Abstract
Malaria control in Somaliland depends on the effective identification of potential malaria vectors, particularly those that may be invasive. The malaria vector Anopheles stephensi has been detected in multiple countries in the Horn of Africa (HOA), but data on its geographic distribution and population genetic diversity are incomplete. We implemented a vector surveillance program and performed molecular analysis of Anopheles in three urban areas in Somaliland. Our study confirmed the presence of both the invasive An. stephensi and the long-established HOA malaria vector Anopheles arabiensis. Further analysis of An. stephensi genetic diversity revealed three cytochrome oxidase I (COI) haplotypes, all of which have been observed in other countries in East Africa and one also observed in South Asia. We also detected the knockdown resistance (kdr) L1014F mutation, which is associated with pyrethroid resistance; this finding supports the need for further assessment of the potential for insecticide resistance. The detection of multiple haplotypes previously observed in other regions of East Africa indicates that An. stephensi is an established population in Somaliland and likely shares its origin with other newly identified An. stephensi populations in East Africa. The detection of genetic diversity in An. stephensi in Somaliland provides a basis for future studies on the history of the species in the region and its dispersal throughout East Africa.
Graphical Abstract

Similar content being viewed by others
Background
Malaria is a major global health threat, with about 215 million cases reported in Africa in 2019 [1]. While significant progress has been made to reduce malaria cases and mortality over the last decade, the recent detection of a new vector, Anopheles stephensi, in the Horn of Africa (HOA) threatens to reverse this progress. Anopheles stephensi, a vector found in South Asia and the Middle East, including large parts of the Arabian Peninsula, was first detected in Djibouti in 2012 [2]. Since then, it has been reported in Ethiopia [3], Sudan [4], Somalia [5] and, most recently, Yemen [5].
The presence of An. stephensi has raised concern about its spread into urban areas and the potential to increase the number of malaria cases [6,7,8], given increased urbanization and human movement in Africa. In Asia, An. stephensi is one of the few Anopheles vectors known to inhabit urban areas [9], likely due to its successful breeding in human-made containers. In addition, studies published between 2019 and 2021 indicate the vector is resistant to pyrethroid-, organophosphate- and carbamate-based insecticides in Ethiopia [10, 11], as observed in established populations (reviewed in Enayati et al. [12]), and is capable of Plasmodium falciparum and Plasmodium vivax transmission [13, 14]. These findings highlight the growing threat of An. stephensi and the need for further investigation of this vector in the Horn of Africa.
Evaluating the risk for malaria transmission by An. stephensi requires accurate data on its present distribution in the HOA. In Somaliland, where no earlier records are available on the presence of An. stephensi, there was an unusual increase in malaria cases in Sahil, increasing from 87 cases reported in 2019 to 1836 cases reported in 2020 (HMIS/DHIS2). To better understand the basis for this increase, more data on the geographic distribution of the vector populations are needed. Anopheles gambiae sensu lato (s.l.) is the major malaria vector in Somaliland, and ecological niche modeling indicates An. arabiensis as the likely candidate from the complex [15], although DNA sequences have not been generated to confirm this identification. Anopheles stephensi was detected in Bossaso City, Puntland State, northern Somalia, in 2019. In March 2020, An. stephensi was detected in Berbera, the main seaport of Somaliland [5], located in the Sahil region. With this initial report of An. stephensi in the region, more information on the distribution of the vector in Somaliland is needed to evaluate its potential impact on malaria transmission. In addition, analysis of the genetic diversity of An. stephensi can elucidate the relationship between the population in Somaliland, new populations in the HOA and long-established populations outside the HOA. Here we present morphological and molecular confirmation of the presence of An. stephensi in Somaliland and a preliminary analysis of its genetic diversity.
Methods
Study sites
As a follow-up to the initial survey conducted in Berbera in March 2020, we conducted additional surveys between September and November 2020 in six urban sites located in semi-arid or desert climate zones (Fig. 1; Table 1). There are two wet seasons in these areas, from March to July and August to November, respectively, with an average precipitation of 370 mm per year [16]. The elevation in the study sites ranged between 96 and 1500 m a.s.l. We surveyed sites in response to the uptick in the number of reported malaria cases (i.e. in the districts of Hargeisa and Berbera). In the Awdal administrative region (western Somaliland), surveys were conducted in the urban sites of Lawyacado and Borama. Other sites included in this report undergo routine surveillance at fixed sentinel sites on the second or third month of each quarter. In the Marodijeh administrative region, we surveyed the urban areas of Hargeisa (two locations), Dacarbudhuq, and Gebliy. Specimens collected during the survey which initially detected An. stephensi in Berbera (Sahil) were also included in the genetic analysis.
