Abstract
Background
Blood-sucking phlebotomine sand flies are vectors of the protozoan parasites Leishmania spp. Although the intestinal microbiota is involved in a wide range of biological and physiological processes and has the potential to alter vector competence, little is known about the factors that modify the gut microbiota composition of sand flies. As a key step toward addressing this issue, we investigated the impact of host species on the gut bacterial composition in Phlebotomus and Lutzomyia sand flies reared under the same conditions.
Methods
Bacterial 16S rRNA gene amplification and Illumina MiSeq sequencing were used to characterize the overall bacterial composition of three laboratory-reared sandflies: Phlebotomus papatasi, Ph. duboscqi, and Lutzomyia longipalpis.
Results
Our results showed that the larvae of the three sand fly species harbored almost the same microbes but had different relative abundances. Adult Ph. papatasi and Ph. duboscqi revealed similar microbiome compositions, which were distinct from that of adult Lu. longipalpis. Furthermore, we showed that Ph. papatasi and Ph. duboscqi are hosts for different bacterial genera. The experiment was repeated twice to improve accuracy and increase reliability of the data, and the same results were obtained even when a distinct composition of the microbiome among the same species was identified probably because of the use of different larvae food batch.
Conclusions
The present study provides key insights into the role of host species in the gut microbial content of different sand fly species reared under the same conditions, which may influence their susceptibility to Leishmania infection.
Graphical Abstract

Similar content being viewed by others
Background
Phlebotomine sand flies (Diptera: Psychodidae) are hematophagous insects that feed on a wide range of hosts and transmit a vast array of pathogens responsible for causing diseases in humans and animals worldwide. Among the more than 1000 sand fly species that have been validated to date, only 10% are known or suspected vectors of different pathogens, including arboviruses and bacteria, but they are well recognized as the main vectors of Leishmania, the causative agent of leishmaniasis, a neglected tropical disease [1, 2].
Sand flies live in groups and interact with diverse microbiota. It has been demonstrated that microbial symbionts influence key aspects of their insect host’s fitness [3,4,5,6]. According to a previous study, Lutzomyia longipalpis flies fed a diet containing rabbit feces were more likely to lay eggs than flies fed sterilized feces of the larvae stage [3]. Additionally, delayed hatching and lower survival rates were observed in larvae fed sterile feces. The reintroduction of eliminated bacteria confirmed the initial findings, suggesting the importance of bacterial presence and specificity for sand fly development. Bacteria belonging to the phylum Proteobacteria participate in insect host nutrition by fixing atmospheric nitrogen [7]. Conversely, the host can also control the microbial composition to some extent, such as by changing nutrient availability through diet choice, host metabolism [8], or by triggering immune factors [9]. It has been demonstrated in this context that sand flies from tropical regions including Lu. longipalpis seemingly bred in soil enriched with decomposed leaves and other detritus, with a preference for tree bases. Additionally, insects tend to mount immune responses to maintain a complex balance between acceptance and rejection, thereby maintaining a peaceful coexistence [9].
The natural gut microbiota is acquired by adult sand flies from various sources, including sugarcane plants and blood from a wide range of hosts or from re-colonization of the gut by microbes ingested by terrestrial-dwelling larval stages [3, 10,11,12,13]. Most larval-stage bacteria undergo biodegradation during the pupal stage, and the microbial charge is immediately and significantly reduced after the emergence of the adult [14, 15]. Female sand flies become infected by ingesting infected cells during blood meals and create an interactive relationship between the microbial community of the gut and the parasite, because the developmental life cycle of Leishmania within the sand fly vector occurs exclusively in the mid- and hindgut in the presence of symbiotic bacteria [16, 17]. The gut bacterial community in sand flies may exert a negative or positive effect on the development of Leishmania depending on the bacterial species [14, 17,18,19,20,21]. Further studies on the link between bacteria deposited during Leishmania-infected sand fly bites and the clinical outcomes of leishmaniasis have suggested that the gut microbes of Lu. longipalpis are egested into the host skin besides Leishmania triggering neutrophil infiltration and facilitating parasitic installation [22]. According to previous studies, particular attention has been given to the interactions occurring between the microbial community of the sand fly gut and parasites, and a pool of symbiotic microbiota has been considered as a potential candidate for paratransgenic or biological approaches for the control of sand fly populations [9, 11]. Previous studies have shown that variations in the insect gut microbiota may be expressed by many factors, including host habitat, diet, developmental stage, and phylogeny, all of which contribute to the composition of the insect gut microbiota [23,24,25,26,27,28]. However, little is known about the host genetic factors that modify the gut microbiota composition of sand flies, and to the best of our knowledge, no research has been conducted on these insects under controlled conditions.
Several species of Phlebotomus and Lutzomyia sand flies, which are geographically distributed across the tropics, have been identified as major vectors of leishmanial parasites [29]. Each sand fly species occupies a specific ecological niche and has a climate-sensitive life cycle, and its population biology is governed by a mixture of abiotic and biotic factors that act independently and through interactions. The sand flies Phlebotomus (Ph.) papatasi and Phlebotomus (Ph.) duboscqi are Old World vectors of Leishmania (L.) major, the etiological agent of cutaneous leishmaniasis. In contrast, Lu. longipalpis sand flies are the major natural vectors of Leishmania (L.) infantum parasites responsible for the transmission of visceral leishmaniasis in the New World. These three phlebotomine sand flies are widely colonized in different international laboratories and are used as live vector models in a diverse array of research projects.
