qPCR results giving circ-Iqsec1, miR-187-3p, and Satb2 expressions in BMSCs after transfection with si-circ-Iqsec1, miR-187-3p inhibitor, or Satb2 overexpression vector alone or in combination. Outputs are expressed as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 vs. NC. ###P < 0.001 vs. si-circ-Iqsec1
Luciferase reporter outcomes validated that miR-187-3p inhibited luciferase activity in WT but not MUT cells (Fig. 5C–D), showing that miR-187-3p was a circ-Iqsec1 target.
Data showed that Satb2 was an miR-187-3p downstream target. MUT or WT 3′-UTR-Satb2 sequences, such as the miR-187-3p binding sequence, were transfected into a luciferase reporter vector to validate correlations between Satb2 and miR-187-3p (Fig. 5E). A luciferase reporter vector was transfected into HEK293 cells with or without miR-187-3p mimic. The luciferase reporter outputs showed that miR-187-3p suppressed luciferase activity in WT but not MUT cells (Fig. 5F), implying that Satb2 was an miR-187-3p target.
RT-qPCR data showed that circ-Iqsec1 expression decreased post-transfection with si-circ-Iqsec1. However, treatment with miR-187-3p inhibitor or Satb2 overexpression vector (Satb2) did not restore circ-Iqsec1 expression in BMSCs (Fig. 5G). This finding suggested that Satb2 and miR-187-3p were circ-Iqsec1 downstream targets. RT-qPCR results revealed that circ-Iqsec1 silencing increased miR-187-3p expression. Satb2 overexpression did not reverse si-circ-Iqsec1-induced miR-187-3p upregulation (Fig. 5H), suggesting that miR-187-3p was located downstream of circ-Iqsec1. The result also showed that circ-Iqsec1 silencing decreased Satb2 expression. miR-187-3p downregulation restored Satb2 expression after si-circ-Iqsec1. After transfection with Satb2 overexpression vector, Satb2 expression increased significantly (Fig. 5I). This suggested that circ-Iqsec1 enhanced Satb2 expression via sponging miR-187-3p.
miR-187-3p downregulation or Satb2 overexpression restores BMSC osteogenic differentiation ability after silencing circ-Iqsec1
RT-qPCR detection showed that RUNX2 and OCN expressions decreased post-circ-Iqsec1 silencing. While miR-187-3p downregulation or Satb2 overexpression restored both RUNX2 and OCN expression (Fig. 6A, B), immunohistochemical staining for ALP and alizarin red staining for calcium showed that miR-187-3p downregulation or Satb2 overexpression restored osteogenic differentiation post-circ-Iqsec1 silencing in BMSCs.
miR-187-3p downregulation or Satb2 overexpression restored BMSC osteogenic differentiation ability after silencing circ-Iqsec1. A, B RT-qPCR data showing RUNX2 (A) and OCN (B) expression. Results are expressed as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 vs. NC. ##P < 0.01, ###P < 0.001 vs. si-circ-Iqsec1. C, D Immunohistochemical staining for ALP and alizarin red staining for calcium showing BMSC osteogenic differentiation ability
Satb2 upregulation restores BMSC osteogenic differentiation ability post-miR-187-3p overexpression
RT-qPCR detection showed that miR-187-3p expression increased in BMSCs post-miR-187-3p mimic transfection. However, overexpression of Satb2 could not reverse miR-187-3p expression after miR-187-3p mimic transfection (Fig. 7A). RT-qPCR data showed that Satb2 expression decreased post-miR-187-3p mimic transfection. However, after transfection with Satb2, Satb2 expression significantly increased (Fig. 7B). RT-qPCR detection showed that RUNX2 and OCN expressions decreased post-miR-187-3p overexpression. However, overexpression of Satb2 restored both RUNX2 and OCN expression (Figs. 7C, 7D). Immunohistochemical staining for ALP and alizarin red staining for calcium illustrated that Satb2 overexpression restored osteogenic differentiation post-miR-187-3p upregulation in BMSCs (Fig. 7E, F).
