Abstract
Background
The genus Borrelia is composed of two well-defined monophyletic groups, the Borrelia burgdorferi sensu lato complex (Bb) and the relapsing fever (RF) group borreliae. Recently, a third group, associated with reptiles and echidnas, has been described. In general, RF group borreliae use rodents as reservoir hosts; although neotropical bats may also be involved as important hosts, with scarce knowledge regarding this association. The objective of this study was to detect the presence of Borrelia spp. DNA in bats from the department of Córdoba in northwest Colombia.
Methods
During September 2020 and June 2021, 205 bats were captured in six municipalities of Córdoba department, Colombia. Specimens were identified using taxonomic keys and DNA was extracted from spleen samples. A Borrelia-specific real-time PCR was performed for the 16S rRNA gene. Fragments of the 16S rRNA and flaB genes were amplified in the positive samples by conventional PCR. The detected amplicons were sequenced by the Sanger method. Phylogenetic reconstruction was performed in IQ-TREE with maximum likelihood based on the substitution model TPM3+F+I+G4 with bootstrap values deduced from 1000 replicates.
Results
Overall, 10.2% (21/205) of the samples were found positive by qPCR; of these, 81% (17/21) and 66.6% (14/21) amplified 16S rRNA and flaB genes, respectively. qPCR-positive samples were then subjected to conventional nested and semi-nested PCR to amplify 16S rRNA and flaB gene fragments. Nine positive samples for both genes were sequenced, and seven and six sequences were of good quality for the 16S rRNA and flaB genes, respectively. The DNA of Borrelia spp. was detected in the insectivorous and fruit bats Artibeus lituratus, Carollia perspicillata, Glossophaga soricina, Phyllostomus discolor, and Uroderma sp. The 16S rRNA gene sequences showed 97.66–98.47% identity with “Borrelia sp. clone Omi3,” “Borrelia sp. RT1S,” and Borrelia sp. 2374; the closest identities for the flaB gene were 94.02–98.04% with “Borrelia sp. Macaregua.” For the 16S rRNA gene, the phylogenetic analysis showed a grouping with “Candidatus Borrelia ivorensis” and “Ca. Borrelia africana,” and for the flaB gene showed a grouping with Borrelia sp. Macaregua and Borrelia sp. Potiretama. The pathogenic role of the Borrelia detected in this study is unknown.
Conclusions
We describe the first molecular evidence of Borrelia spp. in the department of Córdoba, Colombia, highlighting that several bat species harbor Borrelia spirochetes.
Graphical Abstract

Similar content being viewed by others
Background
Pathogenic species of the genus Borrelia are zoonotic bacteria that cause emerging and re-emerging infectious diseases worldwide and constitute a major threat to public health [1]. This genus is composed of two well-defined monophyletic groups: the Borrelia burgdorferi sensu lato (Bb) complex, which cause Lyme borreliosis and are transmitted by hard ticks of the genus Ixodes, and borreliae of the relapsing fever (RF) group, transmitted mainly by soft ticks of the genus Ornithodoros, some species by ixodid ticks (e.g., Borrelia miyamotoi), and Borrelia recurrentis, transmitted by the clothing louse [2]. Recently, a third group associated with reptiles and echidnas (Tachyglossus aculeatus) has been described [3, 4], and share a common ancestor with the RF clade [3, 4].
Most species of Borrelia have complex transmission cycles interacting with multiple vertebrate hosts and vector ticks [1]. For instance, several members of the order Rodentia are reservoir hosts [5, 6]. Moreover, bats may also be involved as alternative hosts [7,8,9,10,11,12]. Indeed, several studies have reported Borrelia spp. in ticks collected from different bat species [13,14,15,16,17,18,19,20]. Interestingly, new putative taxa of Borrelia spirochetes, namely Borrelia sp. Macaregua and Borrelia sp. Potiretama, were detected in bats roosting in caves from Colombia and Brazil, respectively [10, 21].
Given their intimate relationship with emerging microorganisms that cause serious infections in humans, such as severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS) [22], and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [23], bats have recently been the center of attention for scientists. A remarkable fact is that bats harbor more viruses than rodents [24]. However, bacteria harbored by these mammals are neglected, even though they can represent zoonotic pathogens [25]. For example, RF group spirochetes such as Ca. Borrelia fainii and Ca. Borrelia johnsonii are among those neglected agents [26, 27]. In order to contribute to the understanding of Borrelia spp. associated with chiropterans, we carried out a prospective study to detect DNA of Borrelia in organs of bats derived from a COVID-19 study, collected in the department of Córdoba, Colombia.
