Abstract
Objective
To identify rheumatoid arthritis (RA)-associated susceptibility genes and pathways through integrating genome-wide association study (GWAS) and gene expression profile data.
Methods
A transcriptome-wide association study (TWAS) was conducted by the FUSION software for RA considering EBV-transformed lymphocytes (EL), transformed fibroblasts (TF), peripheral blood (NBL), and whole blood (YBL). GWAS summary data was driven from a large-scale GWAS, involving 5539 autoantibody-positive RA patients and 20,169 controls. The TWAS-identified genes were further validated using the mRNA expression profiles and made a functional exploration.
Results
TWAS identified 692 genes with PTWAS values < 0.05 for RA. CRIPAK (PEL = 0.01293, PTF = 0.00038, PNBL = 0.02839, PYBL = 0.0978), MUT (PEL = 0.00377, PTF = 0.00076, PNBL = 0.00778, PYBL = 0.00096), FOXRED1 (PEL = 0.03834, PTF = 0.01120, PNBL = 0.01280, PYBL = 0.00583), and EBPL (PEL = 0.00806, PTF = 0.03761, PNBL = 0.03540, PYBL = 0.04254) were collectively expressed in all the four tissues/cells. Eighteen genes, including ANXA5, AP4B1, ATIC (PTWAS = 0.0113, downregulated expression), C12orf65, CMAH, PDHB, RUNX3 (PTWAS = 0.0346, downregulated expression), SBF1, SH2B3, STK38, TMEM43, XPNPEP1, KIAA1530, NUFIP2, PPP2R3C, RAB24, STX6, and TLR5 (PTWAS = 0.04665, upregulated expression), were validated with integrative analysis of TWAS and mRNA expression profiles. TWAS-identified genes functionally involved in endoplasmic reticulum organization, regulation of cytokine production, TNF signaling pathway, immune response-regulating signaling pathway, regulation of autophagy, etc.
Conclusion
We identified multiple candidate genes and pathways, providing novel clues for the genetic mechanism of RA.
Similar content being viewed by others
Introduction
Rheumatoid arthritis (RA) is a chronic, inflammatory, autoimmune disease primarily affecting the joints, even probably leading to accumulating joint damage and irreversible disability. Epidemiological studies in RA show that it affects up to 0.5–1% of the general adult population worldwide. Approximately two thirds of cases are characterized by rheumatoid factor or autoantibodies that target various molecules including modified self-epitopes [1]. Some strong genetic components are known to be involved in the development of RA. Twins and family studies offered a strong suggestion that the risk of RA increased in individuals with an RA family history by shared genetic factors [2,3,4]. The recognition of key genetic components and mechanisms will lead to a better understanding of the pathogenesis of RA.
Genome-wide association study (GWAS) represents a powerful approach of understanding the genetic basis of many complex traits in common human diseases. Especially, it was proved extremely well-suited to the identification of common single nucleotide polymorphism (SNP)-based variants with modest to large effects on phenotype. GWAS with fine mapping, candidate gene approaches, and a meta-analysis of GWAS has identified ~ 100 loci across the genome harboring RA susceptibility variants [5]. However, the specific biological mechanisms and functional consequences of many genetic variants identified by GWAS remain largely unknown, in particular, their role on disease severity; that is to say, the GWAS approach is likely to miss expression-trait associations of small effect.
Gene expression is an intermediate phenotype between genetic variant and traits underlying disease susceptibility. Many genetic variants devote their effects on complex traits by modulating gene expression [6]. Unfortunately, large-scale expression-trait associations are hampered by specimen availability and cost, as well as intrinsic factor, small effects. Consequently, transcriptome-wide association study (TWAS) was developed to address these problems, which integrates gene expression with large-scale GWAS [7]. It uses a small set of individuals with both genotype and gene expression data as a reference panel to identify significant expression-trait associations. Through extensive simulations of available GWAS data, TWAS identified candidate genes associated with, schizophrenia [8], calcific aortic valve stenosis [9], nonobstructive azoospermia [10], inflammatory biologic age [11], and other complex traits [12, 13]. Meanwhile, the genetic susceptibility variants associated with RA commonly map to enhancer regions [14], which can regulate one or more genes at distant locations in a cell-type-specific manner. Thus, the improved understanding of gene regulation relationship defining which genes are important in which cell types is vital for the predisposition to RA.