Upregulation of Satb2 restored the osteogenic differentiation ability of BMSCs after overexpression of miR-187-3p. A, B RT-qPCR detection shows the expression of miR-187-3p and Satb2. Data are expressed as the mean ± SD. **P < 0.01, ***P < 0.001 vs NC. ###P < 0.001 vs mimic. C, D RT-qPCR detection shows the expression of RUNX2 (A) and OCN (B). Data are expressed as the mean ± SD. *P < 0.05, ***P < 0.001 vs NC###P < 0.001 vs mimic. E, F Immunohistochemical staining for ALP and alizarin red staining show the osteogenic differentiation ability of BMSCs
miR-187-3p downregulation or Satb2 overexpression restores BMSC osteogenic differentiation ability after silencing circ-Iqsec1 in vivo
To investigate whether circ-Iqsec1 enhances bone formation in the mouse model, BMSCs transfected with si-circ-Iqsec1, miR-187-3p inhibitor, or Satb2 overexpression vectors were loaded onto scaffolds that were implanted in the subcutaneous space in nude mice. After 8 weeks, we harvested the implantation samples, which we embedded in paraffin, sectioned, deparaffinized, and stained with H&E or for OCN and RUNX2. H&E staining showed little bone formation in the si-circ-Iqsec1 group, whereas osteoid formation was restored after miR-187-3p downregulation or Satb2 overexpression (Fig. 8A). Immunohistochemical staining for OCN and RUNX2 showed that miR-187-3p downregulation or Satb2 overexpression restored both OCN (Fig. 8B) and RUNX2 (Fig. 8C) expression after silencing circ-Iqsec1.
Discussion
OP is a well-defined trait that leads to increased mortality and morbidity [18]. Because of the pluralistic differentiation potential, BMSCs function to regulate the microenvironment and bone mass and strength [19, 20]. The present investigation discovered that BMSCs have osteogenic differentiation potential. In addition, circRNA was reported to function importantly in osteogenic differentiation [21]. High-throughput sequencing elucidated that circRNA had abnormal expression in BMSCs during osteogenic differentiation and that circ-Iqsec1 expression increased in BMSCs during osteogenic differentiation. circ-Iqsec1 downregulation inhibited RUNX2 and OCN expression and BMSC osteogenic differentiation. The data suggested that circ-Iqsec1 functioned in BMSC osteogenic differentiation.
High-throughput sequencing for mRNA expression detection showed abnormal mRNA expression in BMSCs regarding osteogenic differentiation. Special AT-rich sequence-binding protein 2 (Satb2) expression was increased in BMSCs during osteogenic differentiation. Enhanced SATB2 has been reported to promote osteogenic differentiation of BMSCs from patients with osteonecrosis induced by ethanol [22, 23]. Previous studies have found that Satb2 was a particular immunohistochemical biomarker of osteoblastic differentiation and has been helpful regarding bone and soft tissue tumors [24, 25]. Satb2 downregulation inhibited RUNX2 and OCN expression and BMSC osteogenic differentiation. This is consistent with former investigations showing that Satb2 functions in BMSC osteogenic differentiation.
Previous studies have shown that circRNAs might regulate gene expression by influencing transcription, mRNA turnover, and translation via sponging RNA-binding proteins and microRNAs [18, 26, 27]. This study aimed to identify the connecting target miRNA for Satb2 and circ-Iqsec1. Bioinformatics analysis and luciferase reporter analysis confirmed that miR-187-3p functioned to link circ-Iqsec1 and Satb2. Former investigations showed that miR-187-3p expression decreased during osteogenic differentiation of human adipose-derived mesenchymal stem cells [28]. miR-187-3p expression upregulation inhibited the osteogenic differentiation of osteoblast precursor cells by inhibiting cannabinoid receptor type 2 [29]. The present investigation verified that miR-187-3p expression decreased in BMSCs during osteogenic differentiation.