Methods
Study area and bat captures
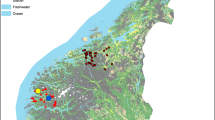
Field trips were carried out during September 2020 and June 2021 in six municipalities of Córdoba Department (Montelíbano, Tierralta, San Antero, Montería, Lorica, and Moñitos) in urban, peri-urban and rural areas with similar environmental conditions: altitude of 12–87 m, 25–28 °C, and average relative humidity of 81% (Fig. 1; Table 2).
Bats were captured using mist nets (6 m × 2 m). Males and females without signs of pregnancy were included in the study. Captured bats were euthanized with an overdose of sodium pentobarbital (200 mg) at a dose of 0.05 mg/g [28]. Following taxonomic identification using dichotomous keys [29], spleen samples were extracted, stored in sterile tubes, and kept at −80 °C until processing. The capture of bats was carried out under the permits of the National Environmental Licensing Authority (ANLA), Resolution No. 00914. All the procedures were approved by the ethics committee of the Faculty of Veterinary Medicine and Zootechnics of the Universidad de Córdoba (No. 003 of December 6, 2019).
Molecular and phylogenetic analyses
DNA extraction was performed in spleen samples using the GeneJET Genomic DNA Purification Kit (Thermo Scientific) following the manufacturer's instructions. A conventional polymerase chain reaction (cPCR) targeting the mammalian ß-actin gene was performed as an internal control for each extraction [30]. Positive samples were then submitted to a real-time PCR (qPCR) specific for the genus Borrelia, [31]. Samples with cycle threshold values ≤ 33 were considered positive and then subjected to conventional nested and semi-nested PCRs to amplify 16S rRNA and flaB genes fragments [32, 33]. The annealing temperature for the 16S rRNA gene in the cPCR was modified in the first round (FD3 [f]/T50 [r]) and second round (Rec4 [f]/Rec9 [r]) from 56 °C to 54 °C (Table 1). Genomic DNA of Borrelia anserina and molecular-grade water were used as positive and negative controls, respectively. Amplicons of the expected size were Sanger-sequenced at Macrogen (Seoul, Korea), sequences assembled with Geneious, and the consensuses were compared in GenBank using BLASTn [34].
The alignments were built in Clustal Omega [35]. For the 16S rRNA and the flaB gene, 70 and 81 sequences downloaded from GenBank were used, respectively [36]. Phylogenetic reconstructions were performed in IQ-TREE with the maximum likelihood method using the TPM3+F+I+G4 nucleotide substitution model with 1000 bootstraps. Trees were visualized and edited with iTOL v5 [37]. Brachyspira pilosicoli was used as root.
Results
Two hundred and five spleen samples from different bat species were processed. Amplicons of the expected size for the ß-actin gene were obtained in all samples, thus confirming successful DNA extraction. Overall, 10.2% (21/205) were positive for the Borrelia 16S rRNA gene by qPCR. In 81% (17/21) and 66,6% (14/21) positive samples, it was possible to amplify 16S rRNA and flaB genes by cPCR. Regarding bat species, Borrelia DNA was detected in 5.4% (11/21) of specimens in the genus Phyllostomus, 2.4% (5/21) in Glossophaga, 1.5% (3/21) in Carollia, 0.5% (1/21) in the genus Artibeus, and 0.5% (1/21) in Uroderma. For each Borrelia gene, nine randomly selected cPCR products were sequenced; however, good quality sequences were obtained in seven 16S rRNA and six flaB samples. Most identical sequences after Blastn comparisons for the 16S rRNA gene matched spirochetes detected in Ornithodoros mimon from Brazil [38]; a Borrelia sp. reported in Amblyomma varanense from Varanus salvator (reptile) from Indonesia [39]; and a Borrelia sp. reported in hard ticks from Portugal [40]. For the flaB gene, sequences were most identical with “Borrelia sp. Macaregua” haplotypes, detected in bats from the Macaregua cave in Santander, Colombia [10] (Table 2).
In the 16S rRNA phylogeny, all the obtained sequences were grouped monophyletically and as a sister lineage to “Candidatus Borrelia ivorensis” and “Candidatus Borrelia africana” out of the RF or Lyme groups (Fig. 2A). The flaB phylogenetic tree places all our sequences into a monophyletic clade together with haplotypes of “Borrelia sp. Macaregua” and Borrelia sp. Potiretama (Fig. 2B).