In this study, we conducted cell/tissue-related TWAS for RA based on the GWAS dataset and gene expression from EBV-transformed lymphocytes (EL), transformed fibroblasts (TF), peripheral blood (NBL), and whole blood (YBL). We subsequently reevaluated the expression of the TWAS-identified genes and made a functional exploration. This is the first time that TWAS is applied to a large-scale GWAS data to detect susceptibility genes associated with RA.
Methods
GWAS summary data of RA
A recent large-scale genome-wide association study meta-analysis of RA was used here [15]. Briefly, the genome-wide summary data was collected from six GWAS collections, 5539 cases and 20,169 controls in total, per-collection: Brigham Rheumatoid Arthritis Sequential Study (483 cases, 1449 controls), Canada (589 cases, 1472 controls), Epidemiological Investigation of Rheumatoid Arthritis (1173 cases, 1089 controls), North American Rheumatoid Arthritis Consortium I (867 cases, 1041 controls) and III (902 cases, 4510 controls), and Wellcome Trust Case Control Consortium (1525 cases, 10,608 controls); typed at 2,556,272 SNPs. Genotyping was conducted using commercial platforms, such as Affymetrix 6.0 array and Illumina 550 K array. RA cases either met the 1987 American College of Rheumatology criteria for the diagnosis of rheumatoid arthritis or were diagnosed by board-certified rheumatologists, with the limitation of anti-cyclic citrullinated peptide (anti-CCP) positive or rheumatoid factor (RF) positive. Detailed information of cohorts, genotyping, imputation, meta-analysis, and quality control approaches can be found in the published studies [15,16,17,18,19,20].
TWAS
FUSION software was applied to the RA GWAS summary data for cell/tissue-related TWAS analysis, including EL, TF, NBL, and YBL. TWAS analysis used pre-computed gene expression weights together with disease GWAS summary statistics to calculate the association of every gene to disease. The genetic values of expression were computed one probe set at a time using SNP genotyping data located 500 kb on either sides of the gene boundary. For this study, the gene expression weights of EL, TF, NBL, and YBL were driven from the FUSION website (http://gusevlab.org/projects/fusion/).
Validating TWAS results by genome-wide mRNA expression profiles of RA
The expression data of RA were downloaded from Gene Expression Omnibus (GEO) DataStets (https://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS3794) and corresponding reference [21]. In the study, a complete genome-wide transcript profiling of peripheral blood mononuclear cells (PBMCs) from 18 RA patients and 15 age- and sex-matched healthy controls were collected. The information about the demographical and clinical characteristics of RA patients and controls was presented in Table S1. The total RNA was isolated from PBMCs using the PAXgene RNA isolation kit (PreAnalytix). The concentration and profiles of total RNA were determined using a NanoDrop ND-1000 (Wilmington) and a Bioanalyzer 2100 (Agilent). Two hundred nanograms of RNA was reverse transcribed. The cDNA was transcribed and cRNA was labeled with biotin-16-UTP. Labeled probe hybridization to Illumina BeadChips human-6v2 was carried out using Illumina’s BeadChip 6v2 protocol. Beadchips were scanned on the Illumina BeadArray 500GX Reader. The preliminary data analysis were performed and normalized by Illumina BeadStudio software. Then the real-time PCR was further performed to detect mRNA expression. Differential analysis per gene was performed with one-way analysis of variance (ANOVA) and P values were adjusted to control the false discovery rate (FDR, 5%).
Functional exploration
The significant genes identified by TWAS were further made a functional exploration that included Gene Ontology (GO), pathway analysis, and PPI network construction using an online analysis tool of Gene Annotation & Analysis Resource, Metascape (http://metascape.org). GO analysis was based on Fisher’s exact test and calculation of P values. Pathway analysis was performed for differentially expressed genes based on the database. The construction of PPI network and associated module analysis was based on GO enrichment analysis using the plugin Molecular Complex Detection (MCODE). MCODE algorithm was then applied to this network to identify neighborhoods where proteins are densely connected.