The data discovered that miR-187-3p downregulation or Satb2 overexpression restored the osteogenic differentiation capability of BMSCs post-silencing circ-Iqsec1 in in vivo and in vitro investigations. Satb2 upregulation restored BMSC osteogenic differentiation capability post-miR-187-3p overexpression.
Conclusion
The present study revealed that circ-Iqsec1 functioned during osteogenic differentiation of BMSCs. circ-Iqsec1 induced BMSC osteogenic differentiation by regulating the miR-187-3p/Satb2/RUNX2/OCN signaling pathway. In addition, the effect of circ-Iqsec1 on osteogenic differentiation may be explored in the near future.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Luo Y, Zhang Y, Miao G, Liu Y, Huang Y. Runx1 regulates osteogenic differentiation of BMSCs by inhibiting adipogenesis through Wnt/beta-catenin pathway. Arch Oral Biol. 2019;97:176–84.
Long X, Duan L, Weng W, Cheng K, Wang D, Ouyang H. Light-induced osteogenic differentiation of BMSCs with graphene/TiO2 composite coating on Ti implant. Colloids Surf B Biointerfaces. 2021;207:111996.
Kanis JA, Cooper C, Rizzoli R, Reginster JY. European guidance for the diagnosis and management of osteoporosis in postmenopausal women. Osteoporos Int. 2019;30(1):3–44.
Ensrud KE, Crandall CJ. Osteoporosis. Ann Intern Med. 2017;167(3):ITC17–32.
Qaseem A, Forciea MA, McLean RM, Denberg TD, Barry MJ, Cooke M, et al. Treatment of low bone density or osteoporosis to prevent fractures in men and women: a clinical practice guideline update from the American College of Physicians. Ann Intern Med. 2017;166(11):818–39.
Zhang D, Ni N, Wang Y, Tang Z, Gao H, Ju Y, et al. CircRNA-vgll3 promotes osteogenic differentiation of adipose-derived mesenchymal stem cells via modulating miRNA-dependent integrin alpha5 expression. Cell Death Differ. 2021;28(1):283–302.
Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4):315–7.
Wang K, Zhao Z, Wang X, Zhang Y. BRD4 induces osteogenic differentiation of BMSCs via the Wnt/beta-catenin signaling pathway. Tissue Cell. 2021;72:101555.
Zhou Y, Qiao H, Liu L, Dong P, Zhu F, Zhang J. miR-21 regulates osteogenic and adipogenic differentiation of BMSCs by targeting PTEN. J Musculoskelet Neuronal Interact. 2021;21(4):568–76.
Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–11.
Wesselhoeft RA, Kowalski PS, Anderson DG. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun. 2018;9(1):2629.
Zheng J, Zhu X, He Y, Hou S, Liu T, Zhi K, et al. CircCDK8 regulates osteogenic differentiation and apoptosis of PDLSCs by inducing ER stress/autophagy during hypoxia. Ann N Y Acad Sci. 2021;1485(1):56–70.
Yu C, Wu D, Zhao C, Wu C. CircRNA TGFBR2/MiR-25-3p/TWIST1 axis regulates osteoblast differentiation of human aortic valve interstitial cells. J Bone Miner Metab. 2021;39(3):360–71.
Yu L, Liu Y. circRNA_0016624 could sponge miR-98 to regulate BMP2 expression in postmenopausal osteoporosis. Biochem Biophys Res Commun. 2019;516(2):546–50.
Li X, Zheng Y, Huang Y, Zhang Y, Jia L, Li W. Circular RNA CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells via the miR-7/GDF5/SMAD and p38 MAPK signaling pathway. Stem Cell Res Ther. 2018;9(1):232.
Liu C, Liu AS, Zhong D, Wang CG, Yu M, Zhang HW, et al. Circular RNA AFF4 modulates osteogenic differentiation in BM-MSCs by activating SMAD1/5 pathway through miR-135a-5p/FNDC5/Irisin axis. Cell Death Dis. 2021;12(7):631.