Map of study sites surveyed in Somaliland in 2020. Blue dots represent sites where Anopheles stephensi was detected. Map was created using Adobe Illustrator v. 2019 (Adobe Inc., San Jose, CA, USA) and Pages OS 13 (Apple Inc., Cupertino, CA, USA) based on maps from Google Maps (https://www.google.com/maps)
Collections
Specimens were collected during a larval survey using standard dipping techniques. The surveyed breeding habitats included artificial containers or objects, such as discarded tires, metal and plastic tanks, berkads (concrete containers for water storage), and natural water sources, such as freshwater pools and stream margins. We reared larvae in the field insectary using water taken from the breeding sites and baking yeast for feeding and transferred pupae into adult emergence cages. Anopheles mosquitoes were identified using the updated key of Afrotropical mosquitoes [17]. The identified An. stephensi samples were preserved with silica gel, and a subsample of specimens was sent to Baylor University for molecular analysis.
PCR and sequencing
DNA was extracted from head and thorax using the Qiagen DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany). Species identification was conducted using the ITS2 endpoint assay protocol detailed in [18, 19]. The primer sequences for PCR in the ITS2 endpoint assay are 5.8SB (5′-ATCACTCGGCTCGTGGATCG-3ʹ) and 28SC (5ʹ-GTCTCGCGACTGCAACTG-3ʹ). When the products of the endpoint assay are visualized with gel electrophoresis, a band will be present if the sample contains An. stephensi DNA. For further species identification, portions of the mitochondrial cytochrome oxidase subunit I (COI) locus and internal transcribed spacer 2 (ITS2) locus were PCR amplified and sequenced for subsequent phylogenetic analysis using protocols previously detailed by Carter et al. [20]. The primer sequences for COI are LCO1490F (5ʹ-GGTCAACAAATCATAAAGATATTGG-3ʹ) and HCO2198R (5ʹ-TAAACTTCAGGGTGACCAAAAAATCA-3ʹ) [21]. The primer sequences for the ITS2 sequences are 5.8SB (5ʹ-ATCACTCGGCTCGTGGATCG-3ʹ) and 28SB (5ʹ-ATGCTTAAATTTAGGGGGTAGTC-3ʹ) [19, 22]. The ITS2 and COI sequences were submitted as queries to NCBI BLAST to confirm the correct locus was amplified. Sequences for the haplotypes identified in this study were submitted to NCBI Nucleotide database (Accession nos. ON421572-ON421575).
We also analyzed the pyrethroid resistance-associated voltage-gated sodium channel (vgsc) gene for the knockdown resistance mutation (kdr). The PCR analyses and sequencing were conducted using previously published protocols [10]. Sequences were submitted as queried to NCBI BLAST to confirm correct kdr locus amplification, and kdr mutation detection was performed using alignment to reference sequences from Yared et al. [10] in CodonCode Aligner version 8 (CodonCode Corp., Centerville, MA, USA).
Sequence analysis
For further confirmation of species identification, we performed phylogenetic analysis that incorporated COI sequences from the Anopheles included in this study, sequences of An. stephensi from the HOA, Arabian Peninsula, Middle East and South Asia and representative sequences from the high scoring segment pair sequences retrieved from GenBank via Nucleotide BLAST [23]. Because COI is not able to differentiate the different members of An. gambiae s.l., additional phylogenetic analysis of the ITS2 sequence was performed for An. gambiae s.l. specimens for species level identification. Phylogenetic relationships were inferred using a maximum likelihood approach with RAxML GUI [24] using the GTR model of nucleotide substitutions and gamma model for rate of heterogeneity (GTRGAMMA option). Anopheles implexus was designated as an outgroup for COI analysis and Anopheles christyi as an outgroup for ITS2 analysis. The tree with the highest log likelihood was visualized and formatted in FigTree [25].
Results and discussion
A total of 103 breeding sites across the six sites were inspected during the survey, and larvae were only detected in 28 berkads. Of the six sites surveyed, An. stephensi was detected at three sites: Berbera (March 2020), Hargeisa (September 2020) and Lawyacado (October 2020). Mosquito larvae/pupae collected at these three sites were reared to adulthood. No An. stephensi were detected at Dacarbudhuq, Borama and Gebliy.