Results
TWAS analysis results of RA
Using the GWAS summary data, TWAS identified 8403 genes in total, and 1440, 4224, 2410, and 4628 genes for EL, TF, NBL, and YBL, respectively. In the gene list, there were 692 significant genes with TWAS P values < 0.05 in total (Fig. 1), including 82 genes for EL, 257 genes for TF, 182 genes for NBL, and 317 genes for YBL. Different tissues or cells have their own gene expression profile. In order to find out the most representative genes, we carried out an overlap analysis of genes in different tissues/cells. The Venn diagram (Fig. 2) showed that the number of genes expressed in one or more tissues/cells. For example, there were 82 TWAS-identified significant genes associated with RA in EL; there were 32 significant genes in both EL and TF; there were 5 significant genes in EL, TF, and YBL; and there were 4 significant genes in EL, TF, YBL, and NBL. The four novel TWAS-significant RA susceptibility genes identified in all four tissues/cells were CRIPAK, MUT, FOXRED1, and EBPL, which were located on chromosomes 4, 6, 11, and 13, respectively. Table 1 presented the more detailed information of the four genes, including heritability of genes (HSQ), rsID of the most significant GWAS SNP in the locus (BEST.GWAS.ID), number of SNPs in the locus (NSNP), and TWAS P value (PTWAS).
Manhattan plot of TWAS-identified genes and significantly expressed genes associated with RA (colorful points). Each point represents a single gene, with physical position (chromosome localization) plotted on the x-axis and -log10 (P value) of association between gene and RA plotted on the y-axis. The significant genes in different tissues/cells are highlighted with different colors (red, EL; green, TF; blu, NBL; yellow, YBL; gray, all)
Validating TWAS results by mRNA expression profiles of RA
Genome-wide transcriptional profiles of PBMCs from 18 RA patients and 15 controls measured on microarrays found 339 differently expressed genes, among which 238 were downregulated and 101 were upregulated. Comparing the TWAS results and mRNA expression profile analysis results, 18 common genes were identified as showed in Table 2, which presented the information of the genes supplied by TWAS. In the 18 genes, ANXA5 and RAB24 presented in EL and ATIC and C12orf65 presented in TF; meanwhile, the left 14 genes presented in NBL or/and YBL.
Functional exploration of the TWAS-identified genes associated with RA
Pathway and process enrichment analysis was carried out with the following ontology sources: KEGG Pathway, GO Biological Processes, GO Molecular Functions, Reactome Gene Sets, Canonical Pathways, and CORUM. The total 654 TWAS-identified genes in the four tissues/cells were successfully submitted to Metascape performing GO enrichment analysis. TWAS-identified genes were annotated with an enrichment of biological processes and KEGG pathways involved in endomembrane system organization, endoplasmic reticulum organization, regulation of cytokine production, TNF signaling pathway, and so on (Fig. 3a). The significant terms were then hierarchically clustered, selected a subset of representative terms, and converted them into a network layout (Fig. 3b). Combining the results of Fig. 3a and b, the identified biological pathways with most significance were regulation of cytokine production, TNF signaling pathway, vascular endothelial growth factor (VEGF) receptor signaling pathway, immune response-regulating signaling pathway, regulation of autophagy, and negative regulation of protein serine/threonine kinase. A protein-protein interaction (PPI) network of the TWAS-identified genes was constructed, and module analysis was conducted using the plugin Molecular Complex Detection (MCODE). The PPI network was constructed based on 1122 GO terms (Fig. 4a). The top three GO terms were regulation of TP53 activity through phosphorylation, retrograde transport at the trans-golgi-network, and regulation of TP53 activity. The significant modules from the PPI network formed 9 MCODE clusters with a class of genes (Fig. 4b), for example, MCODE1, MCODE3, and MCODE5 were characterized by MAPK (mitogen-activated kinase-like protein) family genes, ZNF (zinc finger protein) family genes, and NDUF (NADH ubiquinone oxidoreductase subunit) family genes, respectively.