Fernandez-Tussy P, Ruz-Maldonado I, Fernandez-Hernando C. MicroRNAs and Circular RNAs in Lipoprotein Metabolism. Curr Atheroscler Rep. 2021;23(7):33.
Panda AC. Circular RNAs act as miRNA Sponges. Adv Exp Med Biol. 2018;1087:67–79.
Xie Z, Zhang H, Wang J, Li Z, Qiu C, Sun K. LIN28B-AS1-IGF2BP1 association is required for LPS-induced NFkappaB activation and pro-inflammatory responses in human macrophages and monocytes. Biochem Biophys Res Commun. 2019;519(3):525–32.
Li J, Lu L, Liu Y, Yu X. Bone marrow adiposity during pathologic bone loss: molecular mechanisms underlying the cellular events. J Mol Med (Berl). 2022 Feb;100(2):167–83.
Wang Y, Jiang Z, Yu M, Yang G. Roles of circular RNAs in regulating the self-renewal and differentiation of adult stem cells. Differentiation. 2020;113:10–8.
Yu L, Xu Y, Qu H, Yu Y, Li W, Zhao Y, et al. Decrease of MiR-31 induced by TNF-alpha inhibitor activates SATB2/RUNX2 pathway and promotes osteogenic differentiation in ethanol-induced osteonecrosis. J Cell Physiol. 2019;234(4):4314–26.
Yang X, Yang J, Lei P, Wen T. LncRNA MALAT1 shuttled by bone marrow-derived mesenchymal stem cells-secreted exosomes alleviates osteoporosis through mediating microRNA-34c/SATB2 axis. Aging (Albany NY). 2019;11(20):8777–91.
Conner JR, Hornick JL. SATB2 is a novel marker of osteoblastic differentiation in bone and soft tissue tumours. Histopathology. 2013;63(1):36–49.
Jiang A, Wang N, Yan X, Jiang Y, Song C, Chi H, et al. Hsa-circ-0007292 promotes the osteogenic differentiation of posterior longitudinal ligament cells via regulating SATB2 by sponging miR-508-3p. Aging (Albany NY). 2021;13(16):20192–217.
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–91.
Yang Y, Yujiao W, Fang W, Linhui Y, Ziqi G, Zhichen W, et al. The roles of miRNA, lncRNA and circRNA in the development of osteoporosis. Biol Res. 2020;53(1):40.
Gu H, Xu J, Huang Z, Wu L, Zhou K, Zhang Y, et al. Identification and differential expression of microRNAs in 1, 25-dihydroxyvitamin D3-induced osteogenic differentiation of human adipose-derived mesenchymal stem cells. Am J Transl Res. 2017;9(11):4856–71.
Xu A, Yang Y, Shao Y, Wu M, Sun Y. Inhibiting effect of microRNA-187-3p on osteogenic differentiation of osteoblast precursor cells by suppressing cannabinoid receptor type 2. Differentiation. 2019;109:9–15.
Acknowledgements
None.
Funding
This work was supported by the Fellowship of China Postdoctoral Foundation (2020M682194). The sponsors made no substantial contribution to the article.
Author information
Authors and Affiliations
Contributions
LX F and WL W contributed to the study conception and design. All authors collected the data and performed the data analysis. All authors contributed to the interpretation of the data and the completion of figures and tables. All authors contributed to the drafting of the article and final approval of the submitted version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The ethical review committee of Qilu Hospital of Shandong University approved the animal experiments. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Fan, L., Yang, K., Yu, R. et al. circ-Iqsec1 induces bone marrow-derived mesenchymal stem cell (BMSC) osteogenic differentiation through the miR-187-3p/Satb2 signaling pathway. Arthritis Res Ther 24, 273 (2022). https://doi.org/10.1186/s13075-022-02964-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13075-022-02964-x