Phylogenetic analysis performed in this study. Trees are drawn to scale, with the scale bar indicating nucleotide substitutions per site. The position of the detected Borrelia spp. is highlighted in red. A Phylogenetic tree of 16S rRNA gene constructed with 78 sequences. B Phylogenetic tree of the flaB gene built with 88 sequences
Discussion
To date, there are few studies that show the presence of Borrelia in bats. One of the first reports was made by Nicolle and Comte in 1905, with the finding of spirochetes in the blood of Vespertilio kuhli from Tunisia [41]. Then, in 1945, Nájera Angulo demonstrated the susceptibility of four bats species (Miniopterus schreibersii, Myotis myotis, Rhinolophus euryale, and Rhinolophus hipposideros minimus) from Spain to a “Hispanic spirochete” after inoculation of blood from infected guinea pig [42].
Later, in 2006, in the United States, through serological tests it was possible to verify the circulation of borreliae in Eptesicus fuscus bats [17]. Subsequently, in 2009, in England, spirochetes were identified in liver tissue stained with Warthin–Starry in a bat of the genus Pipistrellus, and it was confirmed as an RF group Borrelia, obtaining a 776-base-pair (bp) segment of the flaB gene [7].
In recent years, Ca. Borrelia fainii was reported in Myotis sp. [9], Rhinolophus pusillus, and Myotis davidii bats in China [12]. Regarding Latin America, in Mexico, Colunga-Salas et al. detected two new lineages of Borrelia, one RF and one Bb, in Saccopteryx bilineata, Choeroniscus godmani, Sturnira parvidens, and Lasiurus cinereus [11]. In Colombia, in 1968, Marinkelle and Grose observed spirochetes in a blood smear of Natalus tumidirostris [43] from the Macaregua cave, and in 2020 Muñoz-Leal et al. detected a new putative taxon within the genus Borrelia in Carollia perspicillata captured in the same cave [10]. Likewise, in 2022, Jorge et al. detected Borrelia sp. Potiretama in Desmodus rotundus bats, in the municipality of Potiretama in Brazil [21]. In the current study, we detected Borrelia DNA in five species of bats (C. perspicillata, Phyllostomus discolor, Artibeus lituratus, Glossophaga soricina, and Uroderma sp.) captured in Colombia, reinforcing the fact that bats do harbor Borrelia spirochetes.
In our study, two phylogenetic analyses were performed including Borrelia spp. previously detected in bats from South America, such as Borrelia sp. Macaregua and Borrelia sp. Potiretama [10, 21]. Unfortunately, 16S rRNA sequences for Borrelia sp. Macaregua are not available in GenBank, so it was not possible to perform comparisons with this species using this gene. Interestingly, 16S rRNA phylogeny depicts the group of Borrelia spp. detected in this study as paraphyletic regarding Borrelia sp. Potiretama reported in bats from Brazil, and Ca. Borrelia ivorensis and Ca. Borrelia africana, two species detected in African ticks, as closely related. The fact that some South American borreliae are phylogenetically closer to their African representatives has also been observed for spirochetes characterized in soft ticks from Brazil and Chile [38, 44]. However, any conclusion regarding the phylogenetic position of the Borrelia spp. detected in our study is premature, since we obtained short 16S rRNA gene sequences. Indeed, the fact that the flaB tree showed the Borrelia sequences of our study forming a monophyletic group with Borrelia sp. Potiretama, contradicting the topology of the 16S rRNA tree, could also be explained by the limited data submitted to analysis for this gene (381–435 bp).
A monophyletic group of Borrelia spp. associated with neotropical bats roosting in caves has been recently proposed based primarily on phylogenetic analyses of flaB sequences [10, 21]. Our study supports this hypothesis, adding more genovariants to this group of bat species inhabiting wooded areas. Because of their defense-immune tolerance capacities, bats are considered excellent reservoirs that favor the emergence of novel viruses [45]. The reasons that South American bats seem to harbor such a remarkable diversity of Borrelia haplotypes could relate to their immunological system as well.