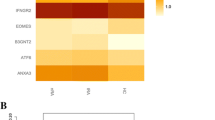
Gene ontology (GO) term analysis of differentially expressed genes. a Heatmap of enriched GO terms. b The network layout of representative GO terms under hierarchical clustering. In the network, each circle node represents a term, where its size is proportional to the number of input genes fall into that term, and its color represents its cluster identity (i.e., nodes of the same color belong to the same cluster). Terms with a similarity score > 0.3 are linked by an edge (the thickness of the edge represents the similarity score)
Discussion
TWAS is a creative and valuable analysis method that can integrate genetic variation with gene expression to identify genes whose cis-regulated expression is associated with complex traits. It captures heterogeneous signals better than individual SNPs or cis-eQTLs and focuses prediction on the genetic component of expression that avoids confounding from environmental differences caused by the trait that may influence expression. What is more, TWAS avoids tissue acquisition challenges that may pose the greatest hurdle for producing larger datasets. Thus, TWAS has been widely applied to yield mechanistic disease insights, yet the first time for RA in this study.
RA is a systemic disease and a variety of immunological events occur not only in joints but also outside the joint at mucosal surfaces and primary lymphoid tissues, especially synovium. Thus, various types of tissues and cells will be attacked by the disease, including the synovium, cartilage, bone, fibroblasts, adipocytes, macrophage, immune cells, and so on. In this work, we conducted cell/tissue-related TWAS for RA. TWAS identified a total of 674 genes with transcriptome-wide-significant associations with RA in four tissues/cells. CRIPAK, MUT, FOXRED1, and EBPL, which were collectively expressed in all the four tissues/cells, were novel genes associating with RA. Consistent with the result of TWAS, eighteen genes, ANXA5, AP4B1, ATIC, C12orf65, CMAH, PDHB, RUNX3, SBF1, SH2B3, STK38, TMEM43, XPNPEP1, KIAA1530, NUFIP2, PPP2R3C, RAB24, STX6, and TLR5, have been reported differently expressed in peripheral blood mononuclear cells of RA patients.
Cysteine-rich PAK1 inhibitor (CRIPAK) is an endogenous inhibitor of p21-activated protein kinase 1 (PAK1), which interacts with Pak1 through the N-terminal regulatory and inhibited the enhancement of estrogen receptor transactivation. The decrease in gene expression of CRIPAK could act to promote the accumulation of phosphorylated myosin light chain and its stimulation of actomyosin ATPase activity in laser-captured serotonin neurons from macaques treated with ovarian hormones [22]. There are few reports about the role and mechanism of CRIPAK in diseases, especially no reports in RA. However, PAK1 has been extensively studied. PAK1, a potential mediator of Rac1/Cdc42 signaling pathway, is involved in regulating the migration, invasion, proliferation, and inflammation of fibroblast-like synoviocytes from rheumatoid arthritis patients [23, 24]. These studies indirectly support the potential role of CRIPAK in rheumatoid arthritis.
Methylmalonyl-CoA mutase (MUT) encodes the mitochondrial enzyme methylmalonyl coenzyme A mutase. In humans, the gene encoded enzyme catalyzes the isomerization of methylmalonyl-CoA to succinyl-CoA, while this enzyme may have different functions in other species. FAD-dependent oxidoreductase domain containing 1 (FOXRED1) encoded protein that is localized to the mitochondria and whose function is involved in assembly, stability, and/or correct functioning of complex I [25, 26]. Numerous processes involved in mitochondrial function are related to RA. For example, oxidative stress impairs energy metabolism in primary cells and synovial tissue of RA patients [27]; the interaction of abnormal cellular metabolism, mitochondrial dysfunction, hypoxia, and the proinflammatory signaling pathways in synovial cells is contributed to synovial invasiveness of RA [28]; and rare/low-frequency variants of the mitochondria respiratory chain-related proteins were aggregated RA patients [29]. Meanwhile, The top three GO terms in MCODE 5 of PPI network were complex I biogenesis, NADH dehydrogenase complex assembly, and mitochondrial respiratory chain complex I assembly. Therefore, we predict the two genes may play roles in the pathology of RA via affecting mitochondrial function.