The present study aimed to test whether host species affect the composition of the gut microbiota in the three sand fly species cited above reared under the same conditions, minimizing the differences brought about by diet and environmental factors. Because the features of each species differ within Phlebotomus and Lutzomyia sand flies, we predicted that there would also be alterations in their microbiomes.
Methods
Origin and maintenance of sand flies
Three laboratory-reared sand fly species were used: two Old World species [Phlebotomus papatasi from Jordan (PPJO) and Ph. duboscqi from Mali (PDMA)] and one New World species [Lu. longipalpis from Jacobina (LLJB)]. These strains were obtained from the Vector Molecular Biology Section, Laboratory of Malaria and Vectors, USA, and maintained in the Division of Medical Zoology at Jichi Medical University, Japan. They were reared for > 5 years under the same laboratory conditions to minimize the potential influence of environmental factors and diet. In the laboratory, the sand flies were maintained in net cages (30 × 30 × 30 cm) at 26 ± 1 °C, 80–90% humidity, and a 12:12 (light: dark) photoperiod. The sand flies had access to a piece of cotton soaked in 30% sucrose solution. Non-blood-fed females were exposed to BALB/c mice (Japan SLC, Shizuoka, Japan) as the blood source. The blood-fed females were transferred individually to vials for oviposition. Eggs were placed in a 150-ml polystyrene container filled with 2 cm of plaster of Paris at its bottom (oviposition container) and were kept in the dark at 26 ± 1 °C, 80–90% humidity. Just before hatching, a very small amount of food was sprinkled on several spots near the eggs. Larval food was prepared by mixing fresh rabbit feces and rabbit chow in a 1:1 ratio, which was fermented at 26 °C for 4 weeks in a dark chamber, air dried, and ground to a powder [30]. Notably, two different food batches were prepared separately at two time different intervals, which may have caused change in bacterial communities.
Experimental infections of sand flies
Female sand flies were experimentally infected by feeding through a chick-skin membrane on heat-inactivated blood containing 106 L. major promastigotes per milliliter. The parasite viability was checked after blood feeding. Engorged sand flies were then separated out and kept at 26 °C under standard conditions [30]. The promastigotes are seen upon microscopic examination of gut specimens at 3 days’ post blood meal.
Sample preparation and bacterial DNA extraction
Bacterial DNA was extracted from all eight larvae at stage L4, and a pool of eight dissected midguts of adults from the three laboratory-reared species of sand flies at different time intervals: 1 week after release, before feeding, and 3 and 7 days post-feeding. The same sample collection protocol was followed to extract DNA from infected and uninfected blood-fed sand flies at 3 days post-feeding. Whole sand fly samples were first washed with distilled water, followed by 70% ethanol, and rinsed thrice with sterile phosphate buffered saline (PBS) and finally with double-distilled water. After washing, sand fly guts were gently dissected under a stereomicroscope on sterilized single-use slide covers using sterile insect needles.
Pooled samples were homogenized in 0.5 ml Micrewtube® containing 250 µl of phosphate-buffered saline (PBS) or saline. Approximately 0.5 g of 0.1 mm-diameter zirconia/silica beads (BioSpec Products, Bartlesville, OK, USA) were added to the extraction tubes to mechanically crush microbial cells using a ShakeMan6 bead crusher (Bio Medical Science, Tokyo, Japan) [31, 32]. Crushed cells were spun down at high speeds to obtain a precipitate of cell debris, and 200 µl of the supernatant was subjected to DNA isolation. Genomic DNA was purified from sand fly pools using the ReliaPrep DNA Cleanup and Concentration System kit (Promega Corporation, Madison, WI, USA), according to the manufacturer’s instructions. The same protocol was followed to extract bacterial DNA from the larval food.
16S RNA gene-based identification of bacteria from sand flies by PCR and Illumina MiSeq Sequencing
Two PCR steps were performed to amplify the variable region (V3–V4) of the 16S rRNA gene. In the first step, PCR amplification was performed in a thermocycler after DNA extraction using primers targeting the V3 and V4 regions of the 16S rRNA gene. The following primers were used to amplify the hypervariable regions V3-V4 of the 16S rRNA gene: forward primer: (5ʹTCGTCGGCAGCGTCAGATGTGT ATAAGAGACAGCCTACGGGNGGCWGCAG-3ʹ) and reverse primer (5ʹGTCTCGT GGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3ʹ). These regions were approved by the Illumina protocol manual [33] and yielded high-quality sequence data as described previously [34]. PCR amplification was executed with 35 cycles of denaturation (95 °C, 30 s), annealing (55 °C, 30 s), and polymerization (72 °C, 30 s) using AmpliTaq Gold 360 DNA polymerase. Index primers were used to reamplify each portion of the PCR product and generate amplicon libraries for Illumina sequencing [33]. The amplified products were subjected to electrophoresis on a 1.5% agarose gel. Sequencing was performed using the Illumina MiSeq platform with MiSeq Reagent Kit version 3 (Illumina Inc., San Diego, CA, USA).
