Abstract
Background
Granada virus belongs to the genus Phlebovirus within the Naples serocomplex and was detected for the first time in sand flies from Spain in 2003. Seroprevalence studies have revealed that Granada virus may infect humans with most cases being asymptomatic. Moreover, recent studies in vector samples revealed that the related Massilia and Arrabida phleboviruses could be also circulating in Spain. The objective of this study was to develop and assess a new sensitive real-time RT-PCR assay for Granada virus diagnosis able to detect the related phleboviruses Massilia and Arrabida.
Methods
Two specific primers and one unique probe to detect Granada, Massilia and Arrabida viruses, without differentiating between them, were designed targeting the conserved L-segment of their genome. Sensitivity was assessed using 10-fold serial dilutions of quantified in vitro DNA samples. Specificity was evaluated by testing different genomic RNA extracted from other representative phleboviruses. The new assay was used for virus detection in sand flies collected in 2012 from the Balearic Archipelago, a touristic hotspot in the Mediterranean.
Results
The real-time RT-PCR assay exhibited a sensitivity per reaction of 19 copies for Granada and Arrabida, and 16 copies for Massilia. No other related phleboviruses were detected. From the 37 pools of sand fly samples studied from four different Balearic Islands, we detected one positive in the island of Cabrera.
Conclusions
To our knowledge, the method described here is the first real-time RT-PCR designed to detect Granada virus and the related Massilia and Arrabida phleboviruses. The study demonstrated that this is a rapid, robust and reliable assay for the accurate diagnosis of human infections as well as for virus surveillance in vectors.

Similar content being viewed by others
Background
Sand fly-borne viruses of the genus Phlebovirus (family Phenuiviridae) are important emerging pathogens in Europe and are widely distributed [1]. All members of this genus are enveloped and the genome is composed of three negative RNA segments, S (small), M (medium) and L (large). The S segment encodes the nucleocapsid protein (N) and non-structural proteins (NSs); the M segment encodes two surface glycoproteins (G1 and G2) and non-structural proteins (NSm), and the L segment encodes the RNA-dependent RNA polymerase. The coding strategy seems to be common to all members of the family [2]. Until now, ten viral species defined by the sero-neutralization tests as well as several tentative species have been described [3], although their real medical importance has not been fully addressed yet. The sand fly fever Naples (SFNV) and Sicilian (SFSV) viruses, cause a “3-day fever”, characterized by flu-like symptoms, and Toscana virus (TOSV) may produce neuroinvasive infection (encephalitis and meningoencephalitis) [1]. TOSV is endemic in the Mediterranean countries, where the Phlebotomus vectors are present. Most TOSV infections have been reported in Italy, France and Spain and sporadically in Portugal, Greece, Cyprus, Turkey and Tunisia.
Recent reports suggest that other sand fly-transmitted phleboviruses may be involved in human disease [1, 4, 5]. This is the case for Granada virus (GRV), a phlebovirus detected for the first time in sand flies captured in 2003 and 2004 in the province of Granada (southeast Spain). Phylogenetic analysis of the complete genome revealed that GRV belongs to the Sand fly fever Naples Complex (SFNC) and it is probably a natural reassortant of the Massilia virus (MASV), donor of the L and S GRV genome segments, with a yet unidentified phlebovirus as donor of the M segment [6]. MASV was detected and isolated for the first time from Phlebotomus pernicious captured in 2005 in Marseille and Nice [7] and later from P. perniciosus captured in 2007–2008 in Portugal (Arrabida region) [8]. In the same study carried out in Portugal, a new phlebovirus named Arrabida (ARRV) was described [9]. ARRV was also phylogenetically classified as member of the SFNC showing a high nucleotide identity with MASV, GRV and Punique virus (PUNV). Whereas the sequences on the L and S segments of ARRV were closely related to MASV and GRV, the glycoproteins (M segment) were most closely related to PUNV, detected in sand flies from Tunisia in 2008, opening the question whether ARRV can be considered as a reassortant between MASV/GRV and PUNV [9]. Therefore, the closely related MASV, GRV and ARRV shared the L and S segments but differed for the M segment due to a reassortment process with different phleboviruses.