EBPL is an emopamil-binding protein (EBP)-like protein. EBP is a high-affinity binding protein for [H-3] emopamil and belongs to the family of so-called sigma receptors. Mutations disrupted EBP impair cholesterol biosynthesis and cause X-chromosomal dominant chondrodysplasia punctate. The EBPL mRNA was expressed ubiquitously and most abundant in the liver, lung, and kidney. However, EBPL has a yet-to-be-discovered function [30].
In the common RA-associated gene list identified by TWAS, we should pay attention to ATIC, RUNX3, and TLR5. 5-Aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (ATIC) encodes a bifunctional protein, which catalyzes the last two steps of the de novo purine biosynthetic pathway. ATIC plays a crucial role in the mechanisms underlying methotrexate’s anti-inflammatory and antiproliferative effects. ATIC missense variant and gene polymorphism affects response to methotrexate treatment in RA patients [31,32,33]. Runt-related transcription factor 3 (RUNX3) encodes a member of the runt domain-containing family of transcription factors and involves on T cell development, T cells polarization, and T cell selection [34, 35]. RA is characterized by the presence of activated T lymphocytes. It is indicated that RUNX3 may play roles on the mechanisms of T cell activation in RA. Toll-like receptor 5 (TLR5) encodes a member of the TLR family that plays an essential role in pathogen recognition and innate immune response activation. TLR5 agonist, flagellin, can promote monocyte infiltration and osteoclast maturation directly through myeloid TLR5 ligation and indirectly via TNF-alpha production from RA and mouse cells [36]. Angiogenesis in RA is fostered directly by TLR5 ligation and indirectly through interleukin-17 induction [37]. TLR5 is the bridge that interconnects the formation of new blood vessels with the maturation of joint osteoclasts, thereby accelerating the bone destruction process in RA [38]. There have been other researches indicating that TLR5 is involved on RA inflammation, bone destruction, and angiogenesis; thus, TLR5 is a critical element and target for RA mechanism.
In the identified biological pathway, TNF signaling pathway, and regulation of autophagy become areas of our interest. The role of TNF signaling pathway in the pathogenesis and progression of RA is obviously important. The approach of targeting TNF has become a considerably effective treatment of RA [39]. It would be of great significance for the prevention and treatment of RA to establish a genetic link between gene expression and RA susceptibility. In addition, in the results presented in Fig. 3b, TNF signaling pathway is associated with several pathways, such as VEGF receptor signaling, cellular responses to external stimulus, immune response-regulating signaling pathway, and regulation of cytokine production, which suggested a possible new perspective that different cell functions are linked by genetic inheritance and then affect the occurrence, development, and treatment of RA. Autophagy is another biological function that plays pivotal roles in RA. It has been evident that dysregulated autophagy damages the maturation survival and proliferation of various immune and non-immune cells in RA pathogenesis, as well as affects the antigen presentation of immune cells [40]. Distinguishing the genes’ polymorphisms in autophagy or other cellular mechanisms in RA pathology is of great significance in establishing the association of autophagy with RA.
There are two limitations of the study that needed attention. First, despite TWAS has great power and is not confounded by reverse causality (disease → gene expression independent of SNP), instances of pleiotropy (multiple effects by a single SNP or linked SNPs involved on gene expression and RA) are statistically indistinguishable from real susceptibility genes with causal link. Second, this study did not include experimental analysis based on RA patients with different stages, different therapies, and so on, which will be considered in future studies.