Quantification of gut bacteria in sand flies
The gut bacteria in sand flies were quantified by qPCR with TB Green Fast Mix (TOYOBO Co., Ltd., Osaka, Japan) using a Thermal Cycler Dice Real Time System Lite (Takara Bio Inc., Shiga, Japan). The bacterial 16S rRNA gene was used as the target, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the reference. The primer sequences were 5ʹ-ACHCCTACGGGDGGCWGCAG-3ʹ (16S-q-337F) and 5ʹ-GTDTYACCGCGGYTGCTGGCAC-3ʹ (16S-q-514R) for the amplification of bacterial 16S rRNA gene, and 5ʹ-TTCGCAGAAGACAGTGATGG-3ʹ (Lugapdh-q-F) and 5ʹ-CCCTTCATCGGTCTGGACTA-3ʹ (Lugapdh-q-R), and 5ʹ-CGACTTCAACAGCA ACTCCCACTCTTCC-3ʹ (Phgapdh-q-F) and 5ʹ-TGGGTGGTCCAGGGTTTCTTACT CCTT-3ʹ (Phgapdh-q-R) for gapdh gene [35, 36]. Relative quantities of 16S rRNA genes were determined by ∆∆Ct method using GAPDH as the reference.
ITS1-based identification of fungus from sand flies by PCR and Illumina MiSeq Sequencing
The fungal ITS1 region was independently amplified using two PCR steps as described above for bacterial DNA. In view of the different kinds of biases (specificity to fungi, mismatches, length, and taxonomy), different primer combinations targeting different parts of the ITS region were used simultaneously, as suggested by the Illumina protocol manual [37]. The ITS regions were sequenced using the Illumina MiSeq platform and MiSeq reagent kit version 3 (Illumina).
Data processing and analysis
The reads generated from the MiSeq platform (600 cycles, paired-end format) were exported as FASTQ files for importing into Quantitative Insights Into Microbial Ecology 2 (QIIME2) (version 2020.2.0) [38]. The paired-end reads were trimmed and merged using the DADA2 program in the QIIME2 Plugin. Sequences were clustered in amplicon sequences variants (ASVs) by the QIIME2 program. The OTUs were annotated based on the SILVA version 138 dataset [39] with 99% sequence identity.
Principle coordinate analysis (PCoA) on Bray-Curtis dissimilarity was used to display the beta diversity indices of bacterial communities among sand flies [40]. The group significance beta diversity indexes were calculated with QIIME2 plugins using a permutational multivariate analysis of variance (PERMANOVA), respectively. A P-value < 0.05 was considered significant.
Results
Microbiome profile of laboratory-reared sand flies
In this study, we characterized the microbiota content of three sand fly species reared under the same conditions, minimizing differences caused by diet and environmental factors, to test whether host species affect bacterial diversity. To characterize the bacterial lineages present in the midgut microbiota, we performed multiplex Illumina sequencing of the V3 and V4 hypervariable regions of the 16S rRNA gene. The experiment was repeated twice (experiment 1 and 2) to obtain accurate and reliable data, and family level analysis was used when the identification of bacterial genera failed. The most dominant sequences in the two repeated experiments belonged to two bacterial families, Rhizobiaceae and Yersiniaceae, belonging to Proteobacteria phylum.
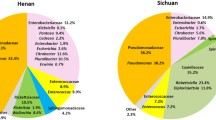
The taxa of the microbiota at the family and genus levels are shown in Fig. 1a (Experiment 1). The larvae of Phlebotomus and Lutzomyia species harbored similar microbes, which were represented by different relative abundances. Based on the relative abundance data and beta diversities analysis, Ph. papatasi and Ph. duboscqi were close to each other and distant from Lu. longipalpis (Additional file 1: Fig. S1a; PERMANOVA, Df = 2, P = 0.347). Lutzomyia longipalpis contains more Sumerlaea and less Sporosarcina than Ph. papatasi and Ph. duboscqi, which belong to Sumerlaeaceae and Planococcaceae, respectively (Fig. 1a). Moreover, two main bacterial genera were identified in used larval food, including Pseudomonas and Nocardiopsis (belonging to Pseudomonadaceae and Nocardiopsaceaea families, respectively), which were present only in some sand fly microbiota. Additionally, bacteria with low percentages grew slightly in some larval and adult sand flies (Tissierella, Sporosarcina, Corynebacterium, Xanthomonadaceae). The gut bacterial communities of adult Ph. papatasi and Ph. duboscqi were similar at 3 and 7 days post-feeding and were dominated by Rhizobiaceae (99.3% for Ph. papatasi and 95.3% for Ph. duboscqi) and Serratia belonging to Yersiniaceae family (99.1% for Ph. papatasi and 99.9% for Ph. duboscqi) (Fig. 1a), respectively. The Rhizobiaceae family was shared across the two freshly released Ph. papatasi (99.4%) and Ph. duboscqi (44.2%) 7 days after release and before feeding, whereas Ph. duboscqi comprised individuals of particular genera [Tissierella (9.8%), Corynebacterium (10.1%), and Paracoccus (12.4%) belonging to the Tissierellaceae, Corynebacteriaceae, and Rhodobacteraceae families, respectively]. In this study, the microbial communities in species of the genus Phlebotomus were close to each other, whereas those in Lu. longipalpis were distant from each other. Three main bacterial genera were identified in the gut of the last sand fly species, freshly released, and 3 and 7 days post-feeding, including Achromobacter (84.3%), Staphylococcus (46.9%), and Tsukamurella (96.2%), respectively [belonging to Alcaligenaceae, Staphylococcaceae, and Tsukamurellaceae families, respectively].