Forty-eight of the collected specimens were sent to Baylor University for molecular analysis, of which morphological characterization identified 36 as An. stephensi and 12 as An. gambiae s.l. All An. gambiae s.l. specimens were from Hargeisa and were included to confirm that any An. stephensi specimen could be correctly distinguished morphologically from the native vector. Molecular analysis of extracted DNA was successful in identifying the species of 45 of the 48 mosquitoes; DNA extraction was unsuccessful in three morphologically identified An. stephensi, and thus molecular analysis was not possible for these three mosquitoes. The species identity of the 33 morphologically-identified An. stephensi was confirmed based on the presence of the characteristic band in the ITS2 endpoint assay. All 12 morphologically-identified An. gambiae s.l. were confirmed not to be An. stephensi based on the absence of the bands in the ITS2 endpoint assay. Phylogenetic analysis of COI further confirmed An. stephensi (bootstrap = 94) and An. gambiae s.l. (bootstrap = 99) identifications (Fig. 2). Phylogenetic analysis of the ITS2 sequence in An. gambiae s.l. further revealed that the An. gambiae s.l. specimens are An. arabiensis (bootstrap = 99, Additional file 1: Figure S1). These results are the first genetic confirmation of these species in Somaliland. These findings also confirm successful implementation of the new morphological key and support the continued use of the key for distinguishing An. stephensi from An. gambiae s.l.
Phylogeny of COI sequences based on maximum likelihood approach. Tree with the highest likelihood score (Final ML Optimization Likelihood Optimization: − 1753.729080) is shown. Anopheles stephensi sequences are shown in color, indicating location of capture, with red indicating taxa in Lawyacado, orange indicating taxa in Berbera and pink indicating taxa in Hargeisa. For An. gambiae sensu lato, blue indicates Hargeisa. Bootstrap values > 70 for notable species clades are shown at nodes. Nodes without numbers had a value < 70. Anopheles stephensi COI haplotypes found in Somaliland (Hap 2–4) are identified with brackets. Abbreviations: COI, Mitochondrial cytochrome oxidase subunit I locus; Hap, haplotype; ML, maximum likelihood
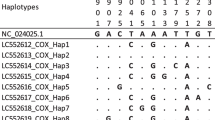
Analysis of the An. stephensi COI sequence data indicated three different haplotypes. These haplotypes have been reported in other countries in East Africa, including Ethiopia (Hap 2–3) [14, 26], Djibouti (Hap 2–4) [27] and Sudan (Hap 2) [4]. The multiple shared haplotypes with Ethiopia and Djibouti may indicate a shared origin of An. stephensi populations or a pattern of movement of An. stephensi between these regions, as would be likely for geographic neighbors. Hap 2 has also been observed outside of the HOA, specifically in Pakistan [28]. The most predominant haplotype was Hap 3 (17/37), followed by Hap 2 (15/37) and Hap 4 (5/37). The distribution of the haplotypes appears to vary by site, although larger sample sizes are needed to confirm this. Notably, COI-Hap 4 was only observed in Lawyacado, the most northwestern site and approximately 18 km from Djibouti city. We also noted that Lawyacado has the highest number of observed haplotypes (n = 3). This follows the trend observed in Ethiopia, where the highest diversity was observed in the northern sites and those most proximal to the eastern border [29]. Preliminary evidence of differences in the level of diversity and genetic differentiation between sites should be followed up with population genomic analysis.
To obtain preliminary insight into the evolution of the insecticide resistance mutations in An. stephensi in this region, we genotyped the kdr locus associated with pyrethroid resistance. Of the 32 mosquitoes with available kdr sequence data, eight carried the kdr L1014F mutation, all as heterozygotes. The highest frequency of kdr variants was in Lawyacado (7/10, 70%). Only one of 12 specimens carried the kdr L1014F in Hargeisa, and none were observed in Berbera (0/10, 0%). There were no kdr L1014S mutations detected in this sample set. The frequency of kdr mutations was found to be higher in Lawyacado than what has been reported in other An. stephensi populations (e.g. 0–16.7% in eastern Ethiopia) [10, 30], which may reflect differences in local vector control approaches. The high-frequency site, which is close to the Djibouti border, may also relate to heightened insecticide-based vector control in nearby Djibouti. However, the sample size is small in this study, and tests for pyrethroid resistance are needed to determine phenotypic resistance. Additional surveillance is critical to determine the extent of insecticide resistance in An. stephensi in Somaliland and to track how An. stephensi populations are responding to changes in control approaches.