With TWAS in this study, we found that the mRNA expression of some genes in human tissues/cells can be affected by SNPs, further associating with RA susceptibility. For example, four genes (CRIPAK, MUT, FOXRED1, and EBPL) in four distinct loci (rs3755963, rs6458697, rs602735, and rs1198329) were associated with RA susceptibility. We carried out a TWAS strategy to pinpoint RA-associated genes; both genomics and transcriptomics were combined, and cis-heritable genes were explored and evaluated efficiently. The study provides a potential functional mechanism of how genetic variants on chromosome may increase RA susceptibility.
Conclusions
In summary, the TWAS study identifies novel and common susceptibility genes for rheumatoid arthritis. Beyond specific mechanistic findings for RA, this work outlines a systematic approach to identify functional mediators of complex disease.
Availability of data and materials
The gene expression datasets generated during the study are available in the FUSION website (http://gusevlab.org/projects/fusion/).
Abbreviations
- RA:
-
Rheumatoid arthritis
- GWAS:
-
Genome-wide association study
- TWAS:
-
Transcriptome-wide association study
- EL:
-
EBV-transformed lymphocytes
- TF:
-
Transformed fibroblasts
- NBL:
-
Peripheral blood
- YBL:
-
Whole blood
- SNPs:
-
Single nucleotide polymorphisms
- Anti-CCP:
-
Anti-cyclic citrullinated peptide
- RF:
-
Rheumatoid factor
- GEO:
-
Gene expression omnibus
- PBMCs:
-
Peripheral blood mononuclear cells
- ANOVA:
-
Analysis of variance
- FDR:
-
False discovery rate
- GO:
-
Gene ontology
- MCODE:
-
Molecular complex detection
- CHR:
-
Chromosome
- HSQ:
-
Heritability of the gene
- BEST.GWAS.ID:
-
rsID of the most significant GWAS SNP in locus
- NSNP:
-
Number of SNPs in the locus
- P TWAS :
-
TWAS P value
- VEGF:
-
Vascular endothelial growth factor
- PPI:
-
Protein-protein interaction
- MAPK:
-
Mitogen-activated kinase-like protein
- ZNF:
-
Zinc finger protein
- NDUF:
-
NADH ubiquinone oxidoreductase subunit
- CRIPAK:
-
Cysteine-rich PAK1 inhibitor
- PAK1:
-
P21-activated protein kinase 1
- MUT:
-
Methylmalonyl-CoA mutase
- FOXRED1:
-
Oxidoreductase domain containing 1
- EBP:
-
Emopamil-binding protein
- RUNX3:
-
Transcription factor 3
- TLR5:
-
Toll-like receptor 5
References
Klareskog L, Catrina AI, Paget S. Rheumatoid arthritis. Lancet. 2009;373(9664):659–72.
MacGregor AJ, Snieder H, Rigby AS, Koskenvuo M, Kaprio J, Aho K, et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 2000;43(1):30–7.
Kuo C-F, Grainge MJ, Valdes AM, See L-C, Yu K-H, Shaw SWS, et al. Familial aggregation of rheumatoid arthritis and co-aggregation of autoimmune diseases in affected families: a nationwide population-based study. Rheumatology (Oxford). 2017;56(6):928–33.
Hemminki K, Li X, Sundquist J, Sundquist K. Familial associations of rheumatoid arthritis with autoimmune diseases and related conditions. Arthritis Rheum. 2009;60(3):661–8.
Smolen JS, Aletaha D, Barton A, Burmester GR, Emery P, Firestein GS, et al. Rheumatoid arthritis Nat Rev Dis Primers. 2018;4:18001.
Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16(4):197–212.
Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BWJH, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48(3):245–52.
Gusev A, Mancuso N, Won H, Kousi M, Finucane HK, Reshef Y, et al. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat Genet. 2018;50(4):538–48.
Theriault S, Gaudreault N, Lamontagne M, Rosa M, Boulanger M-C, Messika-Zeitoun D, et al. A transcriptome-wide association study identifies Palmd as a susceptibility gene for calcific aortic valve stenosis. Nat Commun. 2018;9:988.
Jiang T, Wang Y, Zhu M, Wang Y, Huang M, Jin G, et al. Transcriptome-wide association study revealed two novel genes associated with nonobstructive azoospermia in a Chinese population. Fertil Steril. 2017;108(6):1056–62.