a Microbiota taxa at family and genus levels of three laboratory-reared sand flies in experiment 1. b Microbiota taxa at family and genus levels of three laboratory-reared sand flies in experiment 2. P, Phlebotomus papatasi; D, Ph. duboscqi; L, Lutzomyia longipalpis; L4, larval stage 4; F, food; BBF, before blood feeding; 3d ABF, 3 days after blood feeding; 7d ABF, 7 days after blood feeding
The gut microbiota was quantified by qPCR to determine the changes in bacterial communities in different sand flies (Experiment 1). Our results showed that the microbial charge markedly reduced after the emergence of the adults in Ph. papatasi (≃ six times). However, the bacterial number increased significantly after feeding (on blood) in Ph. papatasi and Ph. duboscqi (≃ three and two times, respectively), and almost the same bacterial charge was retained 7 days after blood feeding (Additional file 2: Fig. S2a).
The taxa of the bacterial communities at the family and genus levels are shown in Fig. 1b (Experiment 2). Based on the diversity data, bacterial communities were very similar among the three larval sand fly species. As recorded in experiment 1, adult Ph. papatasi and Ph. duboscqi were close to each other and distant from adult Lu. Longipalpis (Additional file 1: Fig. S1b; PERMANOVA, Df = 2, P = 0.004). Compared to the first experiment, a complete change in bacterial composition was observed in the larval foods of the two batches linked probably because of bacterial contamination during processing and handling. Xanthomonadaceae and Sphingobacteriaceae families were the main bacteria identified in the food, but were not present in the sand fly microbiota. However, low percentages of bacteria grew in some larvae and adult sand flies (Glycomyces, Rhizobiaceae). In the adults, Rhizobiaceae and Yersiniaceae (Serratia) were the dominant microbial families in experiment 2. Rhizobiaceae were highly abundant in all adult Ph. papatasi and Ph. duboscqi and completely absent in Lu. longipalpis, in which Yersiniaceae (99.3% and 99.8% at 3 and 7 days post-feeding, respectively) and Stenotrophomonas belonging to Xanthomonadaceae (96.7% in sand flies before feeding) constituted almost all the microbiota communities.
In experiment 2, the microbial charge significantly reduced after the emergence of the adults in Ph. papatasi (≃ 6 times). However, the bacterial number increased (≃2, 18, and 7 times in Ph. papatasi, Ph. duboscqi, and Lu. longipalpis, respectively) 3 days after blood feeding and decreased again to reach a very low bacterial quantity on day 7 post-feeding (Additional file 2: Fig. S2b).
Bacterial communities in uninfected and infected sand flies
The diversity of the sand fly gut microbiome was compared between the control and infected sand flies 3 days after blood feeding (Fig. 2). As stated above, a fraction of the bacterial genera and families were shared by the two closely related Phlebotomus species, whereas a special composition was observed in Lu. longipalpis. In addition, we found that the diversity and abundance of the gut bacterial communities between Leishmania-infected and uninfected sand flies were quite different in Phlebotomus and Lutzomyia genera, respectively (Fig. 2).
Most bacteria present in the gut of the Ph. papatasi host belonged to Rhizobiaceae, which decreased from 94.3% in the control group to 12.2% in the infected group. However, Anaplasmataceae increased dramatically from 1.6% in uninfected individuals to 86.8% in infected individuals. Three bacterial genera belonging to two families (Fig. 2) were identified in Ph. duboscqi. The relative abundance of Rhizobiaceae (undetermined and Ochrobactrum genera) increased from 22.8% in uninfected individuals to 70.6% in infected individuals, while that of Yersiniaceae decreased from 77.1% to 29.12%. In contrast, the bacterial composition of Lu. longipalpis changed completely between the control and infected groups, except for Staphylococcus genus (Staphylococcaceae), which maintained almost the same abundance (14.2% and 25.7% in the uninfected and infected groups, respectively) (Fig. 2).
Because the gut microbiota may affect parasite development, quantitative changes in the bacterial communities were assessed. The gut microbiota was quantified by qPCR for uninfected and infected sand flies at separate times. Our results showed that the bacterial load could change slightly or markedly between both sand fly groups, suggesting a quantitative interaction with Leishmania parasites (Additional file 3: Fig. S3).
Fungal profile of laboratory-reared sand flies
As shown in Fig. 3, Chaetomium and Microascus were the main fungal genera in the larvae of sand fly species. The fungal composition of the larval food was dominated by Circinella (32.7%), Cephaliophora (30.2%), and Aspergillus (19.4%) but not sand fly fungi. Additionally, Meyerozyma genus is the only fungus found in all adult sand flies, regardless of the species. However, the fungal composition of the control and infected sand flies was checked, and no difference was found. Meyerozyma was the only genus identified in both groups.