Finally, isolating Borrelia spp. circulating in bats and sequencing their genomes should now be the focus in order to clearly elucidate the phylogenetic relationships. The study of Borrelia in bats is important because several species associated with these mammals, such as Ca. Borrelia fainii and Ca. Borrelia johnsonii, have recently been detected in humans in Africa and the United States, respectively [9, 12, 26, 27]. Therefore, the pathogenic roles of spirochetes detected in neotropical bats should be further investigated.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- Bb:
-
Borrelia burgdorferi sensu lato
- RF:
-
Borreliae of the relapsing fever group
References
Oppler Z, Keeffe K, McCoy K, Brisson D. Evolutionary genetics of Borrelia. Curr Issues Mol Biol. 2021;42:97–112. https://doi.org/10.21775/cimb.042.097.
Margos G, Gofton A, Wibberg D, Dangel A, Marosevic D. The genus Borrelia reloaded. PLoS ONE. 2018;13:1–14. https://doi.org/10.1371/journal.pone.0208432.
Loh SM, Gillett A, Ryan U, Irwin P, Oskam C. Molecular characterization of ‘Candidatus Borrelia tachyglossi’ (family spirochaetaceae) in echidna ticks, Bothriocroton concolor. Int J Syst Evol Microbiol. 2017;67:1075–80. https://doi.org/10.1099/ijsem.0.001929.
Takano A, Goka K, Une Y, Shimada Y, Fujita H, Shiino T, et al. Isolation and characterization of a novel Borrelia group of tick-borne borreliae from imported reptiles and their associated ticks. Environ Microbiol. 2010;12:134–46. https://doi.org/10.1111/j.1462-2920.2009.02054.x.
Sánchez RST, Santodomingo AMS, Muñoz-Leal S, Silva-De la Fuente MC, Llanos-Soto S, Salas LM, et al. Rodents as potential reservoirs for Borrelia spp. in northern Chile. Rev Bras Parasitol Vet. 2020;29:1–10. https://doi.org/10.1590/S1984-29612020029.
Schotthoefer AM, Frost HM. Ecology and epidemiology of Lyme borreliosis. Clin Lab Med. 2015;35:723–43. https://doi.org/10.1016/j.cll.2015.08.003.
Evans NN, Kevin B, Timofte D, Simpson V, Birtles R. Fatal borreliosis in bat caused by relapsing fever spirochete, United Kingdom. Emerg Infect Dis. 2009;15:1331–3. https://doi.org/10.3201/eid1508.090475.
Benerjee A, Baid K, Byron T, Yip A, Ryan C. Seroprevalence in bats and detection of Borrelia burgdorferi in bat ectoparasites. Microorganisms. 2020;8:1–8. https://doi.org/10.3390/microorganisms8030440.
Hui-Ju H, Jian-Wei L, Hong-Ling W. Pathogenic new world relapsing fever Borrelia in a Myotis bat, Eastern China, 2015. Emerg Infect Dis. 2020;26:3083–5. https://doi.org/10.3201/eid2612.191450.
Muñoz-Leal S, Faccini-Martínez ÁA, Pérez-Torres J, Chala-Quintero SM, Herrera-Sepúlveda MT, Cuervo C, et al. Novel Borrelia genotypes in bats from the Macaregua Cave, Colombia. Zoonoses Public Health. 2021;68:12–8. https://doi.org/10.1111/zph.12789.
Colunga-Salas P, Sánchez-Montes S, León-Paniagua L, Becker I. Borrelia in neotropical bats: detection of two new phylogenetic lineages. Ticks Tick Borne Dis. 2021;12:101642. https://doi.org/10.1016/j.ttbdis.2020.101642.
Ze-Min L, Xiao X, Chuan-Min Z, Liu J-X. Human-pathogenic relapsing fever Borrelia found in bats from Central China phylogenetically clustered together with relapsing fever borreliae reported in the New World. PloS Negl Trop Dis. 2021;15:e10009113. https://doi.org/10.1371/journal.pntd.0009113.
Gill J, Ullmann A, Loftis A, Schwan T, Raffel S, Schrumpf M, et al. Novel relapsing fever spirochete in bat tick. Emerg Infect Dis. 2008;14:522–3. https://doi.org/10.3201/eid1403.070766.
Jaenson T, Wilhelmsson P. First record of suspected human-pathogenic Borrelia species in populations of the bat tick Carios vespertilionis in Sweden. Microorganisms. 2021;9:1–12. https://doi.org/10.3390/microorganisms9051100.
Loftis AD, Gill JS, Schriefer ME, Levin ML, Eremeeva ME, Gilchrist MJR, et al. Detection of Rickettsia, Borrelia, and Bartonella in Carios kelleyi (Acari: Argasidae). J Med Entomol. 2005;42:473–80. https://doi.org/10.1093/jmedent/42.3.473.