In Spain, a few studies conducted in sand flies have revealed the presence and co-circulation of several phleboviruses belonging to the SFNC (such as TOSV, GRV and ARRV) and Salehabad virus species Complex (such as Arbia virus). As mentioned above, GRV has been detected in Granada but sequences resembling the L-segment of GRV and MASV have been reported in other areas of Spain such as the Balearic Archipelago (Ibiza island) and Catalonia. However, without further sequence information for the M segment, they cannot be unequivocally assigned to GRV or MASV strains. Furthermore, similar sequences of viruses belonging to the Salehabad virus species Complex have been described in sand flies from Granada [6]. TOSV has also been detected in sand flies in Barcelona and Granada [6, 10]. In a recent study carried out in Madrid, six viral isolates were obtained from P. perniciosus captured in 2012 and 2013 [11]. The phylogenetic analysis and serological assays allowed the identification of two isolates of TOSV B genotype, three isolates strongly related to Italian Arbia virus (ARBV), and one isolate of a novel putative phlebovirus related to the recently characterized ARRV in South Portugal, tentatively named Arrabida-like virus.
Despite the increasing evidence that these viruses circulate in Spain, only human disease has been directly associated with TOSV in different regions (Murcia, Madrid, Catalonia, Andalusia and the Mediterranean coast) [12,13,14]. Two human serological studies carried out in Granada (Spain) have reported the GRV infective capacity. Collao et al. [6] revealed the presence of specific GRV-neutralizing antibodies in sera from healthy individuals collected in 2003 (10.8% of IFA positives, 1.8% of the total analyzed samples). In a different study, GRV seroprevalence was found to be 15.8% by IFA with 2.8% of neutralizing antibodies (18% of IFA positives). Moreover, anti-GRV IgM antibodies were detected (6.6%) in sera from patients with symptoms of unknown etiology [5]. Therefore, the authors concluded that GRV may infect humans but that most cases would probably be asymptomatic. Occasionally, mild GRV infections may occur, usually as self-limited febrile illnesses accompanied by other signs and symptoms (exanthema and/or acute respiratory infection) [5]. These infections most probably take place during the warm months of the year, which occurs with infections caused by other phleboviruses in Mediterranean countries. Whether GRV or the related MASV and ARRV could be circulating in other areas or countries and/or causing mild febrile diseases needs to be determined. Until now only a real-time RT-PCR designed in the nucleoprotein gene (S segment) has been published to detect MASV specifically [7].
All these data strongly support the need for new molecular assays that allow the specific detection of GRV, MASV and ARRV to improve current diagnostic methods and to be used in vector-borne diseases surveillance programmes for these phleboviruses. Therefore, the main objective of this study was to develop the first real-time RT-PCR assay able to detect GRV with high sensitivity, that at the same time might also detect the related phleboviruses MASV and ARRV. This assay will be very useful in studies about the possible implications of these viruses in human disease and in virus surveillance programmes.
Methods
Primer and probe design
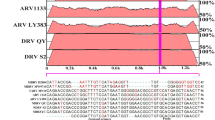
Representative sequences from the three phleboviruses genomic segments (S, M and L) were obtained from the NCBI GenBank database and were aligned using the Clustal W program [15]. Primers and probes were designed using the Primer Express software v2.0 (Applied Biosystems, Foster City, CA, USA), to target a highly conserved region of the L segment of the GRV, MASV and ARRV genomes. We designed two different TaqMan probes with minor groove binder (MGB) and non-fluorescent quencher (NFQ) at the 3′-end, one for GRV-MASV-ARRV (Ph_Probe) and another for the internal control (IC_Probe), both of them were labeled with different fluorophores at the 5′-end (FAM and NED, respectively) (Table 1).