Conclusion
Overall, our study confirms the presence of An. stephensi in multiple sites in Somaliland. Detected An. stephensi showed similar breeding sites as previously reported in other studies in East Africa. Preliminary analysis of COI sequence data reveals shared haplotypes within East Africa and abroad, suggesting An. stephensi may have spread across country borders. Further genomic analyses will reveal the pattern and direction of spread within the HOA to inform programs aimed at controlling and/or eliminating An. stephensi in Somaliland and to prevent further spread in Africa.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and as supplemental files. Sequences are deposited in the NCBI GenBank database (Accession # ON421572-ON421575).
References
WHO. World malaria report 2020. 2020. https://www.who.int/publications/i/item/9789240015791.
Faulde MK, Rueda LM, Khaireh BA. First record of the Asian malaria vector Anopheles stephensi and its possible role in the resurgence of malaria in Djibouti Horn of Africa. Acta Trop. 2014;139:39–43.
Carter TE, Yared S, Gebresilassie A, Bonnell V, Damodaran L, Lopez K, et al. First detection of Anopheles stephensi Liston, 1901 (Diptera: culicidae) in Ethiopia using molecular and morphological approaches. Acta Trop. 2018;188:180–6.
Ahmed A, Khogali R, Elnour MB, Nakao R, Salim B. Emergence of the invasive malaria vector Anopheles stephensi in Khartoum State, Central Sudan. Parasit Vectors. 2021;14:511.
WHO. WHO malaria threats map. https://apps.who.int/malaria/maps/threats/. Accessed 1 Sept 2021.
Hamlet A, Dengela D, Tongren JE, Tadesse FG, Bousema T, Sinka M, et al. The potential impact of Anopheles stephensi establishment on the transmission of Plasmodium falciparum in Ethiopia and prospective control measures. medRxiv. 2021;18:396.
Takken W, Lindsay S. Increased threat of urban malaria from Anopheles stephensi mosquitoes. Africa Emerg Infect Dis. 2019;25:1431–3.
Sinka ME, Pironon S, Massey NC, Longbottom J, Hemingway J, Moyes CL, et al. A new malaria vector in Africa: predicting the expansion range of Anopheles stephensi and identifying the urban populations at risk. Proc Natl Acad Sci USA. 2020;117:24900–8.
Sinka ME, Bangs MJ, Manguin S, Chareonviriyaphap T, Patil AP, Temperley WH, et al. The dominant Anopheles vectors of human malaria in the Asia-Pacific region: occurrence data, distribution maps and bionomic precis. Parasit Vectors. 2011;4:89.
Yared S, Gebressielasie A, Damodaran L, Bonnell V, Lopez K, Janies D, et al. Insecticide resistance in Anopheles stephensi in Somali Region, eastern Ethiopia. Malaria J. 2020;19:180. https://doi.org/10.1186/s12936-020-03252-2.
Balkew M, Mumba P, Yohannes G, Abiy E, Getachew D, Yared S, et al. An update on the distribution, bionomics, and insecticide susceptibility of Anopheles stephensi in Ethiopia, 2018–2020. Malar J. 2021;20:263.
Enayati A, Hanafi-Bojd AA, Sedaghat MM, Zaim M, Hemingway J. Evolution of insecticide resistance and its mechanisms in Anopheles stephensi in the WHO Eastern Mediterranean Region. Malar J. 2020;19:258.
Seyfarth M, Khaireh BA, Abdi AA, Bouh SM, Faulde MK. Five years following first detection of Anopheles stephensi (Diptera: Culicidae) in Djibouti, Horn of Africa: populations established-malaria emerging. Parasitol Res. 2019;118:725–32.
Tadesse FG, Ashine T, Teka H, Esayas E, Messenger LA, Chali W, et al. Anopheles stephensi Mosquitoes as Vectors of Plasmodium vivax and falciparum, Horn of Africa, 2019. Emerg Infect Dis. 2021;27:603.
Drake JM, Beier JC. Ecological niche and potential distribution of Anopheles arabiensis in Africa in 2050. Malar J. 2014;13:213.
Ullah S. Territorial diagnostic report of the land resources of Somaliland. 2016. Food and Agriculture Organization of the UN/Somalia Water and Land Information Management (FAO/FAO-SWALIM). 2016. https://www.faoswalim.org/resources/site_files/L%20-21%20Land%20diagnostic%20report%20.pdf. Accessed 1 Jan 2021.
Coetzee M. Key to the females of Afrotropical Anopheles mosquitoes (Diptera: Culicidae). Malar J. 2020;19:70.
Balkew M, Mumba P, Dengela D, Yohannes G, Getachew D, Yared S, et al. Geographical distribution of Anopheles stephensi in eastern Ethiopia. Parasit Vectors. 2020;13:1–8.