Discussion
Microbiota communities were determined in Phlebotomus and Lutzomyia sand flies reared under the same conditions, and the effects of host species on bacterial structure were explored in the present study. Previous studies on beetles and mosquitoes revealed that microbiomes vary depending on the diet or sampling location of their hosts [26, 41]. The sand flies used in this study had lived for > 5 years under the same conditions, and it was therefore thought that other variables, such as diet, temperature, and humidity, would not strongly affect the microbiome. Notably, females of Ph. papatasi and Ph. duboscqi are Old World species that are morphologically and genetically closely related, and both are known to be vectors of L. major [42]. In contrast, Lu. longipalpis is a New World species and natural vector of Leishmania infantum parasites. The separation between primitive Phlebotomus and Lutzomyia occurred approximately 200 million years ago [43].
Previous studies have shown that host phylogeny is a strong driver of microbiota structure in the insect gut [27, 44, 45]. Recently, it has been suggested that the host genotype is the major modulator of the wild population of Phlebotomus sand fly gut microbiota [28]. In this context, we explored the role of host species in the microbiota diversity of Leishmania free Old and New World sand fly species reared under the same conditions. These results indicate that adult Ph. papatasi and Ph. duboscqi have a similar microbiome composition, which is distinct from that of adult Lu. longipalpis. Furthermore, we showed that Ph. papatasi and Ph. duboscqi are hosts for different bacterial genera. It has been demonstrated that microbiome composition is positively correlated with phylogenetic relatedness between hosts, that is, closely related host species tend to possess more bacterial clades in common than distantly related hosts [46]. These findings support our hypothesis that host species determine the composition of the prokaryotic microbiota in sand flies.
Our results show that Proteobacteria was the main phylum in the studied samples. These findings are consistent with the data from a recent meta-analysis of sand fly associated bacteria, which revealed that > 57% and 47% of the identified bacteria belonged to the Proteobacteria phylum in New and Old World sand fly species, respectively [47]. The distinct composition of the bacterial communities of Ph. papatasi and Ph. duboscqi was driven by two ASVs belonging to Rhizobiaceae and Yersiniaceae (genus Serratia). However, Lu. longipalpis was characterized by a specific bacterial structure dominated by Achromobacter, Tsukamurella, Staphylococcus, and Stenotrophomonas. Further studies are required to investigate the contributions of different bacterial taxa to the life traits of sand fly species, considering the contrasting evidence for the role of endosymbiotic bacteria in the infectivity and survival of Leishmania [14, 17].
A few bacteria from sand fly food grew slightly in some larvae and adults. The observed differences in microbiota communities between the gut and used food may be explained by insect behavioral adaptations, which further promote the dominant gut bacterial taxa, including coprophagy (eating of feces), trophallaxis (the transfer of food or other gut fluids through mouth-to-mouth or anus-to-mouth feeding), and maternal smearing of the bacterial gut on the eggshell, which is subsequently consumed by the offspring [48, 49]. Additionally, host sand flies can influence the physicochemical conditions of the gut, which may result in differential bacterial growth [50]. Based on our results, the modes of bacterial acquisition and growth appear to be similar among Phlebotomus species, which are distinct from Lu. longipalpis.
This experiment was repeated twice to accurately test our bacterial host phylogeny hypothesis, and a distinct composition of the microbiome among the same species was identified. The use of different batches of larval foods containing different bacterial contents may explain the present finding, even if few bacteria from the food grew slightly in some larvae and adults. The latter may interact with the host microorganisms and completely change the microbiota structure. Our findings are in agreement with those of previous studies suggesting a role for diet in gut bacterial composition in insects [26]. It has been demonstrated that the interspecific interactions among microorganisms, by which microbes compete for nutrients and space, may change the microbiota content associated with insects [51, 52]. Theoretical synthesis research has proposed the importance of host control (that is, partner choice and fidelity) in the maintenance of mutualistic associations [53, 54]. However, it is necessary to mention that the remarkable divergence in microbiota structure between the two biological replicates may also reveal a large individual variation in the bacterial composition of the studied sand fly species, and we further propose that the bacterial communities are only loosely and temporarily associated with these sand flies. Because we pooled samples in our experiment, more studies should examine the microbial communities of individual sand flies to assess changes between individuals.
Females of two sand fly genera, Phlebotomus and Lutzomyia, are of medical importance as the only established vectors of Leishmania species that are pathogenic to humans [55]. In the present study, bacterial communities in Leishmania-uninfected and -infected sand flies were identified and compared using Illumina MiSeq sequencing. The gut microbiome has previously been reported to either enhance or inhibit parasite activity depending on the species of bacteria and thus has the potential to alter vector competence [56]. The present study showed that Serratia genus decreased significantly and disappeared completely in infected Ph. duboscqi and Ph. papatasi, possibly favoring L. major development. Indeed, this bacterial genus has been demonstrated to negatively affect L. infantum and Leishmania braziliensis by inducing lysis of the parasite cell membrane and co-infected with Lu. longipalpis in vitro [57, 58]. The antagonistic interaction between bacteria and Leishmania parasites acted as a shaping force of the community assembly. Very recent work has focused on how resident microbiota can affect the Leishmania infection of the vector with surprising results. Basically, it was found that the sand fly microbiota is fundamental for Leishmania development and transmission. One paper suggests that the removal of the microbiota alters the osmolarity of the intestinal environment and is thus deleterious for the Leishmania development [59]. Since we do not know exactly which mechanisms are responsible for this interaction/dependence between Leishmania and microbiota in vitro cultures and in vivo, this may be considered an open field of research.