Lin H, Lunetta KL, Zhao Q, Rong J, Benjamin EJ, Mendelson MM, et al. Transcriptome-wide association study of inflammatory biologic age. Aging-Us. 2017;9(11):2288–301.
Harris SE, Riggio V, Evenden L, Gilchrist T, McCafferty S, Murphy L, et al. Age-related gene expression changes and transcriptome wide association study of physical and cognitive aging traits in the Lothian birth cohort 1936. Aging-Us. 2017;9(12):2489–503.
Mancuso N, Shi H, Goddard P, Kichaev G, Gusev A, Pasaniuc B. Integrating gene expression with summary association statistics to identify genes associated with 30 complex traits. Am J Hum Genet. 2017;100(3):473–87.
Trynka G, Sandor C, Han B, Xu H, Stranger BE, Liu XS, et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat Genet. 2013;45(2):124–30.
Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42(6):508–14.
Plenge RM, Cotsapas C, Davies L, Price AL, Bakker PIW, Maller J, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39(12):1477–82.
Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, et al. Traf1-C5 as a risk locus for rheumatoid arthritis - a genomewide study. N Engl J Med. 2007;357(12):1199–209.
Raychaudhuri S, Remmers EF, Lee AT, Hackett R, Guiducci C, Burtt NP, et al. Common variants at Cd40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40(10):1216–23.
Gregersen PK, Amos CI, Lee AT, Lu Y, Remmers EF, Kastner DL, et al. Rel, encoding a member of the Nf-kappa B family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat Genet. 2009;41(7):820–3.
Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78.
Teixeira VH, Olaso R, Martin-Magniette M-L, Lasbleiz S, Jacq L, Oliveira CR, et al. Transcriptome analysis describing new immunity and defense genes in peripheral blood mononuclear cells of rheumatoid arthritis patients. PLoS One. 2009;4(8):e6803.
Bethea CL, Reddy AP. Effect of ovarian hormones on genes promoting dendritic spines in laser-captured serotonin neurons from macaques. Mol Psychiatry. 2010;15(10):1034–44.
Fu D, Yang Y, Xiao Y, Lin H, Ye Y, Zhan Z, et al. Role of P21-activated kinase 1 in regulating the migration and invasion of fibroblast-like Synoviocytes from rheumatoid arthritis patients. Rheumatology (Oxford). 2012;51(7):1170–80.
Huang M, Wang L, Zeng S, Qiu Q, Zou Y, Shi M, et al. Indirubin inhibits the migration, invasion, and activation of fibroblast-like synoviocytes from rheumatoid arthritis patients. Inflamm Res. 2017;66(5):433–40.
Fassone E, Duncan AJ, Taanman J-W, Pagnamenta AT, Sadowski MI, Holand T, et al. Foxred1, encoding an Fad-dependent oxidoreductase complex-I-specific molecular chaperone, is mutated in infantile-onset mitochondrial encephalopathy. Hum Mol Genet. 2010;19(24):4837–47.
Formosa LE, Mimaki M, Frazier AE, McKenzie M, Stait TL, Thorburn DR, et al. Characterization of mitochondrial Foxred1 in the assembly of respiratory chain complex I. Hum Mol Genet. 2015;24(10):2952–65.
Balogh E, Veale DJ, McGarry T, Orr C, Szekanecz Z, Ng C-T, et al. Oxidative stress impairs energy metabolism in primary cells and synovial tissue of patients with rheumatoid arthritis. Arthritis Res Ther. 2018;20(1):95.
Fearon U, Canavan M, Biniecka M, Veale DJ. Hypoxia, mitochondrial dysfunction and synovial invasiveness in rheumatoid arthritis. Nat Rev Rheumatol. 2016;12(7):385–97.
Mitsunaga S, Hosomichi K, Okudaira Y, Nakaoka H, Suzuki Y, Kuwana M, et al. Aggregation of rare/low-frequency variants of the mitochondria respiratory chain-related proteins in rheumatoid arthritis patients. J Hum Genet. 2015;60(8):449–54.