Michalik J, Wodecka B, Liberska J, Dabert M, Postawa T, Piksa K, et al. Diversity of Borrelia burgdorferi sensu lato species in Ixodes ticks (Acari: Ixodidae) associated with cave-dwelling bats from Poland and Romania. Ticks Tick Borne Dis. 2020;11:101300. https://doi.org/10.1016/j.ttbdis.2019.101300.
Reeves WK, Streicker DG, Loftis AD, Dasch GA. Serologic survey of Eptesicus fuscus from Georgia, U.S.A for Rickettsia and Borrelia and laboratory transmission of a Rickettsia by bat ticks. J Vector Ecol. 2006;31:386–9. https://doi.org/10.3376/1081-1710(2006)31[386:ssoeff]2.0.co;2.
Sándor A, Mihalca A, Domsa C, Péter A, Hornok S. Argasid ticks of palearctic bats: distribution, host selection, and zoonotic importance. Front Vet Sci. 2021;8:684737. https://doi.org/10.3389/fvets.2021.684737.
Schwan TG, Raffel SJ, Schrumpf ME, Gill JS, Piesman J. Characterization of a novel relapsing fever spirochete in the midgut, coxal fluid, and salivary glands of the bat tick Carios kelleyi. Vector-Borne Zoonotic Dis. 2009;9:643–7. https://doi.org/10.1089/vbz.2008.0177.
Socolovschi C, Kernif T, Raoult D, Parola P. Borrelia, Rickettsia, and Ehrlichia species in bat ticks, France, 2010. Emerg Infect Dis. 2012;18:1966–75. https://doi.org/10.3201/eid1812.111237.
Jorge FR, Muñoz-leal S, Oliveira GMB De, Serpa MCA, Magalhães MML, Oliveira LMB De, et al. Novel Borrelia Genotypes from Brazil Indicate a New Group of Borrelia spp. Associated with South American Bats. J Med Entomol. 2022;1–5. https://doi.org/10.1093/jme/tjac160%0A.
Drexler JF, Corman VM, Drosten C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res. 2014;101:45–56. https://doi.org/10.1016/j.antiviral.2013.10.013.
Kirtipal N, Bharadwaj S, Gu S. From SARS to SARS-CoV-2, insights on structure, pathogenicity and immunity aspects of pandemic human coronaviruses. Infect Genet Evol. 2020;85:15. https://doi.org/10.1016/j.meegid.2020.104502.
Mattar S, González M. The amazing bats: friends, enemies or allies? Rev MVZ Córdoba. 2016;22:6177–9. https://doi.org/10.21897/rmvz.1125.
Mühldorfer K. Bats and bacterial pathogens: a review. Zoonoses Public Health. 2013;60:93–103. https://doi.org/10.1111/j.1863-2378.2012.01536.x.
Kingry LC, Anacker M, Pritt B, Bjork J, Respicio-Kingry L, Liu G, et al. Surveillance for and discovery of Borrelia species in US patients suspected of tickborne illness. Clin Infect Dis. 2018;66:1864–71. https://doi.org/10.1093/cid/cix1107.
Qiu Y, Nakao R, Hangombe BM, Sato K, Kajihara M, Kanchela S, et al. Human borreliosis caused by a new world relapsing fever borrelia-like organism in the old world. Clin Infect Dis. 2019;69:107–12. https://doi.org/10.1093/cid/ciy850.
Leary S, Underwood, W. Anthony R, Cartner S, Grandin T, Greenacre, C. Gwaltney-Brant S. McCrackin MA, Meyer R, et al. AVMA guidelines for the euthanasia of animals. Am Vet Med. 2020; 1–111.
Díaz MM, Solari S, Aguirre LF, Aguiar L, Barquez RM. Clave de identificación de los murciélagos de Sudamérica Publicación Especial Nro, 2. 2016.
Dean D, Rothschild J, Ruettger A, Prasad R, Sachse K. Zoonotic Chlamydiaceae species associated with trachoma, Nepal. Emerg Infect Dis. 2013;19:1948–55. https://doi.org/10.3201/eid1912.130656.
Parola P, Diatta G, Socolovschi C, Mediannikov O, Tall A, Bassene H, et al. Tick-borne relapsing fever borreliosis, rural Senegal. Emerg Infect Dis. 2011;17:883–5. https://doi.org/10.3201/eid1705.100573.