Construction of internal control plasmid and DNA standards
An internal control (IC) was used to exclude the presence of inhibition of the amplification step in the real-time RT-PCR (qRT-PCR). Overlapping primers IC_F and IC_Re were designed to construct the IC and a 68-bp DNA fragment was amplified by an end-point PCR. Primers designed in this technique were used to amplify part of the L gene of GRV (strain GR25; GenBank: GU135606). Due to the lack of reference standards for MAS and ARR viruses in our laboratory, DNA quantified standards were artificially constructed. For both viruses, 136-bp DNA fragments were obtained by an end-point PCR carried out with overlapping primers (Table 2). To obtain the IC and DNA standards, PCRs were performed using AMpliTaq DNA Polymerase with Buffer II (Applied Biosystems, Branchburg, NJ, USA) in a PCT-200, Peltier Thermal Cycler (MJ Research, Watertown, MA, USA), in a final volume of 50 μl, of a reaction mix containing: 5 μl 10× Buffer II, 2.5 mM MgCl2, 2.5 U of Taq enzyme, 0.5 μM of dNTPs (GE Health Care Life Science, Freiburg, Germany), 1 μM of each primer IC-F/IC-Re and RNAse-free H2O. PCR conditions were: 5 min 95 °C, 40 cycles of 30 s at 95 °C, 1 min at 72 °C, followed by a final extension step of 10 min at 72 °C. For the IC, the PCR product was purified and cloned using TOPO TA Cloning Kit (Invitrogen, MA, USA) according to manufacturer’s instructions and sequenced to confirm the absence of mutations. DNA concentration was determined spectrophotometrically measuring the optical density at 260 nm (OD260), using a NanoDrop instrument and 10-fold serial dilutions of the quantified DNAs were prepared in RNase-free H2O. Finally, for the IC, the plasmid was linearized using the Not I restriction enzyme. Known amounts of each DNA control were used for optimizing the conditions of the qRT-PCR.
Real-time RT-PCR assay
The real-time RT-PCR (qRT-PCR) was carried out using a commercial kit (Quantitec multiplex RT-PCR Kit; Qiagen, Hilden, Germany). For the assay, 5 μl of RNA sample was mixed with 20 μl of a reaction mix containing: 12.5 μl of 2× Buffer, 1.2 μM of each primer, 0.4 μM of virus probe, 0.2 μM of IC probe, 10 copies of IC plasmid, 0.3 µl of RT mix and nuclease-free water to reach the final volume of 25 µl per tube. Amplification conditions consisted of an initial retrotranscription step of 30 min at 50 °C, followed by an incubation step at 95 °C for 15 min, and 45 cycles of 45 s at 95 °C and 1 min at 60 °C. qRT-PCR was carried out in a 7500 Sequence Detection System from Applied Biosystems. The fluorescence emitted for FAM and NED was measured simultaneously at the end of each cycle. Quantification cycle (Cq) values were measured as the point at which the sample fluorescence signal crossed a predetermined threshold value.
Sensitivity and specificity assessment
Sensitivity was determined according to the detection limit using the quantified DNA standards, which were diluted in the range of 107–10−1 copies/µl and then tested by the qRT-PCR. Each concentration was assayed eight times in two consecutive days and the assay detection limit was calculated using the formula of Reed-Muench [16] for each virus. The analytical specificity is ensured by the thorough selection of the oligonucleotides (primers and probes), which were checked by sequence comparison analysis against publicly available sequences and evaluated by testing 20 different viral genomes from other phleboviruses. To determine the applicability of the assay for virus surveillance, insect samples collected in the field were tested.
Study in field samples
Thirty-seven sand fly pools were collected in several Balearic islands in September 2012 using BG-Sentinel traps with BG-lure and carbon dioxide. The field work was focused on collecting hematophagous insects in Ibiza from 5th to 8th September (5 localities), in Formentera from 9th to 14th September (3 localities), in Cabrera from 15th to 20th September (4 localities) and in Dragonera from 22th to 24th September (4 localities) for a total of 269 trap/nights. Insects were identified and pooled according to species, sex, locality and date. Sand fly wings and body of individuals of the same locality and date were stored separately for molecular analysis. The head and genitalia were transferred individually into 1.5 ml Eppendorf tubes with 90% ethanol. For the identification, they were cleared in Marc-André solution (chloral hydrate/acetic acid) and mounted in chloral gum. Specimen identification was individually verified based on the morphology of the pharynges and/or the male genitalia or female spermathecae. In total 372 sand fly individuals were collected (6 in Ibiza, 105 in Formentera, 227 in Cabrera and 34 in Dragonera); these were divided into 33 pools of Phlebotomus perniciosus and 4 pools of Sergentomyia minuta. Pools were homogenized in 0.5 ml minimum essential medium supplemented with 10 % of fetal bovine serum and antibiotics (50 U/ml penicillin and 50 µg/ml streptomycin) before nucleic acid extraction. RNAs were extracted using QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) and analyzed for phleboviruses detection using a generic RT-nested-PCR [17] and specifically for GRV, MASV and ARRV using the new assay described in this paper.