Moebius FF, Fitzky BU, Wietzorrek G, Haidekker A, Eder A, Glossmann H. Cloning of an emopamil-binding protein (Ebp)-like protein that lacks sterol delta (8)-delta (7) isomerase activity. Biochem J. 2003;374:229–37.
Kurzawski M, Malinowski D, Szarmach N, Nowak A, Goryniak A, Pawlik A, et al. Atic missense variant affects response to methotrexate treatment in rheumatoid arthritis patients. Pharmacogenomics. 2016;17(18):1971–8.
Lima A, Monteiro J, Bernardes M, Sousa H, Azevedo R, Seabra V, et al. Prediction of methotrexate clinical response in portuguese rheumatoid arthritis patients: implication of Mthfr Rs1801133 and Atic Rs4673993 polymorphisms. BioMed Res Int. 2014;2014:368681.
Lee YH, Bae S-C. Association of the Atic 347 C/G polymorphism with responsiveness to and toxicity of methotrexate in rheumatoid arthritis: a meta-analysis. Rheumatol Int. 2016;36(11):1591–9.
Woolf E, Xiao CY, Fainaru O, Lotem J, Rosen D, Negreanu V, et al. Runx3 and Runx1 are required for Cd8 T cell development during thymopoiesis. Proc Natl Acad Sci U S A. 2003;100(13):7731–6.
Steinke FC, Yu S, Zhou X, He B, Yang W, Zhou B, et al. Tcf-1 and Lef-1 act upstream of Th-Pok to promote the Cd4(+) T cell fate and interact with Runx3 to silence Cd4 in Cd8(+) T cells. Nat Immunol. 2014;15(7):646–56.
Kim S-J, Chen Z, Chamberlain ND, Essani AB, Volin MV, Amin MA, et al. Ligation of Tlr5 promotes myeloid cell infiltration and differentiation into mature osteoclasts in rheumatoid arthritis and experimental arthritis. J Immunol. 2014;193(8):3902–13.
S-j K, Chen Z, Chamberlain ND, Volin MV, Swedler W, Volkov S, et al. Angiogenesis in rheumatoid arthritis is fostered directly by toll-like receptor 5 ligation and indirectly through interleukin-17 induction. Arthritis Rheum. 2013;65(8):2024–36.
Bernard NJ. Inflammation: a Tlr5-Tnf positive feedback loop in rheumatoid arthritis. Nat Rev Rheumatol. 2014;10(11):637.
Radner H, Aletaha D. Anti-TNF in rheumatoid arthritis: an overview. Wien Med Wochenschr. 2015;165(1–2):3–9.
Karami J, Masoumi M, Khorramdelazad H, Bashiri H, Darvishi P, Sereshki HA, et al. Role of autophagy in the pathogenesis of rheumatoid arthritis: Latest evidence and therapeutic approaches. Life Sci. 2020;254:117734.
Acknowledgements
We would like to thank the participants and the investigators for generating the publicly available dataset.
Funding
This study was partially supported by the National Natural Science Foundation of China (82073495, 81673112) and Chey Institute for Advanced Studies (International Scholar Exchange Fellowship 2018–2019).
Author information
Authors and Affiliations
Contributions
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Prof. Feng Zhang has full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Cuiyan Wu, Feng Zhang, and Xiong Guo are involved on the study conception and design. Ping Li, Bolun Cheng, Sijia Tan, Peilin Li, Wenyu Li, Huan Liu, Feng’e Zhang, Sen Wang, Yujing Ning, and Yan Wen contributed to the acquisition of data. Cuiyan Wu drafted the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The institutional review board at each site approved the study, and informed consent was obtained from all the participants.
Consent for publication
Not applicable.
Competing interests
None declared.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wu, C., Tan, S., Liu, L. et al. Transcriptome-wide association study identifies susceptibility genes for rheumatoid arthritis. Arthritis Res Ther 23, 38 (2021). https://doi.org/10.1186/s13075-021-02419-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13075-021-02419-9