Djadid ND, Gholizadeh S, Aghajari M, Zehi AH, Raeisi A, Zakeri S. Genetic analysis of rDNA-ITS2 and RAPD loci in field populations of the malaria vector, Anopheles stephensi (Diptera: Culicidae): implications for the control program in Iran. Acta Trop. 2006;97:65–74.
Carter TE, Yared S, Hansel S, Lopez K, Janies D. Sequence-based identification of Anopheles species in eastern Ethiopia. Malar J. 2019;18:135.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–9.
Collins FH, Mendez MA, Rasmussen MO, Mehaffey PC, Besansky NJ, Finnerty V. A ribosomal RNA gene probe differentiates member species of the Anopheles gambiae complex. Am J Trop Med Hyg. 1987;37:37–41.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Edler D, Klein J, Antonelli A, Silvestro D. raxmlGUI 20: a graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol Evol. 2021;12:373–7.
Rambaut A. Figtree Version. 1.4. 4. 2018. http://tree.bio.ed.ac.uk/software/figtree.
Carter TE, Gebresilassie A, Hansel S, Damodaran L, Montgomery C, Bonnell V, et al. Analysis of the knockdown resistance locus (kdr) in Anopheles stephensi, An. arabiensis, and Culex pipiens s.l. for insight Into the evolution of target-site pyrethroid resistance in eastern Ethiopia. Am J Trop Med Hyg. 2022;106:632–8.
de Santi VP, Khaireh BA, Chiniard T, Pradines B, Taudon N, Larreche S, et al. Role of Anopheles stephensi mosquitoes in malaria outbreak, Djibouti, 2019. Emerg Infect Dis. 2021;27:1697–700.
Ashfaq M, Hebert PD, Mirza JH, Khan AM, Zafar Y, Mirza MS. Analyzing mosquito (Diptera: Culicidae) diversity in Pakistan by DNA barcoding. PLoS ONE. 2014;9:e97268.
Carter TE, Yared S, Getachew D, Spear J, Choi SH, Samake JN, et al. Genetic diversity of Anopheles stephensi in Ethiopia provides insight into patterns of spread. Parasit Vectors. 2021;14:1.
Samake JN, Yared S, Getachew D, Mumba P, Dengela D, Yohannes G, et al. Detection and population genetic analysis of kdr L1014F variant in eastern Ethiopian Anopheles stephensi. Infect Genet Evol. 2022;99:105235.
Acknowledgements
The authors would like to thank Dr. Abdi Abdillahi of Somaliland NMCP/MOHD for his support. The authors would also like to thank Drs. Samira Al-Eryani (Technical Officer WHO/EMRO-MVC), Ghasem Zamani (Regional Advisor WHO/EMRO-MVC) and Jamal Amran (Medical Officer WHO Country Office, Somalia) for their contribution to the conceptualization of the project, technical support for implementation of surveillance and review of the manuscript, and Drs. Seth Irish and Sarah Zohdy (U.S. President’s Malaria Initiative) for their helpful discussions. Finally, the authors would like to thank Dr. Bouh Abdi Khaireh (United Nations Development Program, The Global Fund to Fight AIDS, Tuberculosis and Malaria, Djibouti, Republic of Djibouti) and Drs. Sébastien Briolant and Vincent Pommier de Santi (Aix Marseille University, IHU-Méditerranée Infection, French Armed Forces Center for Epidemiology and Public Health) for sharing Djibouti An. stephensi sequence data.
Disclaimer
The authors alone are responsible for the views expressed in this article and they do not necessarily represent the views, decisions or policies of the institutions with which they are affiliated.
Funding
This work is supported by GF/UNICES and WHO country Office Somalia for the entomological surveillance and shipment of the samples. Molecular research reported in this publication was supported by Baylor University.
Author information
Authors and Affiliations
Contributions
TC and SA contributed to the conceptualization of the project. TC and SA contributed to the design of the study. SA organized the mosquito collection protocol and processing. TC organized the molecular analysis protocol and processing. JNS and JS generated the molecular data. TC, JNS and JS conducted sequence analysis. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
Phylogenetic analysis of ITS2 using the maximum likelihood approach. Tree with the highest likelihood score (Final ML Optimization Likelihood: -981.332474) is shown. Anopheles gambiae s.l. from Somaliland are highlighted in blue. Bootstrap values > 70 for notable species clades are shown at nodes. Nodes without numbers had a value < 70.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ali, S., Samake, J.N., Spear, J. et al. Morphological identification and genetic characterization of Anopheles stephensi in Somaliland. Parasites Vectors 15, 247 (2022). https://doi.org/10.1186/s13071-022-05339-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-022-05339-y