Results
Design of primers and probes
Primers and probes were designed to target a highly conserved fragment of the L segment of GRV and the related MASV and ARRV amplifying a 126-bp sequence. An in silico test was performed using a partial nucleotide sequence alignment in the L-segment of representative phleboviruses to check the specificity and to identify possible cross-reactions with other different phleboviruses. After that, only one degenerate nucleotide was necessary to introduce in the phleboviruses probe sequence to detect the three viruses (Table 1). The amount of the IC included in the assay was fixed at 10 copies per reaction without affecting the qRT-PCR sensitivity.
Viral RNA detection, qRT-PCR sensitivity and specificity
The detection limit was assessed using serial dilutions of the quantified GRV, MASV and ARRV DNA standards. Nineteen copies per reaction for GRV and ARRV and 16 copies for MASV were the detection limit estimated using eight replicates amplifying with a quantification cycle (Cq) value under 40 (Table 3). The standard curves and mean Cq values obtained in different assays indicated that the method is highly robust and reproducible.
The qRT-PCR assay was found to be specific for the detection of GRV, MASV and ARRV without differentiating between them and no cross-reactivity was observed with any other sand fly borne phleboviruses (Table 4). The presence of phlebovirus genomes was confirmed using a generic RT-nested-PCR [17].
Screening of phleboviruses in natural sand fly populations from Spain
Thirty-three P. perniciousus pools and four Sergentomyia minuta pools were analyzed. These samples were previously analyzed using a generic RT-nested-PCR for phleboviruses developed by our laboratory that targets a 200-bp fragment of the L-segment. Two positive pools from P. perniciousus were detected, one in Cabrera (MB202) and the other one (MB173) in Formentera. The phylogenetic analysis of the sequences revealed that one sequence (MB202) is related to MASV, GRV and ARRV and the other one (MB173) is related to Alcube virus (Fig. 1). Using the novel qRT-PCR assay developed in this study, only one pool (MB202) was positive corresponding to the MASV-like pool detected with the generic RT-nested-PCR. An additional fragment of 903 bp of this sample was amplified in the M segment with an unpublished in-house RT-PCR, and the sequence obtained showed 83% of homology with MASV strain W (GenBank: EU725772) detected in sand flies from France. Based on these results, the qRT-PCR was shown to be specific and no false-positives were detected.
Phylogenetic tree based on 200 nucleotides of the partial L segment of the phleboviruses detected in the Balearic Islands (Spain). The phylogenetic tree was constructed using Neighbor-Joining method analysis with 1000 bootstrap replications. Sequence information correspond to virus, country detection, year, host and GenBank accession number. The sequences of viruses detected in this study are indicated in bold. The GenBank accession number are MN894575 (MB202) and MN901504 (MB173)
Discussion
Phleboviruses are distributed across much of the Old World, including the Mediterranean basin. Recent investigations suggest that the diversity of this group of viruses is higher than initially suspected, but data on prevalence, geographical distribution and pathogenesis remain unknown [1, 8, 11, 18]. This could be due to a lack of sensitive and specific methods for virus detection and quantification, but also to the lack of studies in natural populations. This is the case of the related phleboviruses GRV, MASV and ARRV that, until now, have been detected in sand flies from Spain, France and Portugal.