Our results show that the microbial charge was markedly reduced after adult emergence and increased significantly after blood feeding. These findings are in line with previous data indicating that the microbial charge immediately and markedly decreases after adult emergence [14, 15] and increases after blood feeding [15, 17]. Additionally, the difference in bacterial load between uninfected and infected sand flies suggests a quantitative interaction with Leishmania parasites. It has already been demonstrated that any manipulation that reduces the size/diversity of natural microbiota affects the ability of Leishmania to establish infections in sand flies [17, 60]. However, studies investigating the bacterial load separately did not find any significant impact on infection rates [34].
Conclusions
This is the first report on the gut bacterial microbiomes of Phlebotomus and Lutzomyia sand flies reared under the same conditions for many generations. Our analysis showed that adult Ph. papatasi and Ph. duboscqi have a similar microbiome composition, which is distinct from that of adult Lu. longipalpis, indicating the role of phylogeny in the composition of insect gut microbiota. The experiment was repeated twice, and the same host phylogeny conclusions were obtained even when a distinct composition of the microbiome among the same species was identified. However, differences in the bacterial diversity and abundance of gut bacterial communities have been reported between uninfected and infected sand flies. Based on these results, future studies should focus on the role of these microorganisms in the biology of sand fly species, considering the contrasting evidence for the role of the detected bacteria in the infectivity and survival of Leishmania.
Availability of data and materials
The microbiome sequences data in this study were registered in NCBI/GenBank/DDBJ database at SAMN35318066–SAMN35318107.
References
Maroli M, Feliciangeli MD, Bichaud L, Charrel RN, Gradoni L. Phlebotomine sandflies and the spreading of leishmaniases and other diseases of public health concern. Med Vet Entomol. 2013;27:123–47.
Akhoundi M, Kuhls K, Cannet A, Votypka J, Marty P, Delaunay P, et al. Historical overview of the classification, evolution, and dispersion of Leishmania parasites and sandflies. PLoS Negl Trop Dis. 2016;10:e0004349.
Peterkova-Koci K, Robles-Murguia M, Ramalho-Ortigao M, Zurek L. Significance of bacteria in oviposition and larval development of the sand fly Lutzomyia longipalpis. Parasit Vectors. 2012;5:145.
Gould AL, Zhang V, Lamberti L, Jones EW, Obadia B, Korasidis N, et al. Microbiome interactions shape host fitness. Proc Nat Acad Sc. 2018;115:E11951–60.
Walters AW, Hughes RC, Call TB, Walker CJ, Wilcox H, Petersen SC, et al. The microbiota influences the Drosophila melanogaster life history strategy. Mol Ecol. 2019;29:639–53.
West AG, Waite DW, Deines P, Bourne DG, Digby A, McKenzie VJ, et al. The microbiome in threatened species conservation. Biol Conserv. 2019;229:85–98.
Gurung K, Wertheim B, Falcao SJ. The microbiome of pest insects: it is not just bacteria. Entomol Exp Appl. 2019;167:156–70.
Alencar RB, de Queiroz RG, Barrett TV. Breeding sites of phlebotomine sand flies (Diptera: Psychodidae) and efficiency of extraction techniques for immature stages in terra-firme forest in Amazonas State. Brazil Acta Trop. 2011;118:204–8.
Telleria EL, Martins-Da-Silva A, Tempone AJ, Traub-Cseko YM. Leishmania, microbiota and sand fly immunity. Parasitol. 2018;145:1336–53.
Dillon RJ, el Kordy E, Shehata M, Lane RP. The prevalence of a microbiota in the digestive tract of Phlebotomus papatasi. Ann Trop Med Parasitol. 1996;90:669–73.
Hillesland H, Read A, Subhadra B, Hurwitz I, McKelvey R, Ghosh K, et al. Identification of aerobic gut bacteria from the kala azar vector, Phlebotomus argentipes: a platform for potential paratransgenic manipulation of sand flies. Am J Trop Med Hyg. 2008;79:881–6.
Mukhopadhyay J, Braig HR, Rowton ED, Ghosh K. Naturally occurring culturable aerobic gut flora of adult Phlebotomus papatasi, vector of Leishmania major in the Old World. PLoS ONE. 2012;7:e35748.
Sant’Anna MRV, Darby AC, Brazil RP, Montoya-Lerma J, Dillon VM, Bates PA, et al. Investigation of the bacterial communities associated with females of Lutzomyia sand Fly species from South America. PLoS ONE. 2012;7:e42531.
Sant’Anna MR, Diaz-Albiter H, Aguiar-Martins K, Al Salem WS, Cavalcante RR, Dillon VM, et al. Colonisation resistance in the sand fly gut: Leishmania protects Lutzomyia longipalpis from bacterial infection. Parasit Vectors. 2014;7:329.
Volf P, Kiewengova A, Nemec A. Bacterial colonization in the gut of Phlebotomus duboscqi (Diptera: Psychodidae): transtadial passage and the role of female diet. Folia Parasitol. 2002;49:73–7.
Guernaoui S, Garcia D, Gazanion E, Ouhdouch Y, Boumezzough A, Pesson B, et al. Bacterial flora as indicated by PCR-temperature gradient gel electrophoresis (TGGE) of 16S rDNA gene fragments from isolated guts of phlebotomine sand flies (Diptera: Psychodidae). J Vector Ecol. 2011;36:S144–7.