Ras N, Lascola B, Postic D, Cutler S, Rodhain F, Baranton G, et al. Phylogenesis of relapsing fever Borrelia spp. Int J Syst Bacteriol. 1996;46:859–65. https://doi.org/10.1099/00207713-46-4-859.
Stromdahl E, Williamson P, Kollars T, Evans S, Barry R, Vince M, et al. Evidence of Borrelia lonestari DNA in Amblyomma americanum (Acari: Ixodidae) removed from Humans. J Clin Microbiol. 2003;41:5557–62. https://doi.org/10.1128/JCM.41.12.5557-5562.2003.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. https://doi.org/10.1016/S0022-2836(05)80360-2.
Sievers F, Higgins DG. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27:135–45.
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank: update. Nucleic Acids Res. 2004;32:D23–6. https://doi.org/10.1093/nar/gkh045.
Letunic I, Bork P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–6. https://doi.org/10.1093/nar/gkab301.
Muñoz-Leal S, Faccini-Martínez ÁA, Teixeira BM, Martins MM, Serpa MCA, Oliveira GMB, et al. Relapsing fever group borreliae in human-biting soft ticks, Brazil. Emerg Infect Dis. 2021;27:321–4. https://doi.org/10.3201/eid2701.200349.
Takano A, Kuwata R, Shimoda H, Hadi UK, Setiyono A, et al. Detection and isolation of tick-borne bacteria (Anaplasma spp., Rickettsia spp., and Borrelia spp.) in Amblyomma varanense ticks on lizard (Varanus salvator). Microbiol Immunol. 2019;63:328–33. https://doi.org/10.1111/1348-0421.12721.
Nunes M, Parreira R, Maia C, Lopes N, Fingerle V, Vieira ML. Molecular identification of Borrelia genus in questing hard ticks from Portugal: phylogenetic characterization of two novel relapsing fever-like Borrelia sp. Infect Genet Evol. 2016;40:266–74. https://doi.org/10.1016/j.meegid.2016.03.008.
Nicolle C, Comte C. Sur une spirollose d’un chéiroptère (Vespertilio kuhli). Ann Inst Pasteur (Paris). 1906;20:311–20.
Najera-Angulo L. Receptividad de los murciélagos cavernícolas españoles (Miniopterus schreibersii, Myotis myotis, Rhinolophus euryale y Rh Hipposideros minimus) al virus de la fiebre recurrente mediterránea. Bol La Real Soc Española Hist Nat. 1945;23:217–28.
Marinkelle CJ, Grose ES. Species of Borrelia from a Colombia Bat (Natalus Tumidirostris). Nature. 1968;218:487. https://doi.org/10.1038/218487a0.
Thompson M, Muñoz-Leal S, Troncoso I, Thomas R, Santodomingo A, Moreno-Salas L, et al. A Borrelia sp. in Ornithodoros octodontus (Argasidae). Syst Appl Acarol. 2021;11:1997–2001.
Irving AT, Ahn M, Goh G, Anderson DE, Wang LF. Lessons from the host defences of bats, a unique viral reservoir. Nature. 2021;589:363–70. https://doi.org/10.1038/s41586-020-03128-0.
Acknowledgements
We thank the University of Córdoba, where the project was carried out, Yeimi López for making the map in Fig. 1, and Marcelo Labruna for providing positive control. SML was funded by Fondecyt Iniciación No. 11220177.
Funding
No funding.
Author information
Authors and Affiliations
Contributions
YL, SeMu, SaMa, and ÁAFM designed the initial study. CM, AC, CG, and JM carried out the fieldwork. CM, AC, CG, and JM performed the taxonomic identification and processing of the bats. YL, CM, KG, MM, and JR performed DNA extraction, PCRs, and sequencing. YL, SeMu, and ÁAFM implemented the phylogenetic analyses. YL, CM, SeMu, and ÁAFM wrote the first draft of the manuscript. All authors contributed to the interpretation and review of the data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The ethics committee of the Faculty of Veterinary Medicine and Zootechnics of the University of Córdoba, through Act No. 003 of December 6, 2019, approved the capture of bats, carried out under the permits of the National Authority of Environmental Licenses [ANLA], Resolution No. 00914.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
López, Y., Muñoz-Leal, S., Martínez, C. et al. Molecular evidence of Borrelia spp. in bats from Córdoba Department, northwest Colombia. Parasites Vectors 16, 5 (2023). https://doi.org/10.1186/s13071-022-05614-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-022-05614-y