In Spain, several phlebovirus species belonging to two antigenic complexes have been described in several geographical areas from the northwest, center and south of the country. Viruses belonging to the Sand fly fever Naples virus species complex, Toscana, Granada, Massilia-like and Arrabida-like have been detected, and Arbia-like virus belonging to the Salehabad virus species complex has been detected [5, 6, 11]. Amongst them, TOSV is the only member recognized to cause human disease. Whereas there is no clear evidence that MASV, GRV, ARRV or ARBV are pathogenic to humans, the last reports of seroprevalence studies carried out in humans in Spain, showed that GRV may infect humans with low prevalence, leading to asymptomatic or self-limited febrile illnesses cases accompanied by other signs and symptoms like exanthema and/or acute respiratory infection [5]. Therefore, despite there being very limited data about the capacity of the latter viruses to cause disease, this possibility cannot be neglected. Given the lack of data about these novel viruses and the possibility that they can infect and cause disease to humans, additional epidemiological and clinical studies on natural populations are strongly needed. Moreover, the viral loads reached in human infections by these viruses are unknown, and for this reason a highly sensitive PCR available for diagnosis is required. Until now, only one real-time RT-PCR designed in the S segment has been published to specifically detect MASV [7], but it has not been validated with the related GRV and ARRV phleboviruses which were described later. Furthermore, new molecular assays to detect these viruses are required that enhance sensitivity and specificity at the same time that are applicable to a broad range of samples.
In this study, we developed a rapid, sensitive and specific qRT-PCR for clinical investigation into GRV, MASV and ARRV infection. Here we show the applicability of a new qRT-PCR assay with enhanced sensitivity and broad range spectrum able to detect the three phleboviruses simultaneously and which we used to detect these pathogens in sand flies collected in the Balearic Archipelago, a touristic hotspot in the Mediterranean. Our findings demonstrate for the first time the detection of MASV-like in P. perniciousus from Cabrera island (Balearic Archipelago), which would support the spread and circulation of this virus in Spain and in neighbouring Mediterranean countries.
Conclusions
The new method developed here is a rapid, robust and reliable assay for the accurate diagnosis of human infection as well as for surveillance in vector populations, providing information about the disease burden of these infections.
Availability of data and materials
All data involved and arising from the study are included in this published article.
Abbreviations
- SFNV:
-
Sand fly fever Naples virus
- SFSV:
-
Sicilian fever Naples virus
- TOSV:
-
Toscana virus
- GRV:
-
Granada virus
- SFNC:
-
Sand fly fever Naples complex
- MASV:
-
Massilia virus
- ARRV:
-
Arrabida virus
- PUNV:
-
Punique virus
- ARBV:
-
Arbia virus
- MGB:
-
minor groove binder
- NFQ:
-
non-fluorescent quencher
- IC:
-
internal control
- qRT-PCR:
-
real-time RT-PCR
References
Moriconi M, Rugna G, Calzolari M, Bellini R, Albieri A, Angelini P, et al. Phlebotomine sand fly-borne pathogens in the Mediterranean Basin: human leishmaniasis and phlebovirus infections. PLoS Negl Trop Dis. 2017;11:e0005660.
Di Bonito P, Mochi S, Grò MC, Fortini D, Giorgi C. Organization of the M genomic segment of Toscana phlebovirus. J Gen Virol. 1997;78:77–81.
https://talk.ictvonline.org/taxonomy/. Accessed 2 Feb 2019.
Alkan C, Moin Vaziri V, Ayhan N, Badakhshan M, Bichaud L, Rahbarian N, et al. Isolation and sequencing of Dashli virus, a novel Sicilian-like virus in sandflies from Iran; genetic and phylogenetic evidence for the creation of one novel species within the Phlebovirus genus in the Phenuiviridae family. PLoS Negl Trop Dis. 2017;11:e0005978.
Navarro-Marí JM, Gómez-Camarasa C, Pérez-Ruiz M, Sanbonmatsu-Gámez S, Pedrosa-Corral I, Jiménez-Valera M. Clinic-epidemiologic study of human infection by Granada virus, a new phlebovirus within the sandfly fever Naples serocomplex. Am J Trop Med Hyg. 2013;88:1003–6.