Kelly PH, Bahr SM, Serafim TD, Ajami NJ, Petrosino JF, Meneses C, et al. The gut microbiome of the vector Lutzomyia longipalpis is essential for survival of Leishmania infantum. Mol Bio. 2017;8:e01121-e1216.
Schlein Y, Polacheck I, Yuval B. Mycoses, bacterial infections and antibacterial activity in sandflies (Psychodidae) and their possible role in the transmission of leishmaniasis. Parasitology. 1985;90:57–66.
Pumpuni CB, Demaio J, Kent M, Davis JR, Beier JC. Bacterial population dynamics in three anopheline species: the impact on Plasmodium sporogonic development. Am J Trop Med Hyg. 1996;54:214–8.
Boulanger N, Lowenberger C, Volf P, Ursic R, Sigutova L, Sabatier L, et al. Characterization of a defensin from the sand fly Phlebotomus duboscqi induced by challenge with bacteria or the protozoan parasite Leishmania major. Infect Immun. 2004;72:7140–6.
Aksoy S, Weiss B, Attardo G. Paratransgenesis applied for control of tsetse transmitted sleeping sickness. In: Aksoy S, editor. transgenesis and the management of vector-borne disease. New York: Springer; 2008. p. 35–48.
Dey R, Joshi AB, Oliveira F, Pereira L, Guimaraes-Costa AB, Serafim TD, et al. Microbes egested during bites of infected sand flies augment severity of leishmaniasis via inflammasome-derived IL-1b. Cell Host Microb. 2018;23:134–43.
Chandler JA, Morgan Lang J, Bhatnagar S, Eisen JA, Kopp A. Bacterial communities of diverse Drosophila species: ecological context of a host–microbe model system. PLoS Genet. 2011;7:e1002272.
Yun JH, Roh SW, Whon TW, Jung MJ, Kim MS, Park DS, et al. Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl Environ Microbiol. 2014;80:5254–64.
Dobson AJ, Chaston JM, Newell PD, Donahue L, Hermann SL, Sannino DR, et al. Host genetic determinants of microbiota-dependent nutrition revealed by genome-wide analysis of Drosophila melanogaster. Nat Commun. 2015;6:6312.
Muturi EJ, Lagos-Kutz D, Dunlap C, Ramirez JL, Rooney AP, Hartman GL, et al. Mosquito microbiota cluster by host sampling location. Parasit Vector. 2018;11:468.
Sanders JG, Powell S, Kronauer DJ, Vasconcelos HL, Frederickson ME, Pierce NE. Stability and phylogenetic correlation in gut microbiota: lessons from ants and apes. Mol Ecol. 2014;23:1268–83.
Papadopoulos C, Karas PA, Vasileiadis S, Ligda P, Saratsis A, Sotiraki S, et al. Host species determines the composition of the prokaryotic microbiota in Phlebotomus sandflies. Pathogens. 2020;9:428.
Bates PA. Transmission of Leishmania metacyclic promastigotes by phlebotomine sand flies. Int J Parasitol. 2007;37:1097–106.
Lawyer P, Killick-Kendrick M, Rowland T, Rowton E, Volf P. Laboratory colonization and mass rearing of phlebotomine sand flies (Diptera, Psychodidae). Parasite. 2017;24:42.
Smith B, Li N, Andersen AS, Slotved HC, Krogfelt KA. Optimising bacterial DNA extraction from faecal samples: comparison of three methods. Open Microbiol J. 2011;5:14–7.
Tabbabi A, Watanabe S, Mizushima D, Caceres AG, Gomez EA, Yamamoto DS, et al. Comparative analysis of bacterial communities in Lutzomyia ayacuchensis populations with different vector competence to Leishmania parasites in Ecuador and Peru. Microorganisms. 2020;29:9–68.
Illumina, I. 16S MetagenOmic Sequencing Library Preparation; 2013. https://support.illumina.com/content/dam/illuminasupport/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf. Accessed 5 Sept 2018.
Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2:1–7.
da Silva GD, Iturbe-Ormaetxe I, Martins-da-Silva A, Telleria EL, Rocha MN, Traub-Csekö YM, et al. Wolbachia introduction into Lutzomyia longipalpis (Diptera: Psychodidae) cell lines and its effects on immune-related gene expression and interaction with Leishmania infantum. Parasit Vectors. 2019;15:12–33.
Amni F, Maleki-Ravasan N, Nateghi-Rostami M, Hadighi R, Karimian F, Meamar AR, et al. Co-infection of Phlebotomus papatasi (Diptera: Psychodidae) gut bacteria with Leishmania major exacerbates the pathological responses of BALB/c mice. Front Cell Infect Microbiol. 2023;26:1115542.
Illumina, I. Fungal MetagenOmic Sequencing Demonstrated Protoco; 2018. https://support.illumina.com/downloads/fungal-metagenomic-sequencing-demonstrated-protocol.1000000064940.html. Accessed 6 July 2020.
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–7.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6.
Mcmurdie P, Holmes S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data (Version 1.30.0); 2013.
Kolasa M, Scibior R, Mazur MA, Kubisz D, Dudek K, Kajtoch L. How hosts taxonomy, trophy, and endosymbionts shape microbiome diversity in beetles. Microb Ecol. 2018;78:995–1013.
Ciháková J, Volf P. Development of different Leishmania major strains in the vector sandflies Phlebotomus papatasi and P. duboscqi. Ann Trop Med Parasitol. 1997;91:267–79.
Gullan PJ, Cranston PS. The insects: an outline of entomology. 2nd ed. London: Wiley-Blackwell; 2000. p. 470.
Colman DR, Toolson EC, Takacs-Vesbach CD. Do diet and taxonomy influence insect gut bacterial communities? Mol Ecol. 2012;21:5124–37.
McLean AHC, Godfrey HCJ, Ellers J, Henry LM. Host relatedness influences the composition of aphid Microbiomes. Environ Microbiol Rep. 2019;11:808–16.
Brucker RM, Bordenstein SR. The roles of host evolutionary relationships (genus: Nasonia) and development in structuring microbial communities. Evolution. 2012;66:349–62.
Fraihi W, Fares W, Perrin P, Dorkeld F, Sereno D, Barhoumi W, et al. An integrated overview of the midgut bacterial flora composition of Phlebotomus perniciosus, a vector of zoonotic visceral leishmaniasis in the Western Mediterranean Basin. PLoS Negl Trop Dis. 2017;11:e0005484.
Brune A. Symbiotic associations between termites and prokaryotes. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The prokaryotes, vol. 1. New York: Springer; 2006. p. 439–74.
Bright M, Bulgheresi S. A complex journey: transmission of microbial symbionts. Nat Rev Microbiol. 2010;8:218–30.
Bäumler A, Fang FC. Host specificity of bacterial pathogens. Cold Spring Harb Perspect Med. 2013;3:a010041.
Guiot HF. Role of competition for substrate in bacterial antagonism in the gut. Infect Immun. 1982;38:887–92.
Wilson KH, Perini F. Role of competition for nutrients in suppression of Clostridium difficile by the colonic microflora. Infect Immun. 1988;56:2610–4.
Bull JJ, Rice WR. Distinguishing mechanisms for the evolution of co-operation. J Theor Biol. 1991;149:63–74.
Sachs JL, Mueller UG, Wilcox TP, Bull JJ. The evolution of cooperation. Q Rev Biol. 2004;79:135–60.
Killick-Kendrick R. The biology and control of phlebotomine sand flies. Clin Dermatol. 1999;17:279–89.
Tabbabi A, Mizushima D, Yamamoto DS, Kato H. Sand Flies and their microbiota. Parasitologia. 2022;2:71–87.
Moraes CS, Seabra SH, Albuquerque-Cunha JM, Castro DP, Genta FA, de Souza W, et al. Prodigiosin is not a determinant factor in lysis of Leishmania (Viannia) braziliensis after interaction with Serratia marcescens D-mannose sensitive fimbriae. Exp Parasitol. 2009;122:84–90.
Campolina TB, Villegas LEM, Monteiro CC, Pimenta PFP, Secundino NFC. Tripartite interactions: Leishmania, microbiota and Lutzomyia longipalpis. PLoS Negl Trop Dis. 2020;14:e0008666.
Louradour I, Monteiro CC, Inbar E, Ghosh K, Merkhofer R, Lawyer P, et al. The midgut microbiota plays an essential role in sand fly vector competence for Leishmania major. Cell Microbiol. 2017;19:e12755.
Vivero RJ, Castañeda-Monsalve VA, Romero LRD, Hurst G, Cadavid-Restrepo G, Moreno-Herrera CX. Gut microbiota dynamics in natural populations of Pintomyia evansi under experimental infection with Leishmania infantum. Microorganisms. 2021;9:1214.
Acknowledgements
Not applicable.
Funding
This study was financially supported by the Ministry of Education, Culture and Sports, Science and Technology (MEXT) of Japan (grant nos. 18K19460, 20H03155, and 21K19193).
Author information
Authors and Affiliations
Contributions
Conceptualization, AT, DM, and HK; methodology, AT, DM, DSY, and HK; Software, DM; Validation, AT, DM, and HK; Formal analysis, DM; Investigation, AT; Resources, DM, DSY, AT, and HK; Writing—original draft preparation, AT and HK; Writing—review and editing, AT, DM, DSY, and HK; Visualization, DM; Supervision, HK; Project administration, AT and HK; Funding acquisition, HK. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All procedures were carried out according to the ethical guidelines approved by the Jichi Medical University (permit numbers: animal experiment, 20003-01; DNA experiment, 21–56).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
a. PCoA plot illustrating beta diversity distance matrices of the Bray-Curtis distance comparing the sample distribution among the three species, experiment 1. b. PCoA plot illustrating beta diversity distance matrices of the Bray-Curtis distance comparing the sample distribution among the three species, experiment 2.
Additional file 2: Figure S2.
a. The relative bacterial quantities of three laboratory-reared sand flies, experiment 1. b. The relative bacterial quantities of three laboratory-reared sand flies, experiment 2.
Additional file 3: Figure S3.
The relative bacterial quantities of uninfected and infected three laboratory-reared sand flies
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Tabbabi, A., Mizushima, D., Yamamoto, D.S. et al. Effects of host species on microbiota composition in Phlebotomus and Lutzomyia sand flies. Parasites Vectors 16, 310 (2023). https://doi.org/10.1186/s13071-023-05939-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-023-05939-2