Collao X, Palacios G, de Ory F, Sanbonmatsu S, Pérez-Ruiz M, Navarro JM, et al. Granada virus: a natural phlebovirus reassortant of the sandfly fever Naples serocomplex with low seroprevalence in humans. Am J Trop Med Hyg. 2010;83:760–5.
Charrel RN, Moureau G, Temmam S, Izri A, Marty P, Parola P, et al. Massilia virus, a novel Phlebovirus (Bunyaviridae) isolated from sandflies in the Mediterranean. Vector Borne Zoonotic Dis. 2009;9:519–30.
Amaro F, Zé-Zé L, Alves MJ, Börstler J, Clos J, Lorenzen S, et al. Co-circulation of a novel phlebovirus and Massilia virus in sandflies, Portugal. Virol J. 2015;12:174.
Amaro F, Hanke D, Zé-Zé L, Alves MJ, Becker SC, Höper D. Genetic characterization of Arrabida virus, a novel phlebovirus isolated in South Portugal. Virus Res. 2016;214:19–25.
Sanchez-Seco MP, Vazquez A, Collao X, Hernandez L, Aranda C, Ruiz S, et al. Surveillance of arboviruses in Spanish wetlands: detection of new flavi- and phleboviruses. Vector Borne Zoonotic Dis. 2010;10:203–6.
Remoli ME, Jimenez M, Fortuna C, Benedetti E, Marchi A, Genovese D, et al. Phleboviruses detection in Phlebotomus perniciosus from a human leishmaniasis focus in South-West Madrid region, Spain. Parasit Vectors. 2016;9:205.
Echevarria JM, de Ory F, Guisasola ME, Sanchez-Seco MP, Tenorio A, Lozano A, et al. Acute meningitis due to Toscana virus infection among patients from both the Spanish Mediterranean region and the region of Madrid. J Clin Virol. 2003;26:79–84.
Martinez-Garcia FA, Moreno-Docon A, Lopez-Lopez M, Albert-Lacal L, Martinez-Toldos MC, Segovia-Hernandez M, et al. A case of meningitis due to Toscana virus in Murcia. Rev Neurol. 2007;45:317–8.
Navarro JM, Fernandez-Roldan C, Perez-Ruiz M, Sanbonmatsu S, de la Rosa M, Sanchez-Seco MP. Meningitis by Toscana virus in Spain: description of 17 cases. Med Clin. 2004;122:420–2.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9.
Reed LJ, Muench H. A simple method of estimating 50% endpoints. 1938;27:493–7.
Sanchez-Seco MP, Echevarria JM, Hernandez L, Estevez D, Navarro-Mari JM, Tenorio A. Detection and identification of Toscana and other phleboviruses by RT-nested-PCR assays with degenerated primers. J Med Virol. 2003;71:140–9.
Calzolari M, Ferrarini G, Bonilauri P, Lelli D, Chiapponi C, Bellini R, et al. Co-circulation of eight different phleboviruses in sand flies collected in the northern Apennine Mountains (Italy). Infect Genet Evol. 2018;64:131–4.
Acknowledgements
Not applicable.
Funding
This study was supported in part by the projects CP13/00114 and PI14CIII/00014 from the Instituto de Salud Carlos III. EGD was supported by Juan de la Cierva and Ramón y Cajal contracts from the Ministerio de Educación y Ciencia, Spain. Financial support was provided by project ERG-PARIS-276838 from the European Commission. JF and DR were supported by project CGL2009-11445 from the Ministerio de Economia y Competitividad, Spain.
Author information
Authors and Affiliations
Contributions
All authors participated in the research and/or article preparation. AV designed the study, participated in data analysis and drafted the paper. DR and EGD carried out the vector captures. LD and LH carried out the optimization of the method and analysis of the data. NL and L.H. carried out the molecular analysis of the vectors. EGD, JF and MPSS participated in data interpretation and drafted the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Davó, L., Herrero, L., Sánchez-Seco, M.P. et al. Real-time RT-PCR assay to detect Granada virus and the related Massilia and Arrabida phleboviruses. Parasites Vectors 13, 270 (2020). https://doi.org/10.1186/s13071-020-04110-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-020-04110-5